RNA is a central component of gene expression and is
the link between genetic information (DNA) and proteins. Multiple
mechanisms are involved in regulating gene expression. One such
mechanism is mediated by chemical modification of RNA either during
or after transcription. More than 100 post-transcriptional
modifications have been found in all RNA species (1). The functions of these modifications are
diverse and depend on the biological context. Among the
modifications, recent studies revealed that N6-methyladenosine
(m6A) modification in eukaryotic mRNAs plays significant roles in
pleiotropic biological processes including stem cell biology,
development, immunology, and cancer biology (2–5). m6A
modification was discovered several decades ago in mouse and human
transcriptomes (6–11). However, its role was unclear until
recently. The discovery that fat mass and obesity-associated
protein (FTO) is an enzyme that demethylates m6A prompted studies
to evaluate the biological consequences of m6A (12). FTO protein was originally recognized
as a regulator of lipid metabolism (13–15). The
availability of m6A-specific antibodies and transcriptome-wide m6A
mapping studies further revealed the unique pattern of m6A
distribution and functions in the regulation of RNA biology
(16,17). Studies demonstrated that aberrant
regulation of m6A turnover is linked to pathophysiological
disorders such as excessive fatty acid deposition, developmental
retardation, type 2 diabetes mellitus, aberrant germ cell
formation, circadian period elongation, and cancers (13–15,18–35).
Among these, emerging evidence suggests that factors metabolizing
m6A are involved in multiple forms of human cancer, including lung
cancer, glioblastoma, breast cancer, and acute myeloid leukemia
(AML). The review discusses recent progress in understanding
m6A-related cancers (23–35).
In the 1970s, several groups detected m6A in human
and mouse cells. The early discovery of m6A modification was made
in noncoding RNAs including snoRNAs, tRNAs, and rRNAs, and mRNAs
(6–11). However, the function of the
modification was unknown because of technical limitations in
functional analysis. In 2012, two independent studies profiled the
transcriptome-wide m6A distribution and found a conserved pattern
of its enrichment in both the human and mouse transcriptome
(16,17). Since then, multiple studies confirmed
similar patterns of the transcriptome-wide distribution of m6A in
some eukaryotes including cow, yeast, and plant (36–38). It is
estimated that more than ≥7,600 coding and ≥300 noncoding RNAs
contain m6A, and 0.1–0.4% of total adenines (As) were methylated in
mammalian transcripts (11,16,17,39). In
general, the studies revealed the following: i) There is a
consensus sequence [(G/A/U)(G>A)m6AC(U>A>C)] marked by m6A
(16,17,39–42). ii)
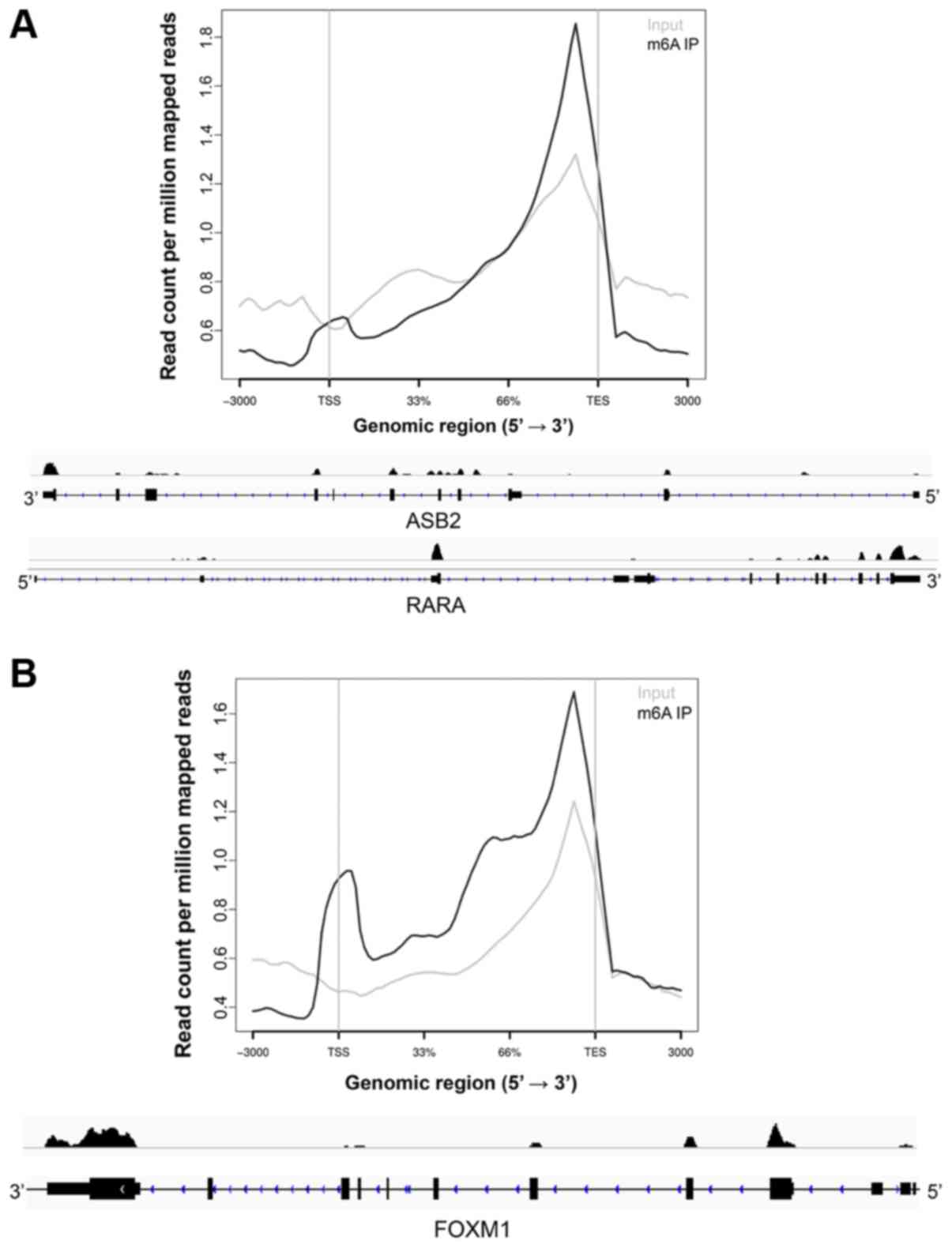
m6A is preferentially enriched around the transcription end site
(TES) in mRNAs or last exon in noncoding RNAs (16,17)
(Fig. 1). Relatively less enriched
m6A peaks are also observed at the transcription start site (TSS)
(43) (Fig.
1). iii) Some m6A-marked genes are conserved in human and mouse
cells (16). For example, ~46% of
conserved m6A peaks between human and mouse ES cells were
identified. The data suggest a conserved function of the
modification in regulating stem cell biology. The unique pattern of
m6A enrichment implicates its functions in translation efficiency,
splicing, mRNA export, and alternative polyadenylation (16,40,44–46).
In general, m6A accumulation accelerates transcript decay (45). In contrast, TSS m6A is known to
accelerate the CAP-independent translation start by changing the 3D
structure of mRNA (47). These
structural changes recruit transcription initiation proteins such
as eIF3 (47). Although early
functional studies of m6A were shown to positively regulate
splicing, a recent study demonstrated that m6A is observed in exons
of newly synthesized transcripts. In contrast to previous
hypotheses, m6As in exon are quite static, as m6A exists in the
exons of chromatin-associated RNAs and the same pattern of m6A
distribution is observed even after translocation into the
cytoplasm (48). Therefore, the data
suggest that m6A plays an independent role in splicing. The study
confirmed that m6A methylation is negatively correlated with the
half-life of the transcript. Ke et al (49) showed that genes with a long last exon
have a higher density of m6A compared to genes with short last
exons. Furthermore, the study suggested that m6A density affects
utilization of the polyA site; if a gene more frequently utilizes a
proximal polyA site, m6A density around the proximal polyA site is
low, whereas if a gene less frequently utilizes a proximal polyA
site, m6A density around the site is high (49). The idea of its role in controlling
splicing was originally proposed based on the observation that m6A
was more observed in intron compared to exon sequences. However,
the study suggests that m6A-mediated regulation of splicing is less
likely.
Several enzymes catalyze m6A formation. The first
protein identified to have this function is methyltransferase-like
protein (METTL) 3, which forms a heterodimeric complex with another
methyltransferase, METTL14 (50).
Although both METTL3 and METTL14 contain conserved catalytic
domains, only METTL3 appears to contain a methyl donor,
S-adenosylmethionine, binding domain (50). METTL3 is highly conserved in most
eukaryotes and broadly expressed, and its enriched subcellular
localization is found in nuclear speckles (51). Genetic ablation of mouse METTL3 or
METTL14 causes aberrant differentiation of embryonic stem cells,
spermatogonial cells, and neuronal lineages with bulk reduction of
m6A levels (2,52,53).
Wilms's tumor protein 1 (WT1)-associated protein (WTAP), known as a
splicing factor and binding partner of human WT1, has shown to be
required for embryonic development and cell cycle progression
(40). WTAP-depleted mice display
lethal at peri-gastrula stages with failure in cell proliferation
(54). Although WTAP does not possess
methyltransferase activity, it appears to modulate m6A methylation
by interacting with the METTL3-METTL14 complex.
FTO protein belongs to a member of Fe(II)- and
α-ketoglutarate-dependent AlkB dioxygenase family and is known to
be linked to childhood and adult obesity (55). FTO was shown to demethylate m6A in
vitro and its loss results in growth retardation and aberrant
metabolism with increased m6A levels (12,56). A
forced overexpression of FTO leads to decreased m6A levels in HeLa
cells (12). FTO has been suggested
as an important player in neuronal development and cancers in
recent studies (26,28,32,57).
ALKBH5 is also a member of the Fe(II)- and
α-ketoglutarate-dependent AlkB dioxygenase family. ALKBH5-depleted
male animals suffer from sterility with accumulation of
polyA-tailed RNAs in the cytoplasm (58). As described above, iron as a cofactor
in the biochemical reaction appears to be essential, as mutations
of amino acid residues in the iron binding pocket lead to loss of
catalytic activity (59).
Recent studies demonstrated that m6A modifications
can be recognized by at least three proteins. The first proteins
identified in immunoprecipitation experiments were YT521-B homology
domain-containing family protein (YTHDF)2, and YTHDF3 (60). The two proteins contain the conserved
YTH domain, which is known to preferentially interact with
single-stranded RNA compared to single-stranded DNA (60). In mouse, there are five known YTH
domain-containing proteins including four cytoplasmic (YTHDF1-3 and
YTHDC2) and one nuclear (YTH domain-containing 1) (61,62).
YTHDF2 binds to m6A and accelerates degradation of m6A-containing
RNAs (61). YTHDF2 depletion causes
female-specific sterility (63).
Furthermore, YTHDF2 was shown to play a critical role in
maternal-to-zygotic transition by accelerating the decay of
maternally inherited mRNAs in zebrafish (64). In contrast, YTHDF3 promotes mRNA
translation (62). Together, the
studies suggest that regulation of the half-life of RNAs by m6A
addition and/or removal plays an important role in mammalian early
development.
Cancer types that are associated with proteins
regulating m6A generation or removal are summarized in Table I.
AML is a devastating disease characterized by
defects in differentiation, increased proliferation, and inhibition
of cell apoptosis (65). WTAP has
been suggested to have an oncogenic role in AML (25). Approximately 32% of AML patients
carrying NPM1 and/or FTL3-ITD mutation(s) exhibit aberrant
overexpression of the WTAP gene (25). Genome-scale analysis revealed that
2–9% of AML patients carry copy number variations (CNV) in m6A
regulatory genes including FTO, ALKBH5, YTHDF1, YTHDF2, METTL3, and
METTL14 (26). Loss of CNV in the
ALKBH5 gene is the most obviously observed in AML patients
(26). In addition, METTL3 depletion
in human hematopoietic progenitor cells and myeloid leukemia cell
lines leads to accelerated cell differentiation with reduced cell
proliferation, as well as delayed leukemic progression after
transplantation into mice, respectively (27). m6A modification enhances the
translation of PTEN, c-MYC, and BCL2 transcripts (28,29). In
contrast, FTO is aberrantly upregulated in AMLs with
t(15;17)/PML-RARA, t(11q23)/MLL, FLT3-ITD, and/or NPM1 mutations
(30). FTO promotes cell
transformation and leukemogenesis. Mechanistically, FTO-mediated
m6A reduction directly inhibits all-trans-retinoic acid-induced AML
cell differentiation by regulating the expression of ASB2 and RARA
in vitro (30). The data
suggests that fine-tuning of m6A formation closely controls
differentiation, survival, and lineage commitment of hematopoietic
cells.
Glioblastoma is a life-threatening brain tumor
showing a median overall survival between 10 and 20 months. The
incidence of glioblastoma is approximately 3–4/100,000, increases
with age, and peaks in patients aged 50–60 years (66,67).
Multiple genes have shown to be involved in causing glioblastoma
(67). A high level of ALKBH5
expression is detected in glioblastoma stem-like cells (GSCs), and
ALKBH5 silencing inhibits the proliferation of patient-derived GSCs
(31). Mechanistically, ALKBH5
upregulates FOXM1 expression by direct m6A demethylation in the
FOXM1 transcript (31). Depletion of
ALKBH5 abolishes GSC tumorigenesis by reducing FOXM1 expression.
Silencing of a methyltransferase, METTL14 or METTL3, promotes human
GSC self-renewal, growth, and tumorigenesis, whereas overexpression
of METTL3 inhibits GSC self-renewal and growth (32,33).
It has been shown that METTL3 is upregulated in lung
adenocarcinoma and promotes the survival, growth, and invasion of
human lung cancer cells by promoting the translation of genes
related to cancer (34). More
recently, METTL3 expression was found to be upregulated in human
hepatocellular carcinoma (HHC) and its overexpression was related
to poor prognosis in patients with HCC (35). Loss of METTL3 function leads to
suppressed HCC tumorigenicity and lung metastasis in mice by
increasing SOCS2 expression. The regulation of SOCS2 expression
appears to be accelerated by binding of YTHDF2 to its mRNAs.
Consistent with the notion, YTHDF2 depletion results in delayed
degradation of the SOCS2 transcript (35).
Epitranscriptomics refers to regulation of gene
expression by post-transcriptional covalent modification of the
transcript (68,69). m6A is now known to be involved in one
of the most important epitranscriptomic mechanisms, is readily
detected transcriptome-wide and controls gene expression by
modulating the biology of RNAs. m6A enrichment displays a unique
pattern. As described above, studies have shown that regulation of
m6A formation is linked to human cancers. Based on these findings,
it is reasonable to speculate that there are unidentified cancer
types caused by dysregulation of m6A modification. Furthermore, m6A
and enzymes may be therapeutic targets for treatment of these
identified cancers. Additionally, screening of chemicals that
potentially regulate m6A formation or removal is an appropriate
approach for therapeutic purposes.
The author would like to thank Ms. Hyunjin Yoo for
editing the manuscript and Mr. Muhammad Arsalan Iqbal for helping
with data analysis.
The present study was supported by the National
Research Foundation of Korea (grant no. NRF-2015M3A9C6028965).
The datasets and codes used for computational
analyses are available from corresponding author upon request.
KH designed and conceived the data analyses and
wrote the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Machnicka MA, Milanowska K, Oglou Osman O,
Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S,
Dunin-Horkawicz S, Rother KM, et al: MODOMICS: A database of RNA
modification pathways-2013 update. Nucleic Acids Res. 41:(Database
Issue). D262–D267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Batista PJ, Molinie B, Wang J, Qu K, Zhang
J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al: m(6)A
RNA modification controls cell fate transition in mammalian
embryonic stem cells. Cell Stem Cell. 15:707–719. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geula S, Moshkovitz Moshitch S,
Dominissini D, Mansour AA, Kol N, Divon Salmon M, Hershkovitz V,
Peer E, Mor N, Manor YS, et al: Stem cells. m6A mRNA methylation
facilitates resolution of naïve pluripotency toward
differentiation. Science. 347:1002–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Connell MA, Mannion NM and Keegan LP:
The epitranscriptome and innate immunity. PLoS Genet.
11:e10056872015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tajaddod M, Jantsch MF and Licht K: The
dynamic epitranscriptome: A to I editing modulates genetic
information. Chromosoma. 125:51–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perry RP and Kelley DE: Existence of
methylated messenger RNA in mouse L cells. Cell. 1:37–42. 1974.
View Article : Google Scholar
|
|
8
|
Adams JM and Cory S: Modified nucleosides
and bizarre 5′-termini in mouse myeloma mRNA. Nature. 255:28–33.
1975. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Furuichi Y, Shatkin AJ, Stavnezer E and
Bishop JM: Blocked, methylated 5′-terminal sequence in avian
sarcoma virus RNA. Nature. 257:618–620. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lavi S and Shatkin AJ: Methylated simian
virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1
cells. Proc Natl Acad Sci USA. 72:2012–2016. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei CM, Gershowitz A and Moss B:
Methylated nucleotides block 5′ terminus of HeLa cell messenger
RNA. Cell. 4:379–386. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Scuteri A, Sanna S, Chen WM, Uda M, Albai
G, Strait J, Najjar SS, Nagarajah R, Orrú M, Usala G, et al:
Genome-wide association scan shows genetic variants in the FTO gene
are associated with obesity related traits. PLoS Genet. 3:e1152007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dina C, Meyre D, Gallina S, Durand E,
Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et
al: Variation in FTO contributes to childhood obesity and severe
adult obesity. Nat Genet. 39:724–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Frayling TM, Timpson NJ, Weedon MN,
Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H,
Rayner NW, et al: A common variant in the FTO gene is associated
with body mass index and predisposes to childhood and adult
obesity. Science. 316:889–894. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dominissini D, Moshkovitz Moshitch S,
Schwartz S, Divon Salmon M, Ungar L, Osenberg S, Cesarkas K, Hirsch
Jacob J, Amariglio N, Kupiec M, et al: Topology of the human and
mouse m6A RNA methylomes revealed by m6A-seq. Nature. 485:201–206.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen F, Huang W, Huang JT, Xiong J, Yang
Y, Wu K, Jia GF, Chen J, Feng YQ, Yuan BF and Liu SM: Decreased
N(6)-methyladenosine in peripheral blood RNA from diabetic patients
is associated with FTO expression rather than ALKBH5. J Clin
Endocrinol Metab. 100:E148–E154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Huang W, Huang JT, Shen F, Xiong
J, Yuan EF, Qin SS, Zhang M, Feng YQ, Yuan BF and Liu SM: Increased
N6-methyladenosine in human sperm RNA as a risk factor for
asthenozoospermia. Sci Rep. 6:243452016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fustin JM, Doi M, Yamaguchi Y, Hida H,
Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I
and Okamura H: RNA-methylation-dependent RNA processing controls
the speed of the circadian clock. Cell. 155:793–806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Daoud H, Zhang D, Mcmurray F, Yu A, Luco
SM, Vanstone J, Jarinova O, Carson N, Wickens J, Shishodia S, et
al: Identification of a pathogenic FTO mutation by next-generation
sequencing in a newborn with growth retardation and developmental
delay. J Med Genet. 53:200–207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Davis W, van Rensburg SJ, Cronje FJ, Whati
L, Fisher LR, van der Merwe L, Geiger D, Hassan MS, Matsha T,
Erasmus RT and Kotze MJ: The fat mass and obesity-associated FTO
rs9939609 polymorphism is associated with elevated homocysteine
levels in patients with multiple sclerosis screened for vascular
risk factors. Metab Brain Dis. 29:409–419. 2014.PubMed/NCBI
|
|
23
|
Zhang C, Samanta D, Lu H, Bullen JW, Zhang
H, Chen I, He X and Semenza GL: Hypoxia induces the breast cancer
stem cell phenotype by HIF-dependent and ALKBH5-mediated
m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA.
113:E2047–E2056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang C, Zhi WI, Lu H, Samanta D, Chen I,
Gabrielson E and Semenza GL: Hypoxia-inducible factors regulate
pluripotency factor expression by ZNF217- and ALKBH5-mediated
modulation of RNA methylation in breast cancer cells. Oncotarget.
7:64527–64542. 2016.PubMed/NCBI
|
|
25
|
Bansal H, Yihua Q, Iyer SP, Ganapathy S,
Proia DA, Penalva LO, Uren PJ, Suresh U, Carew JS, Karnad AB, et
al: WTAP is a novel oncogenic protein in acute myeloid leukemia.
Leukemia. 28:1171–1174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kwok CT, Marshall AD, Rasko JE and Wong
JJ: Genetic alterations of m6A regulators predict poorer
survival in acute myeloid leukemia. J Hematol Oncol. 10:392017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma
D, Lv J, Heng J, Ding Y, Xue Y, et al: m6A modulates
haematopoietic stem and progenitor cell specification. Nature.
549:273–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barbieri I, Tzelepis K, Pandolfini L, Shi
J, Millán-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister
AJ, Han N, et al: Promoter-bound METTL3 maintains myeloid leukaemia
by m6A-dependent translation control. Nature.
552:126–131. 2017.PubMed/NCBI
|
|
29
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme
METTL3 controls myeloid differentiation of normal hematopoietic and
leukemia cells. Nat Med. 23:1369–1376. 2017.PubMed/NCBI
|
|
30
|
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C,
Huang H, Nachtergaele S, Dong L, Hu C, et al: FTO plays an
oncogenic role in acute myeloid leukemia as a
N6-methyladenosine RNA demethylase. Cancer Cell.
31:127–141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A
demethylase ALKBH5 maintains tumorigenicity of glioblastoma
stem-like cells by sustaining FOXM1 expression and cell
proliferation program. Cancer Cell. 31(591–606): e62017. View Article : Google Scholar
|
|
32
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation
regulates the self-renewal and tumorigenesis of glioblastoma stem
cells. Cell Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jin DI, Lee SW, Han ME, Kim HJ, Seo SA,
Hur GY, Jung S, Kim BS and Oh SO: Expression and roles of Wilms'
tumor 1-associating protein in glioblastoma. Cancer Sci.
103:2102–2109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2 dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Horowitz S, Horowitz A, Nilsen TW, Munns
TW and Rottman FM: Mapping of N6-methyladenosine residues in bovine
prolactin mRNA. Proc Natl Acad Sci USA. 81:5667–5671. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schwartz S, Agarwala SD, Mumbach MR,
Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija
R, Ruvkun G, et al: High-resolution mapping reveals a conserved,
widespread, dynamic mRNA methylation program in yeast meiosis.
Cell. 155:1409–1421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo GZ, MacQueen A, Zheng G, Duan H, Dore
LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J and He C: Unique
features of the m6A methylome in Arabidopsis thaliana. Nat Commun.
5:56302014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao X, Yang Y, Sun BF, Shi Y, Yang X,
Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al: FTO-dependent
demethylation of N6-methyladenosine regulates mRNA splicing and is
required for adipogenesis. Cell Res. 24:1403–1419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Smemo S, Tena JJ, Kim KH, Gamazon ER,
Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR,
Wasserman NF, et al: Obesity-associated variants within FTO form
long range functional connections with IRX3. Nature. 507:371–375.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rottman F, Shatkin AJ and Perry RP:
Sequences containing methylated nucleotides at the 5′ termini of
messenger RNAs: Possible implications for processing. Cell.
3:197–199. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mauer J, Luo X, Blanjoie A, Jiao X,
Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q,
et al: Reversible methylation of m6Am in the
5′ cap controls mRNA stability. Nature. 541:371–375. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification destabilizes
developmental regulators in embryonic stem cells. Nat Cell Biol.
16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang Y, Sun BF, Xiao W, Yang X, Sun HY,
Zhao YL and Yang YG: Dynamic m6A modification and its
emerging regulatory role in mRNA splicing. Sci Bull. 60:21–32.
2015. View Article : Google Scholar
|
|
47
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)A promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ke S, Pandya-Jones A, Saito Y, Fak JJ,
Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr and Darnell
RB: m6A mRNA modifications are deposited in nascent
pre-mRNA and are not required for splicing but do specify
cytoplasmic turnover. Genes Dev. 31:990–1006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ke S, Alemu EA, Mertens C, Gantman EC, Fak
JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al:
A majority of m6A residues are in the last exons, allowing the
potential for 3′ UTR regulation. Genes Dev. 29:2037–2053. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bokar JA, Shambaugh ME, Polayes D, Matera
AG and Rottman FM: Purification and cDNA cloning of the
AdoMet-binding subunit of the human mRNA
(N6-adenosine)-methyltransferase. RNA. 3:1233–1247. 1997.PubMed/NCBI
|
|
51
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou
Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, et al:
Mettl3-/Mettl14-mediated mRNA N6-methyladenosine
modulates murine spermatogenesis. Cell Res. 27:1216–1230. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yoon KJ, Ringeling FR, Vissers C, Jacob F,
Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al:
Temporal control of mammalian cortical neurogenesis by
m6A methylation. Cell. 171(877–889): e172017.
|
|
54
|
Horiuchi K, Umetani M, Minami T, Okayama
H, Takada S, Yamamoto M, Aburatani H, Reid PC, Housman DE, Hamakubo
T and Kodama T: Wilms' tumor 1-associating protein regulates G2/M
transition through stabilization of cyclin A2 mRNA. Proc Natl Acad
Sci USA. 103:17278–17283. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gerken T, Girard CA, Tung YC, Webby CJ,
Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill
LA, et al: The obesity associated FTO gene encodes a
2-oxoglutarate-dependent nucleic acid demethylase. Science.
318:1469–1472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fischer J, Koch L, Emmerling C, Vierkotten
J, Peters T, Brüning JC and Rüther U: Inactivation of the Fto gene
protects from obesity. Nature. 458:894–898. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li L, Zang L, Zhang F, Chen J, Shen H, Shu
L, Liang F, Feng C, Chen D, Tao H, et al: Fat mass and
obesity-associated (FTO) protein regulates adult neurogenesis. Hum
Mol Genet. 26:2398–2411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu
W, Tong Y, Cheng C and Chen Z: Crystal structures of the human RNA
demethylase Alkbh5 reveal basis for substrate recognition. J Biol
Chem. 289:11571–11583. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang Z, Theler D, Kaminska KH, Hiller M,
de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM,
Allain FH and Stamm S: The YTH domain is novel RNA binding domain.
J Biol Chem. 285:14701–14710. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li A, Chen YS, Ping XL, Yang X, Xiao W,
Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al: Cytoplasmic
m6A reader YTHDF3 promotes mRNA translation. Cell Res.
27:444–447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ivanova I, Much C, Di Giacomo M, Azzi C,
Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ and
O'Carroll D: The RNA m6A reader YTHDF2 is essential for
the post-transcriptional regulation of the maternal transcriptome
and oocyte competence. Mol Cell. 67(1059–1067): e42017.
|
|
64
|
Zhao BS, Wang X, Beadell AV, Lu Z, Shi H,
Kuuspalu A, Ho RK and He C: m6A-dependent maternal mRNA
clearance facilitates zebrafish maternal-to-zygotic transition.
Nature. 542:475–478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
De Kouchkovsky I and Abdul-Hay M: Acute
myeloid leukemia: A comprehensive review and 2016 update. Blood
Cancer J. 6:e4412016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Touat M, Idbaih A, Sanson M and Ligon KL:
Glioblastoma targeted therapy: Updated approaches from recent
biological insights. Ann Oncol. 28:1457–1472. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lieberman F: Glioblastoma update:
Molecular biology, diagnosis, treatment, response assessment, and
translational clinical trials. F1000Res. 6:18922017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Saletore Y, Meyer K, Korlach J, Vilfan ID,
Jaffrey S and Mason CE: The birth of the Epitranscriptome:
Deciphering the function of RNA modifications. Genome Biol.
13:1752012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hussain S, Aleksic J, Blanco S, Dietmann S
and Frye M: Characterizing 5-methylcytosine in the mammalian
epitranscriptome. Genome Biol. 14:2152013. View Article : Google Scholar : PubMed/NCBI
|