Introduction
Colorectal cancer (CRC) is one of the most common
types of solid tumor, with ~1.2 million new confirmed diagnoses of
CRC annually worldwide and with 55% of cases occurring in
economically developed countries (1).
Changes in lifestyle, population growth and the aging population
have made CRC an increasingly global health problem (2). Patients with CRC have a poor prognosis;
15% of patients present with metastases at the time of initial
diagnosis, and ~35% of patients diagnosed with CRC ultimately
develop metastasis (3,4). Despite the recent use of neoadjuvant
chemotherapy (NCT) or NCT in combination with targeted agents,
which has resulted in an improvement in the overall survival rate,
CRC remains one of the leading causes of cancer mortality, with
>600,000 mortalities occurring due to CRC annually worldwide
(5,6).
Identifying specific molecules associated with CRC may facilitate
the development of diagnostic and prognostic biomarkers, and more
specific targets for therapy, in order to further improve the
clinical outcome of patients with CRC.
The sirtuins (SIRTs) are a conserved family of
nicotinamide adenine dinucleotide (NAD)-dependent histone
deacetylases composed of seven SIRT isoforms (SIRT1-7) in mammalian
genomes (7,8) The nuclear protein SIRT6 is a member of
the SIRT family, which has been demonstrated to exert diverse
cancer-associated functions, including in genomic stability, DNA
repair, gene transcription repression and stress resistance, by
multiple molecular mechanisms (9).
SIRT6 is a chromatin-bound factor that promotes genomic integrity
by de-acetylating histone H3 lysine 9 (H3K9) and 56; SIRT6-mediated
histone de-acetylation contributes to the repair of DNA
double-strand breaks (10–12). SIRT6 may act as a chromatin-bound
factor to protect cells from genomic instability, including in
non-transformed cells (13,14). SIRT6 has also been reported to
function at the gene expression level via the recruitment of
transcription factors, including NF-κB and c-Jun, and may
deacetylate H3K9 at the site of gene promoters (15,16) A
previous study identified SIRT6 as a novel molecular mechanism for
controlling energy metabolism in cancer cells (17). In 2012, the Cancer Cell Line
Encyclopedia reported that the SIRT6 gene is deleted in 35% of the
1,000 human cancer cell lines recorded in their database, including
in 29% of CRC cell lines (18).
However, the role of SIRT6 in CRC and the molecular mechanisms by
which it acts as a regulator of cancer-associated metabolism remain
unknown.
Nicotinamide mononucleotide adenylyltransferase 2
(NMNAT2) is an isoform of NMNAT, which catalyzes a vital step in
the synthesis of NAD (19). NMNAT2 is
a sensitive enzyme marker for NAD levels that reflects the
intracellular redox equilibrium and cellular energy state, acting
as a sensor for cells with a high energy demand, including cancer
cells (20,21). Although the associated biological
mechanism remains largely unknown, there is increasing evidence
that NMNAT2 promotes the survival of cancer cells by accelerating
glycolysis (22). SIRT6 controls the
expression of multiple glycolytic genes, including HIF-1α (23).
Given the evidence of the possible roles for SIRT6
and NMNAT2 in human cancer, including of SIRT6 in the control of
energy metabolism by acting either as a substrate for, or regulator
of, NMNAT2 (24), the present study
was performed to determine the expression of SIRT6 and NMNAT2 in
different grades and stages of CRC.
Materials and methods
Ethical statement
All patients included in this study were informed of
the study protocol and requirements, and provided signed informed
consent to participate in the study. The study, including the
protocols for collection of tissue specimens and analysis of
clinical information, was approved by the Institutional Review
Board of Nanfang Hospital and Zhujiang Hospital of the Southern
Medical University (Guangzhou, China).
Inclusion and exclusion criteria
All patients included in the present study met the
following criteria: i) A histologically confirmed diagnosis of
primary CRC; ii) treatment with radical surgical resection; iii)
adequate tissue specimens available for tumor and normal tissue.
Patients were excluded from the study if they met the following
exclusion criteria: i) A history of other types of malignant
diseases or metastatic CRC; ii) receipt of preoperative
radiotherapy, chemotherapy and other treatments.
Tissue specimens and clinical data
collection
A total of 113 patients with CRC were enrolled in
the study. The patients underwent curative surgical resection at
the Nanfang Hospital and Zhujiang Hospital of the Southern Medical
University between January 2010 and May 2016. From these patients,
it was possible to obtain tissue from 29 fresh samples between
January 2016 and May 2016, from which tissue sections were made and
the extraction of mRNA and proteins was performed.
The 113 patients included 65 males and 48 females
aged between 30 and 91 years, with a median age of 64 years.
Patients were classified into four stages (stage I, 36; stage II,
30; stage III, 29; and stage IV, 18) according to the Union for
International Cancer Control Tumor-Node-Metastasis (TNM) staging
system (25). Tumors were graded
according to the World Health Organization system (26) as well differentiated (n=18),
moderately differentiated (n=87) or poorly differentiated (n=8).
The following clinical data were also recorded and analyzed in the
study: Tumor size, depth of invasion, lymph node metastasis and
distant metastasis. The histological diagnosis, classification and
tissue sampling was performed by two experienced senior
pathologists.
From the surgically resected colectomy specimens,
pathologists sampled tumor tissue as well as adjacent normal
colorectal tissue that was confirmed to be free from infiltration
by CRC and located >5 cm beyond the tumor margin. Fresh tissue
samples that were sampled following resection were frozen in liquid
nitrogen and stored at −80°C until analysis was performed.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of gene
transcription
A total of 29 samples of tissue RNA was extracted
with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and RNA concentration was measured by using a
NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.,
Penfield, NY, USA). PrimeScript RT master mix (Takara Bio, Inc.,
Otsu, Japan) was added to synthesize the first strand of cDNA using
the Reverse Transcription system (Promega Corporation, Madison, WI,
USA) according to the manufacturer's protocol.
RT-qPCR was performed using the 7500 FastReal-Time
Two-step PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the SYBR-Green qPCR master mix (Takara, Tokyo, Japan).
The amplification protocol commenced with a 30-sec pre-denaturation
step at 95°C, followed by 40 cycles of denaturation at 95°C for 5
sec, and annealing and extension at 60°C for 34 sec. Subsequent
verification of product specificity was conducted in the melting
stage, ending with inactivation at 95°C for 15 sec.
Quantitative gene expression data were normalized to
the expression level of GAPDH and the analysis of each sample was
performed in triplicate. The primers for human genes were designed
by PrimerBank public database and purchased from Generay Biotech
Co., Ltd. (Shanghai, China). The human-specific primers for each
gene are listed in Table I. Relative
quantification was performed using the 2−ΔΔCq method
(27).
 | Table I.Gene primers used for the polymerase
chain reaction analysis of SIRT6 and NMNAT2 in colorectal carcinoma
tumors. |
Table I.
Gene primers used for the polymerase
chain reaction analysis of SIRT6 and NMNAT2 in colorectal carcinoma
tumors.
| Gene | Forward primer,
5′-3′ | Reverse primer,
5′-3′ |
|---|
| SIRT6 |
CCCACGGAGTCTGGACCAT |
CTCTGCCAGTTTGTCCCTG |
| NMNAT2 |
TGTCCACGACTCCTATGGAAA |
GTCCGATCACAGGTGTCATGG |
| GAPDH |
GGAGCGAGATCCCTCCAAAAT |
GGCTGTTGTCATACTTCTCATGG |
Western blotting analysis of gene
translation
A total of 29 sample pairs of CRC and adjacent
non-cancer colorectal tissues were lysed in
radioimmunoprecipitation buffer (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China) containing a protease inhibitor cocktail,
phenylmethanesulfonyl fluoride and DL-dithiothreitol
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in order to extract
the protein from the samples. Following quantification with a
bicinchoninic acid protein assay kit (Thermo Fisher Scientific,
Inc.), the proteins were mixed with 5X SDS-PAGE loading buffer
(Invitrogen; Thermo Fisher Scientific, Inc.) in a water bath heated
to 95°C for 5 min to denature the proteins. A total of 30 µg
protein was loaded per lane and separated by 12% SDS-PAGE.
Following SDS-PAGE separation, proteins were transferred to
polyvinyldifluoridine membranes.
Subsequent to blocking with 10% bovine serum albumin
(Gibco; Thermo Fisher Scientific, Inc.) at room temperature for 1
h, the membranes were incubated overnight at 4°C with primary
antibodies, including anti-SIRT6 (cat no. ab62739; dilution, 1:800;
Cell Signaling Technology, Inc., Danvers, MA, USA), anti-NMNAT2
(cat no. sc-515206; dilution, 1:1,000; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) and anti-GAPDH (cat no. ab125247; dilution,
1:2,000, Abcam, Cambridge, MA, USA), followed by washing with TBS-T
buffer [including 10 mM Tris-HCl (Ph 7.4), 150 mM NaCl and 0.05%
Tween-20] and incubation with secondary antibodies [goat anti-mouse
IgG (cat no. ab6789; dilution, 1:5,000) and goat anti-rabbit IgG
(cat no. ab6721; dilution, 1:10,000), obtained from Abcam] at room
temperature for 1 h. GAPDH served as an endogenous protein control.
The immunoreactive bands were visualized with enhanced
chemiluminescence (Beyotime Institute of Biotechnology, Shanghai,
China) and the gray values of each band were detected using
Quantity One software version 4.6.2 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Immunohistochemical detection of
protein expression in tissue sections
FFPE tumor and adjacent tissues were sectioned onto
glass slides as 4-µm tissue sections. These sections were de-waxed
by dipping in xylene twice (10 min each) at room temperature and
rehydrated through a graded alcohol series subsequent to heating
overnight at 60°C. The endogenous tissue peroxidase activity was
blocked with 3% hydrogen peroxide, and pressure- and heat-induced
antigen retrieval was performed in citrate buffer (pH 6.0; Wuhan
Boster Biological Technology Co., Ltd., Wuhan, China) for 3 min.
The tissue sections were exposed to 10% goat serum albumin (OriGene
Technologies, Inc., Rockville, MD, USA) blocking solution (for 30
min at room temperature) prior to incubation in the primary
antibody solution of anti-SIRT6 (dilution, 1:150; Abcam), or
anti-NMNAT2 (dilution, 1:100; Santa Cruz Biotechnology, Inc.)
overnight at 4°C, then the slides were incubated with
Biotin-conjugated goat anti-mouse/rabbit IgG secondary antibody
(cat no. SP-9000; ready-to-use; OriGene Technologies, Inc.) for 1 h
at room temperature. Reactions between antibodies and target
proteins were visualized using 3,3′-diaminobenzidine staining.
Tissue counterstaining was performed with Mayer's hematoxylin
(ready-to-use) for 3 min at room temperature. The incubation of the
tissue sections with 0.01 mol/l PBS instead of primary antibodies
was performed as a negative control.
Light microscopy was performed using an Olympus
microscope (Olympus Corporation, Tokyo, Japan) and
immunohistochemical staining was assessed independently by two
experienced clinical pathologists. Nuclear immunostaining for SIRT6
and cytoplasmic immunostaining for NMNAT2 were considered as
positive staining. Immunostaining intensity and the percentage of
positive cells were evaluated by light microscopy with the
following intensity scoring system: 0 (negative), 1 (weak), 2
(moderate) and 3 (strong). The mean percentage of positive cells
was quantified from five different visual fields at ×200
magnification using the following percentage positive scoring
system: 0, <5%; 1, 5–25%; 2, 25–50%; 3, 50–75%; 4, >75%. The
final score for each tissue section was calculated as the
immunostaining intensity score (0 to 3) plus the positive
percentage score (0 to 4) and immunostaining was considered to be
positive when the final score was ≥3.
Statistical analysis
The data are presented as the count, percentage or
mean ± standard deviation. Statistical analysis was performed with
SPSS version 20.0 (IBM Corp., Armonk, NY, USA). The RT-qPCR and
western blotting data were analyzed by paired t-tests. The
χ2 or Fisher's exact tests were used for the analysis of
immunohistochemistry staining positivity and clinicopathological
parameters. The correlation between SIRT6 and NMNAT2 expression was
assessed with Spearman's non-parametric correlation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Protein expression of SIRT6 and NMNAT2
in CRC
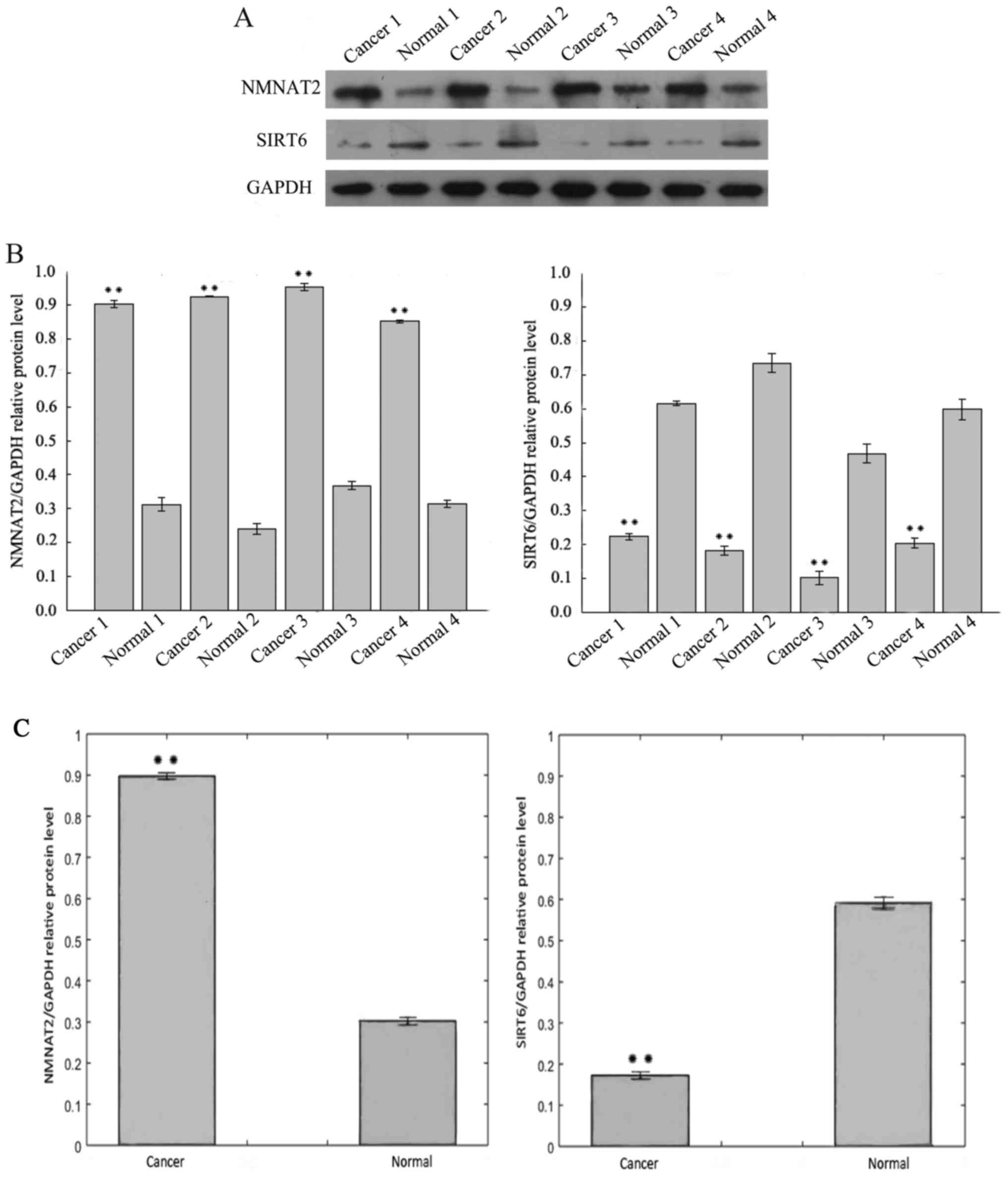
Fig. 1A includes the
results of the western blotting analysis of SIRT6 protein
expression in CRC tissue samples, which was lower than the adjacent
normal tissues from the same specimen. In contrast, the protein
levels of NMNAT2 were increased in CRC when compared with adjacent
tissue. The expression of SIRT6 and NMNAT2 protein differed
significantly between the CRC and adjacent samples (Fig. 1B and C).
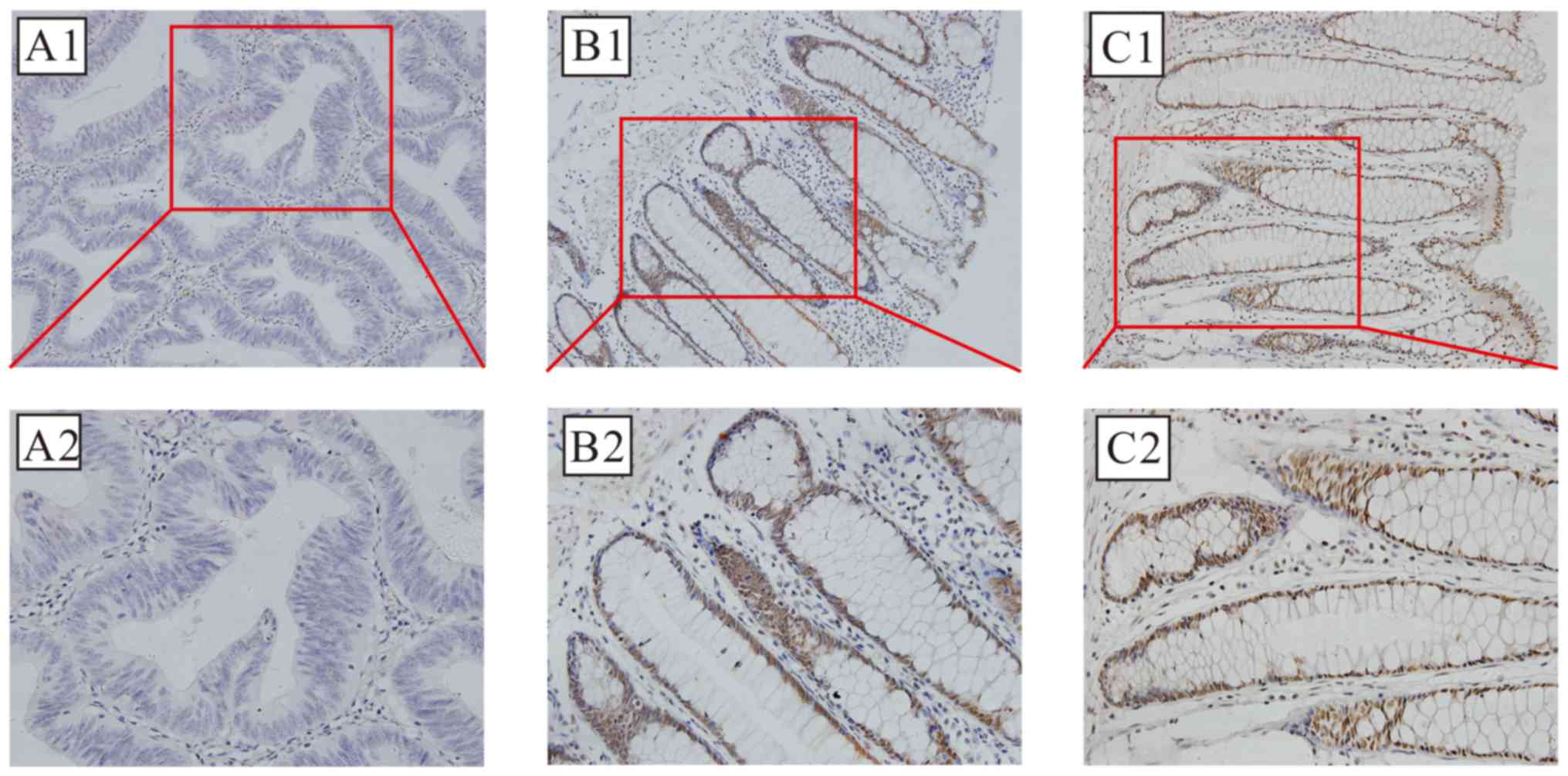
SIRT6 protein expression is localized primarily in
nucleus; brown-stained nuclei were observed in the positive
sections, as demonstrated in Fig. 2.
Positive SIRT6 staining was detected in 25 of 113 cases (22.12%) of
CRC, compared with 91 of 113 (80.53%) adjacent non-cancer
specimens; this difference was statistically significant
(χ2=77.152; P<0.01).
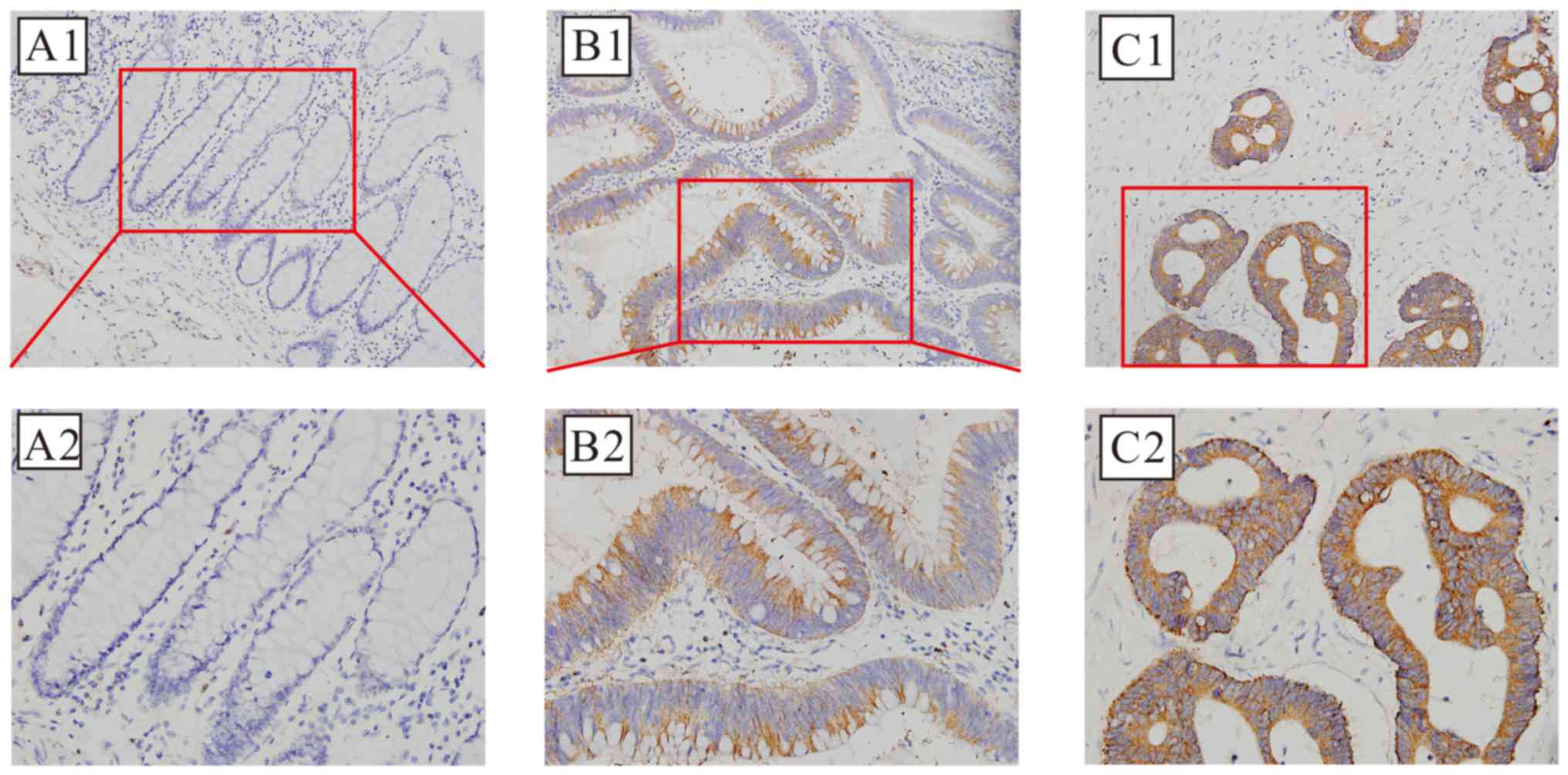
The positive cytoplasmic immunohistochemical
staining for NMNAT2 protein, which was detected in 82 of 113 cases
(72.57%) of CRC, compared with 36 of 113 (31.86%) adjacent
non-cancer specimens is demonstrated in Fig. 3; this difference was also
statistically significant (χ2=37.525; P<0.01).
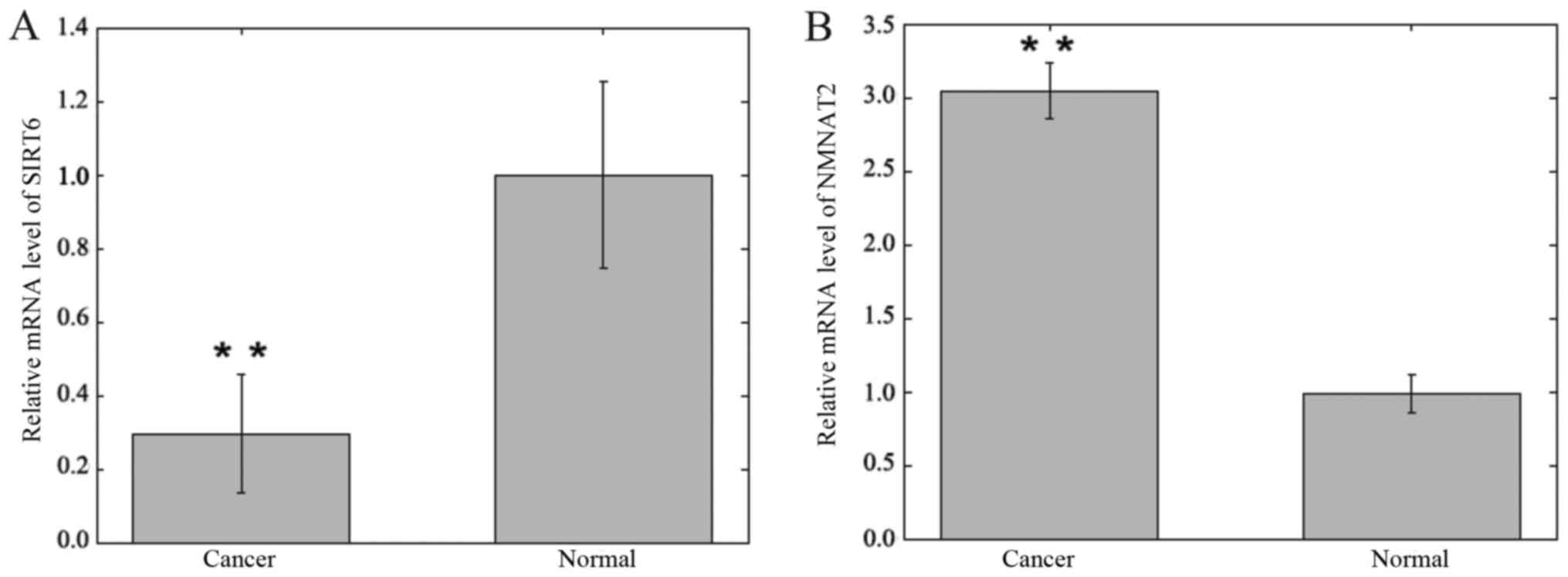
SIRT6 and NMNAT2 mRNA expression
levels in CRC
The mRNA expression levels of SIRT6 and NMNAT2 were
detected in CRC and adjacent samples using RT-qPCR and were
normalized to GAPDH. The expression levels of SIRT6 mRNA in CRC
specimens were significantly lower compared with matched adjacent
non-cancer tissue specimens (0.295±0.161 vs. 1.000±0.252;
P<0.01; Fig. 4). The expression
levels of NMNAT2 mRNA in CRC specimens were significantly increased
compared with matched adjacent non-cancer tissue specimens
(3.046±0.186 vs. 0.978±0.130; P<0.01; Fig. 4).
Correlation between the expression levels of SIRT6
and NMNAT2. The immunohistochemistry data was further analyzed to
confirm the inverse correlation between the protein expression of
SIRT6 and NMNAT2 in CRC tissue (r=−0.246; P<0.01). Furthermore,
NMNAT2-positive CRC tissue was more likely to be SIRT6-negative
than NMNAT2-positive adjacent tissue (P<0.05), whereas a similar
NMNAT2-positive rate was identified between SIRT6-positive CRC and
normal samples (P>0.05). In summary, overexpression of NMNAT2
protein was associated with the reduced expression of SIRT6 protein
in CRC tissue.
Association of clinical parameters
with SIRT6 and NMNAT2 expression in CRC tissue
Table II summarizes
the positive and negative immunohistochemical staining detection of
SIRT6 and NMNAT2, and compares the immunopositivity with the
demographic and clinicopathological parameters of the 113 patients
with CRC from the present study.
 | Table II.Association between the positive
expression of SIRT6 and NMNAT2 in colorectal carcinoma tumors as
determined with immunohistochemistry with clinical and demographic
patient parameters. |
Table II.
Association between the positive
expression of SIRT6 and NMNAT2 in colorectal carcinoma tumors as
determined with immunohistochemistry with clinical and demographic
patient parameters.
|
|
| SIRT6, n (%) |
| NMNAT, n (%) |
|
|---|
|
|
|
|
|
|
|
|---|
| Parameters | Patients | Negative | Positive | P-value | Negative | Positive | P-value |
|---|
| Total | 113 | 88 | 25 |
| 31 | 82 |
|
| Sex |
|
|
| 0.527 |
|
| 0.435 |
|
Male | 65 | 52 (80.0) | 13 (20.0) |
| 16 (24.6) | 49 (75.4) |
|
|
Female | 48 | 36 (75.0) | 12 (25.0) |
| 15 (31.3) | 33 (68.7) |
|
| Age, years |
|
|
| 0.529 |
|
| 0.319 |
|
≤64 | 57 | 43 (75.4) | 14 (24.6) |
| 18 (31.6) | 39 (68.4) |
|
|
>64 | 56 | 45 (80.4) | 11 (19.6) |
| 13 (23.2) | 43 (76.8) |
|
| Tumor size, cm |
|
|
| 0.180 |
|
| 0.332 |
| ≤3 | 50 | 36 (72.0) | 14 (28.0) |
| 16 (32.0) | 34 (68.0) |
|
|
>3 | 63 | 52 (82.5) | 11 (17.5) |
| 15 (23.8) | 48 (76.2) |
|
|
Differentiation |
|
|
| 0.040 |
|
| 0.826 |
|
Well | 18 | 10 (55.6) | 8 (44.4) |
| 6 (33.3) | 12 (66.7) |
|
|
Moderate | 87 | 72 (82.8) | 15 (17.2) |
| 23 (26.4) | 64 (73.6) |
|
|
Poor | 8 | 6 (75.0) | 2 (25.0) |
| 2 (25.0) | 6
(75.0) |
|
| Depth of
invasion |
|
|
| 0.040 |
|
| 0.023 |
| T1 | 7 | 5 (71.4) | 2 (28.6) |
| 4 (57.1) | 3 (42.9) |
|
| T2 | 38 | 24 (63.2) | 14 (36.8) |
| 15 (39.5) | 23 (60.5) |
|
| T3 | 17 | 14 (82.4) | 3 (17.6) |
| 4 (23.5) | 13 (76.5) |
|
| T4 | 51 | 45 (88.2) | 6 (11.8) |
| 8 (15.7) | 43 (84.3) |
|
| Lymph node
metastasis |
|
|
| 0.017 |
|
| 0.063 |
| N0 | 72 | 51 (70.8) | 21 (29.2) |
| 24 (33.3) | 48 (66.7) |
|
|
N1+2 | 41 | 37 (90.2) | 4 (9.8) |
| 7 (17.1) | 34 (82.9) |
|
| Distant
metastasis |
|
|
| 0.353 |
|
| 0.775 |
| M0 | 95 | 72 (75.8) | 23 (24.2) |
| 27 (28.4) | 68 (71.6) |
|
| M1 | 18 | 16 (88.9) | 2 (11.1) |
| 4 (22.2) | 14 (77.8) |
|
|
Tumor-node-metastasis stage |
|
|
| 0.032 |
|
| 0.048 |
| I | 36 | 22 (61.1) | 14 (38.9) |
| 16 (44.4) | 20 (55.6) |
|
| II | 30 | 25 (83.3) | 5 (16.7) |
| 5 (16.7) | 25 (83.3) |
|
|
III | 29 | 25 (86.2) | 4 (13.8) |
| 6 (20.7) | 23 (79.3) |
|
| IV | 18 | 16 (88.9) | 2 (11.1) |
| 4 (22.2) | 14 (77.8) |
|
The positive expression of SIRT6 protein in CRC
tissue was positively associated with the depth of tumor invasion,
tumor differentiation grade, lymph node metastasis status and TNM
stage (P<0.05). The positive expression of SIRT6 protein in CRC
tissue was more frequent in tumor tissue samples of
well-differentiated (low-grade) CRC when compared with moderately
differentiated or poorly differentiated (high-grade) CRC. Compared
with patients in stage I/II, less positive sections were detected
in stage III/IV patients.
The positive expression of NMNAT2 protein in CRC
tissues was positively associated with the depth of tumor invasion
and TNM stage (P<0.05). However, no significant association
between SIRT6 or NMNAT2 positive protein expression and the other
clinical parameters (including age, sex, tumor size and distant
metastasis) were observed in the present study.
Discussion
The initiation and progression of CRC involves
multiple steps, including hyperplasia, metaplasia, pre-invasive
carcinoma formation and ultimately, aggressive and invasive cancer.
Cells gradually acquire tumor characteristics, including unlimited
proliferation, freedom from senescence and apoptosis resistance,
leading to uncontrolled growth and tumorigenesis. Normal cells can
gain the characteristic abilities of cancer cells following the
inactivation of a single tumor suppressor or the activation of a
single carcinogenic gene. For insight into the cellular molecular
pathogenesis of CRC, specific molecular features associated with
colorectal carcinogenesis are in the process of being
identified.
The present study included well-characterized human
CRC tissues and matched normal colorectal tissues to compare the
protein and mRNA expression of SIRT6 and NMNAT2 with western
blotting, immunohistochemistry and RT-qPCR. SIRT6 protein and mRNA
expression levels were significantly reduced, whereas NMNAT2
protein and mRNA expression levels were significantly increased, in
CRC tissue relative to the adjacent tissue (P<0.01).
Immunohistochemistry confirmed a negative correlation between the
expression of SIRT6 and NMNAT2 (r=−0.246, P<0.01). The reduced
expression of SIRT6 and increased expression of NMNAT2 in CRC
tissue were associated with the depth of tumor invasion, TNM stage,
tumor differentiation grade and the presence of lymph node
metastasis (P<0.05).
The findings of the present study are supported by
previously published in vitro and in vivo studies in
animals and human cells. In a mouse model expressing an
adenomatosis polyposis coli mutation, SIRT6 deficiency was
demonstrated to increase the incidence of invasive colonic
adenocarcinoma (17). The knockout of
the SIRT6 gene from mouse embryonic fibroblasts enhanced the
proliferative capability of cells and induced tumor formation
without activating oncogenes in a severe combined immune deficiency
mouse model (16). In human
endometrial and hepatocellular cancer cell lines, the increased
expression of SIRT6 suppressed proliferation and induced apoptosis
by inhibiting the expression of the anti-apoptotic protein,
surviving (28,29). In the present study, it was
demonstrated that in human CRC, increasing tumor grade and stage
was associated with the decreased or deficient expression of SIRT6.
This result may support the hypothesis that SIRT6 acts as a
tumor-suppressor gene in human colorectal tissue. In support of
this view, SIRT6 was previously demonstrated to affect cancer cell
proliferation by suppressing the transcriptional activity of c-Myc,
a known oncogene (30).
However, other studies have indicated that there may
be other metabolic mechanisms involved in the tumor-suppressive
effects of SIRT6 (31,32). The ‘Warburg effect’ is the
reprogramming of the cellular energy metabolism to support
continuous cell growth and proliferation in malignancy (33,34). It
was previously demonstrated that the deletion of SIRT6 may activate
an energy metabolism program that promotes tumorigenesis
independently of any other transforming events (17). The roles of SIRT6 as a metabolic
enzyme and a potential regulator of cancer cell metabolism remain
to be investigated.
NMNAT2 is important in cell metabolism, and so the
increased expression of NMNAT2 in rapidly dividing cancer cells
would be expected (21,22). SIRT3 has been demonstrated to be
necessary for the deacetylation of NMNAT2 in lung cancer cells,
which promotes cell proliferation and growth through affecting
energy metabolism, so it is logical that SIRT6 may have a similar
role in CRC (35). However, the
present study has also demonstrated that the mechanism underlying
SIRT6 as a tumor suppressor in CRC may also involve regulating the
transcription or expression of NMNAT2. The protein and the mRNA
levels of NMNAT2 were upregulated in CRC tissue when compared with
the normal colorectal tissue samples, and a negative correlation
between SIRT6 and NMNAT2 was identified. This may indicate that
NMNAT2 is involved in the tumorigenesis and development of CRC by
maintaining intracellular NAD, fuelling rapid cell growth and
proliferation. Furthermore, the results indicate that the
expression of NMNAT2 may be regulated, at least partly, by SIRT6
through mRNA and protein regulatory mechanisms in CRC.
In the present study, the expression levels of SIRT6
and NMNAT2 in CRC were studied in relation to patient demographics
and clinicopathological parameters using immunohistochemistry, a
technique that is part of routine histopathological diagnosis and
prognostic evaluation. The histological protein expression of SIRT6
and NMNAT2 in human CRC was negatively correlated with the presence
of CRC and with CRC grade (r=−0.246; P<0.01). These
immunohistochemistry findings may support the hypothesis that the
increased expression of NMNAT2 and reduced expression of SIRT6 were
associated with CRC progression, and that the downregulation of
SIRT6 may promote the expression of NMNAT2. Further studies are
recommended to investigate the potential association between SIRT6
and NMNAT2 in CRC, and other malignant tumor types, to provide new
insights into the inhibition of tumor progression by affecting the
tumor cell metabolism.
The present study has potential clinical
implications for CRC and other forms of malignant disease,
indicating that further studies should be undertaken. In patients
with ovarian cancer or hepatocellular carcinoma, the decreased
expression of SIRT6 was previously associated with accelerated
tumor cell growth and poor clinical prognosis (14,36).
Previous studies have demonstrated an oncogenic role for activated
metabolic enzyme adaptations during cancer progression, including
the serine biosynthetic rate-limiting enzyme D-3-phosphoglycerate
dehydrogenase, and overexpression of the glycine-metabolizing
enzyme glycine decarboxylase (37–39). There
is evidence that the inhibition of aberrant glycolysis may be a
potential treatment strategy for cancer, with the enzymes
associated with the dysfunction of NAD metabolism being suggested
as treatment targets for obesity, diabetes and cancer (40). Poly(ADP) ribose polymerase inhibitors
have also been investigated for chemoprevention, radiosensitization
and as anticancer agents (41). This
supports the potential for NMNAT2 as a diagnostic and therapeutic
target in CRC.
The present study had several limitations. It should
be acknowledged that this was a preliminary study that investigated
a small number of tumor samples from a limited number of colorectal
cancer surgical units. Although tissue sampling was performed with
the assistance of experienced pathologists, it was not possible to
exclude the possibility that the ‘normal’ tissues were inflamed,
adenomatous or dysplastic, or contained micrometastases, as
sampling was performed visually. The tissues sampled were collected
and stored according to a standard protocol to ensure that the
integrity of tissue proteins and mRNA were preserved. However, in a
busy surgical and surgical pathology unit, delays can occur between
the surgical removal of a specimen and tissue sampling that cannot
easily be controlled. Furthermore, the immunohistochemical staining
of the tumor and normal cells was evaluated by skilled
pathologists, but the scoring was performed ‘by eye’, which may be
subject to grading bias; this limitation could be overcome in
future studies with the use of image analysis and objective
immunohistochemical quantification techniques.
In conclusion, the findings of this preliminary
study demonstrated that the increased expression of NMNAT2 and
reduced expression of SIRT6 were associated with the progression of
CRC. It is possible that the downregulation of SIRT6 may promote
the expression of NMNAT2. Further studies are required to
characterize the role of NMNAT2 and SIRT6 as potential diagnostic
and prognostic biomarkers and as targets for therapy in CRC, and
other types of malignant tumor.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Guangdong Province, China (grant no., 2014
a030313295).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
JQ was responsible for designing the present study,
for conducting the main experiments and compiling the manuscript.
QD was involved in preparing the sample sections and for protein
extraction. LW collated and analyzed the data. RC, DZ and LX
contributed to the sample collection. CC and JY were involved in
designing the study and revising the manuscript critically for
important intellectual content.
Ethics approval and consent to
participate
The present study, including the protocols for
collection of tissue specimens and analysis of clinical
information, was approved by the Institutional Review Board of
Nanfang Hospital and Zhujiang Hospital of the Southern Medical
University (Guangzhou, China).
Patient consent for publication
All patients included in this study were informed of
the study protocol and requirements, and provided signed informed
consent to participate in the study.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
NAD
|
nicotinamide adenine dinucleotide
|
|
NMNAT2
|
nicotinamide mononucleotide
adenylyltransferase 2
|
|
SIRT6
|
sirtuin 6
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rabeneck L, Horton S, Zauber AG and Earle
C: Colorectal Cancer. In: Cancer: Disease Control Priorities, Third
Edition (Volume 3)Gelband H, Jha P, Sankaranarayanan R and Horton
S: The International Bank for Reconstruction and Development/The
World Bank (C) 2015 International Bank for Reconstruction and
Development/The World Bank. Washington (DC): 2015
|
|
6
|
Zhu AX, Kudo M, Assenat E, Cattan S, Kang
YK, Lim HY, Poon RT, Blanc JF, Vogel A, Chen CL, et al: Effect of
everolimus on survival in advanced hepatocellular carcinoma after
failure of sorafenib: The EVOLVE-1 randomized clinical trial. JAMA.
312:57–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bruzzone S, Parenti MD, Grozio A,
Ballestrero A, Bauer I, Del Rio A and Nencioni A: Rejuvenating
sirtuins: The rise of a new family of cancer drug targets. Curr
Pharm Des. 19:614–623. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lerrer B, Gertler AA and Cohen HY: The
complex role of SIRT6 in carcinogenesis. Carcinogenesis.
37:108–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Michishita E, McCord RA, Berber E, Kioi M,
Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL,
Barrett JC, et al: SIRT6 is a histone H3 lysine 9 deacetylase that
modulates telomeric chromatin. Nature. 452:492–496. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tennen RI and Chua KF: Chromatin
regulation and genome maintenance by mammalian SIRT6. Trends
Biochem Sci. 36:39–46. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toiber D, Erdel F, Bouazoune K, Silberman
DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor
B, Giacosa S, et al: SIRT6 recruits SNF2H to DNA break sites,
preventing genomic instability through chromatin remodeling. Mol
Cell. 51:454–468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cea M, Cagnetta A, Adamia S, Acharya C,
Tai YT, Fulciniti M, Ohguchi H, Munshi A, Acharya P, Bhasin MK, et
al: Evidence for a role of the histone deacetylase SIRT6 in DNA
damage response of multiple myeloma cells. Blood. 127:1138–1150.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang G, Liu Z, Qin S and Li K: Decreased
expression of SIRT6 promotes tumor cell growth correlates closely
with poor prognosis of ovarian cancer. Eur J Gynaecol Oncol.
36:629–632. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu SS, Cai Y, Ye JT, Pi RB, Chen SR, Liu
PQ, Shen XY and Ji Y: Sirtuin 6 protects cardiomyocytes from
hypertrophy in vitro via inhibition of NF-κB-dependent
transcriptional activity. Br J Pharmacol. 168:117–128. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sundaresan NR, Vasudevan P, Zhong L, Kim
G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam
V, et al: The sirtuin SIRT6 blocks IGF-Akt signaling and
development of cardiac hypertrophy by targeting c-Jun. Nat Med.
18:1643–1650. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sebastian C, Zwaans BM, Silberman DM,
Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber
D, et al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The Cancer Cell Line Encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mayer PR, Huang N, Dewey CM, Dries DR,
Zhang H and Yu G: Expression, localization, and biochemical
characterization of nicotinamide mononucleotide adenylyltransferase
2. J Biol Chem. 285:40387–40396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jayaram HN, Kusumanchi P and Yalowitz JA:
NMNAT expression and its relation to NAD metabolism. Curr Med Chem.
18:1962–1972. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mouchiroud L, Houtkooper RH and Auwerx J:
NAD+ metabolism: A therapeutic target for age-related
metabolic disease. Crit Rev Biochem Mol Biol. 48:397–408. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong L, D'Urso A, Toiber D, Sebastian C,
Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD,
Nir T, et al: The histone deacetylase Sirt6 regulates glucose
homeostasis via Hif1alpha. Cell. 140:280–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bauer I, Grozio A, Lasigliè D, Basile G,
Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A, et al:
The NAD+-dependent histone deacetylase SIRT6 promotes cytokine
production and migration in pancreatic cancer cells by regulating
Ca2+ responses. J Biol Chem. 287:40924–40937. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Resch A and Langner C: Lymph node staging
in colorectal cancer: Old controversies and recent advances. World
J Gastroenterol. 19:8515–8526. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Van Cutsem E and Ducreux M: Colorectal and
gastric cancer in 2015: The development of new agents and molecular
classifications. Nat Rev Clin Oncol. 13:69–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fukuda T, Wada-Hiraike O, Oda K, Tanikawa
M, Makii C, Inaba K, Miyasaka A, Miyamoto Y, Yano T, Maeda D, et
al: Putative tumor suppression function of SIRT6 in endometrial
cancer. FEBS Lett. 589:2274–2281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang ZG and Qin CY: Sirt6 suppresses
hepatocellular carcinoma cell growth via inhibiting the
extracellular signal-regulated kinase signaling pathway. Mol Med
Rep. 9:882–888. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q,
Kong S, Ye J, Gao B and Fang D: USP10 antagonizes c-Myc
transcriptional activation through SIRT6 stabilization to suppress
tumor formation. Cell Rep. 5:1639–1649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kleszcz R, Paluszczak J and Baer-Dubowska
W: Targeting aberrant cancer metabolism-The role of sirtuins.
Pharmacol Rep. 67:1068–1080. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiong X, Wang G, Tao R, Wu P, Kono T, Li
K, Ding WX, Tong X, Tersey SA, Harris RA, et al: Sirtuin 6
regulates glucose-stimulated insulin secretion in mouse pancreatic
beta cells. Diabetologia. 59:151–160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vander Heiden MG, Locasale JW, Swanson KD,
Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G,
Rabinowitz JD, Asara JM and Cantley LC: Evidence for an alternative
glycolytic pathway in rapidly proliferating cells. Science.
329:1492–1499. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:703–707. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li H, Feng Z, Wu W, Li J, Zhang J and Xia
T: SIRT3 regulates cell proliferation and apoptosis related to
energy metabolism in non-small cell lung cancer cells through
deacetylation of NMNAT2. Int J Oncol. 43:1420–1430. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marquardt JU, Fischer K, Baus K, Kashyap
A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, et al:
Sirtuin-6-dependent genetic and epigenetic alterations are
associated with poor clinical outcome in hepatocellular carcinoma
patients. Hepatology. 58:1054–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Locasale JW, Grassian AR, Melman T,
Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen
T, Sharfi H, et al: Phosphoglycerate dehydrogenase diverts
glycolytic flux and contributes to oncogenesis. Nat Genet.
43:869–874. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Possemato R, Marks KM, Shaul YD, Pacold
ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et
al: Functional genomics reveal that the serine synthesis pathway is
essential in breast cancer. Nature. 476:346–350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang WC, Shyh-Chang N, Yang H, Rai A,
Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al: Glycine
decarboxylase activity drives non-small cell lung cancer
tumor-initiating cells and tumorigenesis. Cell. 148:259–272. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cantó C, Houtkooper RH, Pirinen E, Youn
DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux
PA, Cettour-Rose P, et al: The NAD(+) precursor nicotinamide
riboside enhances oxidative metabolism and protects against
high-fat diet-induced obesity. Cell Metab. 15:838–847. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mégnin-Chanet F, Bollet MA and Hall J:
Targeting poly(ADP-ribose) polymerase activity for cancer therapy.
Cell Mol Life Sci. 67:3649–3662. 2010. View Article : Google Scholar : PubMed/NCBI
|