Introduction
Colorectal cancer (CRC) is the most prevalent type
of cancer and the second most important cause of cancer-associated
mortality in Western countries (1),
and the incidence of CRC is increasing in certain countries of
Eastern Europe and Asia (2). The
progression from normal colonic mucosa to malignant tumor is a
multistep process, which is involved in many genetic alternations.
Chromosomal instability, primarily including adenomatous polyposis
coli (APC) gene mutation, and the inactivation of the mismatch
repair gene system are the potential molecular mechanisms that lead
to CRC (3,4). Proteinase-activated receptors (PARs), a
subfamily of G-protein-coupled receptors, composed of four members,
PAR1, PAR2, PAR3 and PAR4 (5), have
been identified to be involved in the progression of various types
of cancer (6–10). PAR1 and PAR2 were widely expressed in
adenocarcinomas, melanomas, osteosarcomas, glioblastomas,
meningiomas, leukemias, and breast and colon cancer. PAR1 and PAR2
promote cancer cell proliferation, motility and metastasis
(11–13). PAR3 was expressed in kidney and liver
cancer (14,15). In human pancreatic adenocarcinoma
PANC-1 cells, knockdown of PAR3 markedly enhanced cell migration
and invasion (16). PAR4 was also
expressed in multiple tumors. In prostate cancer, PAR4 expression
was increased compared with the normal glands; however, no
correlation between PAR4 expression and Gleason score was
identified (17,18). PAR4 mRNA was identified in 10/14 (71%)
human colon cancer cell lines. PAR4 protein expression was absent
from normal colon mucosa, but appeared as evident staining in the
dysplastic and colon cancerous mucosa (19). In our previous study, we identified
that PAR4 expression was increased in colon cancer compared with
the associated normal tissue, and the upregulated expression was
associated with lymph node invasion and cell differentiation
(6). However, the effect of PAR4 on
CRC cell proliferation, survival and migration, and the potential
molecular mechanism involved in the functions have, to the best of
our knowledge, not been investigated. We hypothesized that PAR4
promoted the proliferation and migration of CRC cells. Therefore,
PAR4 was overexpressed in CRC LoVo cells or knocked down in HT-29
cells, and the effect of PAR4 on the proliferation and migration of
CRC cells, and the phosphorylation level of
extracellular-signal-regulated kinase (ERK)1/2 were investigated.
The results of the present study indicated that overexpression of
PAR4 promoted proliferation, survival and migration of CRC cells,
and increased the phosphorylation level of ERK1/2. These
observations suggested that PAR4 is involved in the progression of
CRC.
Materials and methods
Ethics statement
The present study was reviewed and approved by The
Second Affiliated Hospital of Kunming Medical University. All
animals were raised according to the protocols approved by the
Kunming Medical Experimental Animal Care Commission.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
RNA extraction, first-strand cDNA synthesis and the
design of primers were as described previously (7). The primers used for qPCR were as
follows: GAPDH (107 bp product; internal control),
5′-TGATGACATCAAGAAGGTGGTGAAG-3′ (forward) and
5′-TCCTTGGAGGCCATGTGGGCCAT-3′ (reverse); PAR4 (147 bp product),
5′-CCTTCATCTACTACTACTACGTGTCG-3′ (forward) and
5′-ACTGGAGCAAAGAGGAGTGG-3′ (reverse). qPCR for PAR4 and GAPDH were
performed using an SYBR Green Real-Time PCR kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with the conditions as follows:
Initial denaturation at 95°C for 1 min, followed by 40 cycles of
95°C for 15 sec and 60°C for 1 min. Each sample was run three
times. Products were analyzed using a continuous fluorescence
detector with Opticon Monitor 3.0 software (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). No-template controls (no cDNA in PCR)
were included to detect non-specific or genomic amplification and
primer dimerization. Relative quantitative evaluation of PAR4 was
performed using the E-method (6,7) and
expressed as a ratio of the transcript of PAR4 to GAPDH in the
different cell lines. The identities of qPCR products were
confirmed by DNA sequencing.
Western blot analysis
Mouse anti-human PAR4 monoclonal antibody (cat. no.
sc-130078; 1:1,000) and mouse anti-human β-actin monoclonal
antibody (cat. no. sc-517582; 1:1,000) were from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA), and mouse anti-human
phospho-ERK1/2 monoclonal antibody (cat. no. 9106; 1:2,000) and
rabbit anti-human ERK1/2 polyclonal antibody (cat. no. 9102;
1:1,000) were from Cell Signaling Technology, Inc. (Danvers, MA,
USA). Cells (1×104) were washed twice with ice-cold PBS
and were lysed with radioimmunoprecipitation lysis buffer (Thermo
Fisher Scientific, Inc.) containing a protease and phosphatase
inhibitor cocktail (Roche Diagnostics, Basel, Switzerland). The
protein concentration was determined using a Quick Start™ Bradford
protein assay kit (cat. no. 5000201; Bio-Rad Laboratories, Inc.).
Samples (20 µg protein) were separated by SDS-PAGE (30% acrylamide)
and then transferred onto a polyvinyl difluoride membrane. The
membrane was subsequently blocked with 3% bovine serum albumin
diluted in TBS with 0.1% Tween-20 (1:1,000; cat. no. T8220; Beijing
Solarbio Science and Technology Co., Ltd., Beijing, China) and
incubated with the primary antibody and the horseradish
peroxidase-labeled goat anti-rabbit (cat. no. SE134; 1:1,000) and
anti-mouse (cat. no. SE131; 1:1,000) (both from Beijing Solarbio
Science and Technology Co., Ltd.). Proteins were visualized using
Super Signal reagents (Pierce; Thermo Fisher Scientific, Inc.) with
the ChemiDoc XRS imaging system (Bio-Rad Laboratories, Inc.) and
analyzed using the Image Lab 5.2 software (Bio-Rad Laboratories,
Inc.).
Cell culture
Human CRC LoVo and HT-29 cell lines were from the
American Type Culture Collection (Manassas, VA, USA). LoVo cells
were cultured in Ham's F-12K (Kaighn's) medium (cat. no. 21127022;
Pierce; Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v)
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 U/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.), and HT-29 cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% (v/v) FBS, 100 U/ml penicillin and 100 U/ml
streptomycin. Cells were cultured at 37°C in a humidified
atmosphere containing 5% CO2.
Generation of LoVo cells
overexpressing PAR4
cDNA encoding human PAR4 was inserted into
bicistronic retroviral vector to obtain pBMN-PAR4-NEO.
pBMN-PAR4-NEO was used to produce retrovirus containing PAR4, and
LoVo cells were infected using the retrovirus. In brief, retrovirus
expressing PAR4 was generated by transfecting pBMN-PAR4-NEO plasmid
into Phoenix cells (Pierce; Thermo Fisher Scientific, Inc.). At 48
h after transfection, the retrovirus supernatant was collected,
centrifuged (1,000 × g for 3 min at room temperature) and filtered
through a 0.45 µm filter. LoVo/PAR4 cells were produced using the
retrovirus supernatant with PAR4, and LoVo/vector cells were
generated using the retrovirus supernatant with pBMN-I-NEO only.
G418 (600 µg/ml) was used to select the stable overexpression of
PAR4 (LoVo/PAR4) and the empty vector (LoVo/vector).
Knockdown of PAR4 in HT-29 cells
Since PAR4 was expressed in CRC HT-29 cells, short
hairpin RNA (shRNA) was used to knock down PAR4 in HT-29 cells.
shRNA targeting PAR4 was from GE Healthcare Dharmacon, Inc.
(Lafayette, CO, USA). Briefly, lentivirus expressing PAR4 shRNA was
generated by co-transfecting PAR4 shRNA plasmids with pCMV-dR8.2
dvpr and pCMV-VSVG packaging plasmids into 293FT cells (Shanghai
Genechem Co., Ltd., Shanghai, China.). At 48 h after transfection,
the lentiviral supernatant was collected, centrifuged (1,000 × g
for 3 min at room temperature) and filtered through a 0.45 µm
filter. HT-29 cells were infected with collecting lentivirus
supernatant with shPAR4. The lentiviral vector pGIPZ, containing
puromycin and green fluorescent protein (GFP) selection markers,
was used to knock down PAR4. HT-29/pGIPZ cells were generated using
the lentivirus supernatant with pGIPZ only as control. HT-29 cells
transduced with pGIPZ only or pGIPZ-shPAR4 were selected using 5
µg/ml puromycin to generate HT-29/pGIPZ or HT-29/shPAR4,
respectively.
Cell proliferation assay
Cell proliferation was determined by viable cell
counting in Dulbecco's modified Eagle's (high-glucose) medium
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS.
LoVo/vector or LoVo/PAR4 cells were seeded in a 6-well plate with
each containing 1×105 cells in 3 ml culture medium
individually The number of cells was determined daily for 5 days
using a hemocytometer and the fold increase in cell proliferation
was calculated.
Cell survival assay
The soft agar colony assay was performed as
described by Yu et al (20).
In brief, 2×104 LoVo/vector or LoVo/PAR4 cells were
seeded in the 0.35% top agarose in culture medium with 5% FBS, and
the base agarose concentration was 0.7% in the same medium in a
6-well plate, and the cells were fed every 3 days with culture
medium containing 5% FBS. The colony number was quantified in 10
randomly selected visual fields when the cells were cultured for 2
weeks.
In vivo tumorigenicity assay
The animal protocol was designed to minimize pain or
discomfort to the animals. The female BALB/c nude mice with 4–6
weeks old were obtained from Experimental Animal Center of Kunming
Medical University (Kunming, China), and were randomly divided into
2 groups (n=4/group). The animals (weight, 21.19±1.45 g) were
acclimatized to laboratory conditions (23°C, 12-h light/12-h dark
cycle, 50% humidity, ad libitum access to food and water)
for 2 weeks prior to experimentation. A total of 1×106
LoVo/PAR4, LoVo/vector cells, HT-29/pGIPZ, HT-29/shPAR4#4 or
HT-29/shPAR4#5 cells in 50 µl PBS were injected subcutaneously into
the two sides of the male nude mice. Tumor growth was monitored
weekly by determining the tumor volume using the formula
V=(W2 × L)/2, where V is volume; W is width and L is
length. After 5 weeks, the mice were sacrificed, and the tumors
were isolated and weighed. Animals were sacrificed immediately on
presentation of signs of pain, distress, suffering or impending
mortality.
Cell migration assay
Migratory activities of LoVo/PAR4, LoVo/vector
cells, HT-29/pGIPZ, HT-29/shPAR4#4 or HT-29/shPAR4#5 cells in
vitro were determined using a Matrigel insert migration assay.
Following starvation for 24 h, 3×105 cells in 300 ml
serum-free medium were seeded into a FluoroBlok™ Cell Culture
insert (Corning Incorporated, Corning, NY, USA). The lower chamber
of a 24-well plate contained 500 ml pre-warmed culture medium
containing 10% FBS. At 16 h after seeding, the non-migrating cells
remaining in the insert were scraped off using a cotton swab and
the migrated cells in the bottom part of the insert were labeled
with calcein acetoxymethyl ester (Invitrogen; Thermo Fisher
Scientific, Inc.) in culture medium containing 10% FBS. Cells that
had migrated through the membranes were quantified by determining
the number of cells in five randomly selected visual fields.
Statistical analysis
Results are expressed as the mean ± standard
deviation. Differences between the groups were evaluated using a
paired Student's t-test or one-way analysis of variance.
Statistical analyses were performed using SPSS software for Windows
(version 21.0; IBM Corp., Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
PAR4 increases LoVo cell proliferation
and survival
To investigate the effect of PAR4 on CRC cell
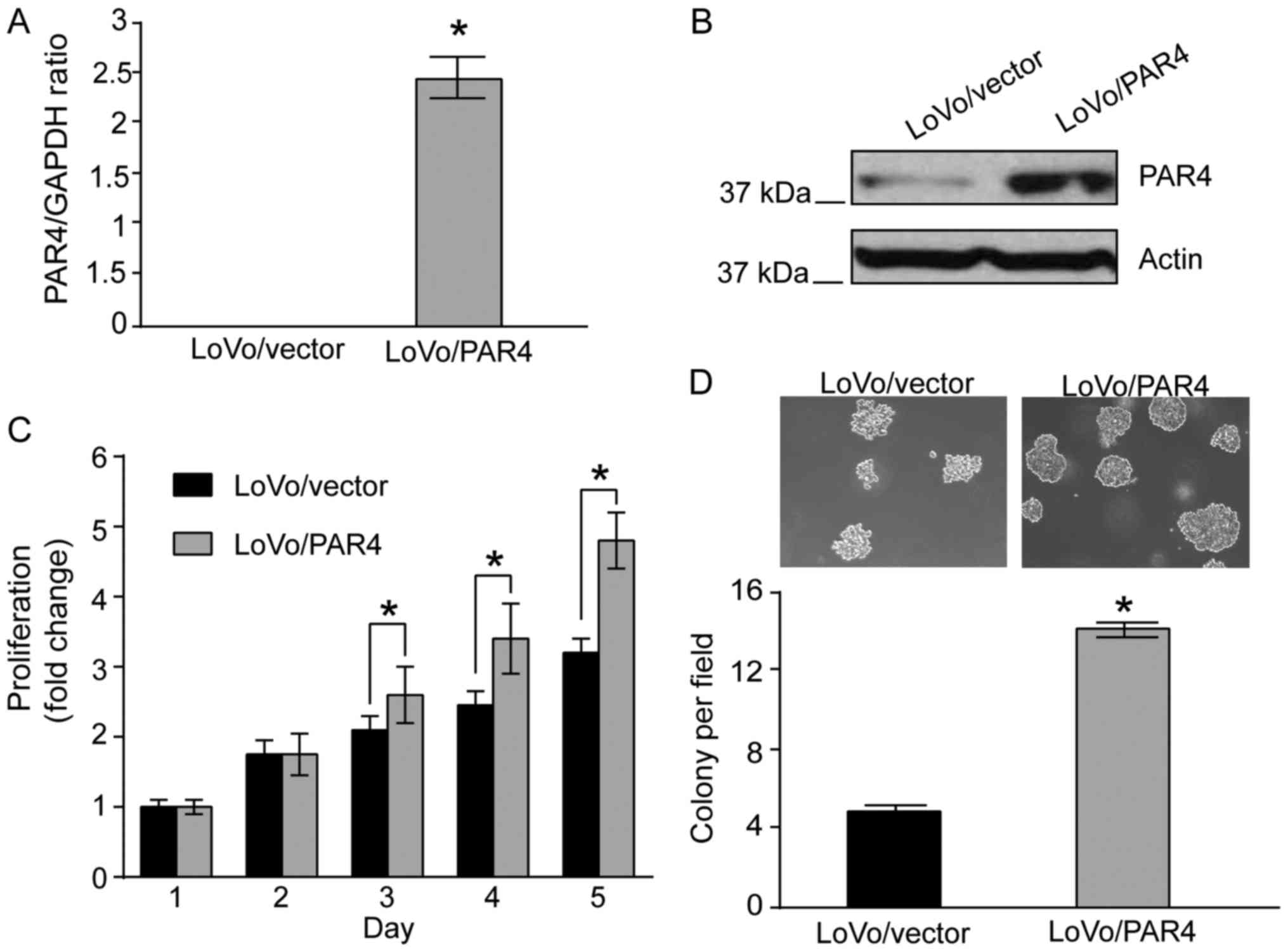
proliferation, PAR4 was overexpressed in LoVo cells. qPCR analysis
indicated that mRNA expression of PAR4 was increased significantly
in LoVo/PAR4 cells compared with in LoVo/vector cells (Fig. 1A). Western blot analysis revealed that
PAR4 protein was increased in LoVo/PAR4 cells compared with in the
LoVo/vector cells (Fig. 1B). The
effect of overexpression of PAR4 on LoVo cells was investigated. In
culture medium containing 10% FBS, from day 3 onwards, a
significant difference in cell proliferation activity between
LoVo/PAR4 cells and the control LoVo/vector cells was identified
(Fig. 1C), indicating that PAR4
promotes LoVo cell proliferation under standard culture conditions.
It was next investigated whether the PAR4-mediated proliferation
leads to increased anchorage-independent growth in soft agar. When
LoVo/vector cells were plated in soft agar, none or few colonies
were observed when the colonies were cultured for 2 weeks, whereas
a significant number of LoVo/PAR4 cells colonies was observed.
Quantification indicated that expression of PAR4 led to a
>3-fold increase in colony formation of LoVo cells (Fig. 1D). Taken together, these results
indicated that increased expression of PAR4 enhances proliferation
and survival of LoVo cells.
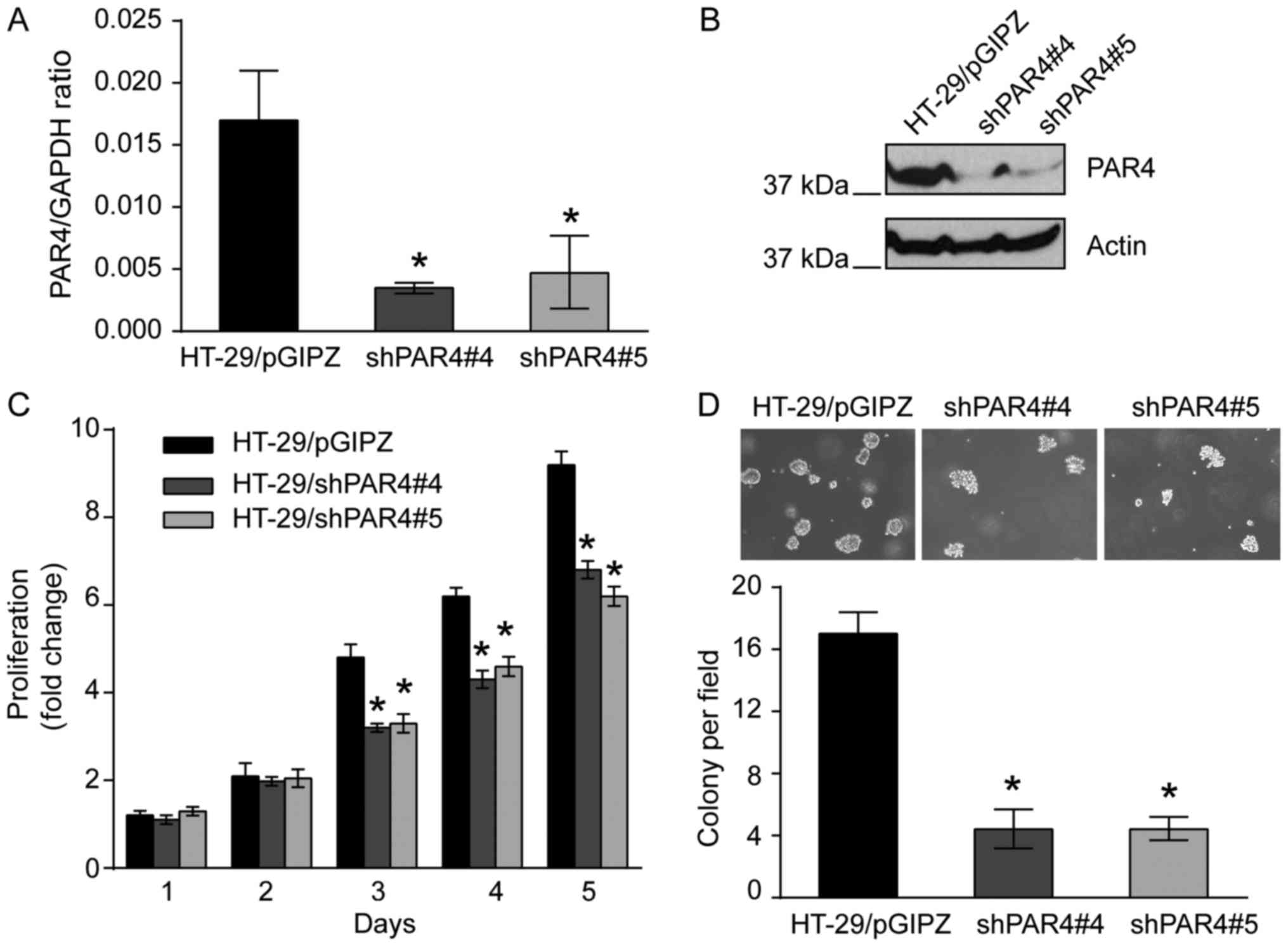
Knockdown of PAR4 decreases
proliferation and anchorage-independent growth of HT-29 cells
Since overexpression of PAR4 promotes LoVo cell
proliferation, the effect of proliferation of knocking down PAR4 on
CRC cells as investigated. Since PAR4 was expressed in CRC HT-29
cells (21), lentiviral shRNA was
used to knock down PAR4 in HT-29 cells. The lentiviral vector
pGIPZ, containing puromycin and GFP selection markers, was used to
knock down PAR4. HT-29 cells with knockdown of PAR4 were generated
with 5 µg/ml puromycin resistance selection. HT-29 cells
transfected with pGIPZ vector were selected using 5 µg/ml puromycin
and were used as a control. qPCR indicated that PAR4 mRNA
expression was decreased in HT-29/shPAR4#4 (P<0.05) or
HT-29/shPAR4#5 (P<0.05) clones compared with HT-29/pGIPZ control
cells (Fig. 2A). Western blotting
using PAR4-specific antibody was used to confirm the knockdown of
PAR4 in these cell lines. As expected, the protein expression level
of PAR4 in HT-29/shPAR4#4 and HT-29/shPAR4#5 cells was markedly
decreased compared with HT-29/pGIPZ control cells (Fig. 2B).
The effect of knockdown of PAR4 on HT-29 cell
proliferation was investigated. As presented in Fig. 2C, the proliferative activities of
HT-29/shPAR4#4 and HT-29/shPAR4#5 clones were significantly
decreased compared with HT-29/pGIPZ control in standard culture
medium containing 10% FBS. Similarly, in the colony formation
assay, when these cells were plated in agarose with culture medium
containing 5% FBS and cultured for 2 weeks, colony formation of
HT-29/shPAR4#4 or HT-29/shPAR4#5 cells in soft agar was decreased
significantly compared with HT-29/pGIPZ cells after 2 days.
Determination of the colony numbers indicated that knockdown of
PAR4 in HT-29 cells led to a 4-fold decrease in colony formation
compared with the HT-29/pGIPZ control cells (P<0.05; Fig. 2D). The results suggested that
knockdown of PAR4 in HT-29 cells significantly decreased the
proliferation and survival of CRC HT-29 cells.
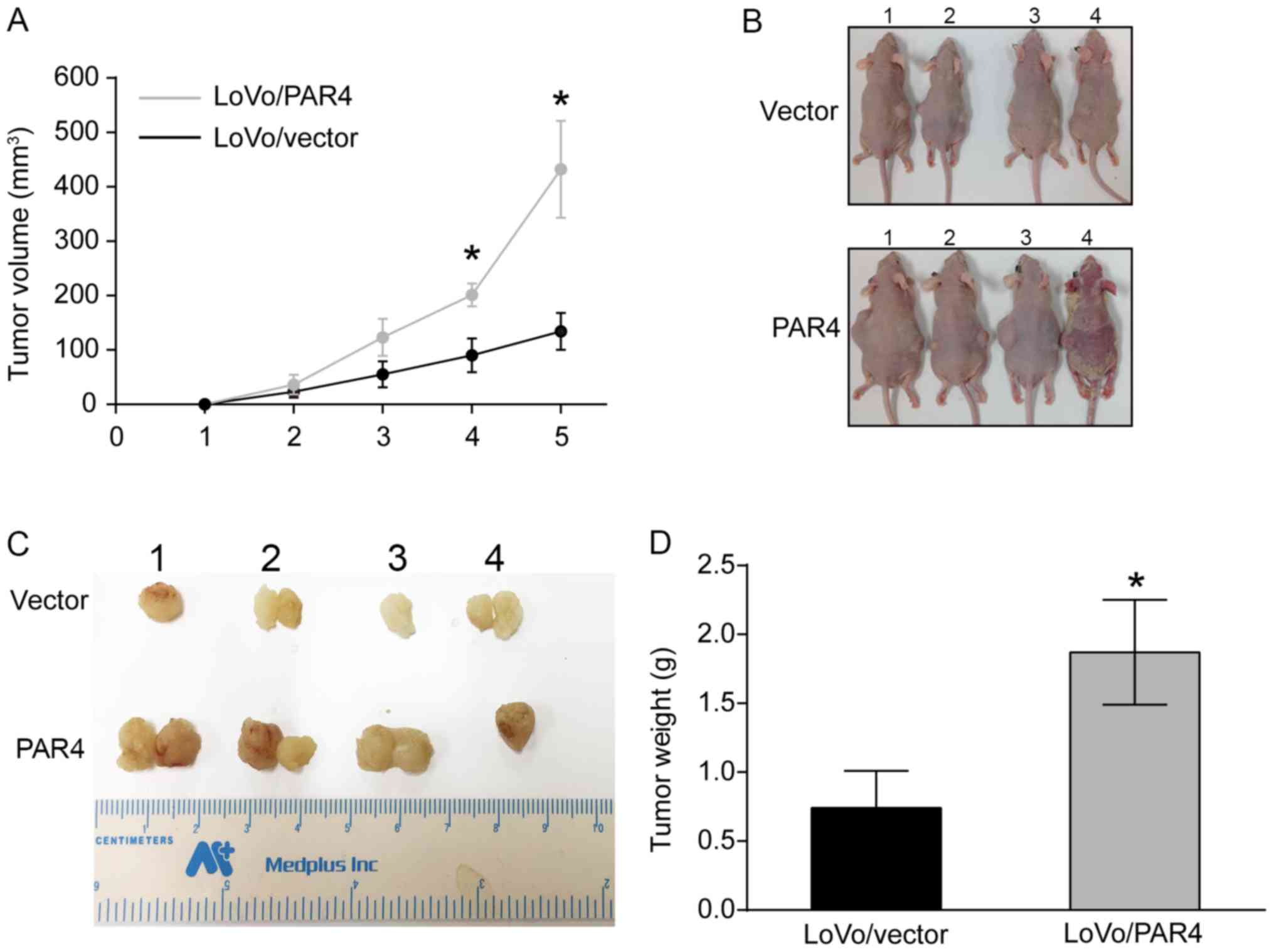
Overexpression of PAR4 increases LoVo
cell tumorigenesis
Since PAR4 overexpression increased LoVo cell
proliferation and survival, the effect of PAR4 on tumorigenesis was
investigated. LoVo/vector and LoVo/PAR4 cells were injected into
the two sides of each nude mouse subcutaneously, and the tumor
volume was determined weekly. As presented in Fig. 3A, quantification of the tumor volume
over 5 weeks indicated that the LoVo/PAR4 tumor volume increased
more rapidly compared with the LoVo/vector tumor volume, and there
was as significant difference between LoVo/PAR4 and LoVo/vector
from 3 weeks onwards. When the mice were sacrificed in week 5, it
was identified that there were six tumors present in the
LoVo/vector group, but seven tumors present in the LoVo/PAR4 group.
The tumor of mouse no. 1 in PAR4/LoVo group exhibited the longest
diameter (1.83 cm). Multiple tumors was observed for mouse no. 2
(two tumors; 0.3 and 0.5 cm), no. 3 (two tumors; 0.2 and 0.5 cm)
and no. 4 (two tumors; 0.2 and 0.3 cm) in the control group, and
no. 1 (two tumors; 1.8 and 0.5 cm), no. 2 (two tumors; 0.2 and 0.7
cm) and no. 3 (two tumors; 1.7 and 0.2 cm) in the LoVo/PAR4 group
(Fig. 3B). The average tumor weight
for the LoVo/vector group was 0.675±0.12 g, and for the LoVo/PAR4
group was 1.875±0.09 g, which suggested that there was significant
difference between the tumor weight between LoVo/vector and
LoVo/PAR4 group (Fig. 3C and D). The
weights of the animals at the time of sacrifice were 1.87, 1.89,
1.19 and 1.96 g for the LoVo/PAR4 group. The tumor burden of mouse
no. 1 and no. 3 in the LoVo/PAR4 group was >10% of its body
weight, and the maximum tumor burden was 11.6%. Furthermore, a
‘cornmeal-like appearance’ in mouse #4 (but no other mouse) of the
LoVo/PAR4 group may have been caused by an infection by
Corynebacterium bovis, which healed after 6 days, and the
mouse did not exhibit signs of pain or suffering during the
experimental period. The alteration in the skin of this mouse had
little effect on the results of the present study. The results
indicated that expression of PAR4 promoted colorectal tumor growth,
possibly through an increase in proliferation and survival of CRC
cells. However, when HT-29/pGIPZ, HT-29/shPAR4#4 or shPAR4#5 cells
were each injected into nude mice subcutaneously, there was no
significant difference in tumor growth among them (data not
shown).
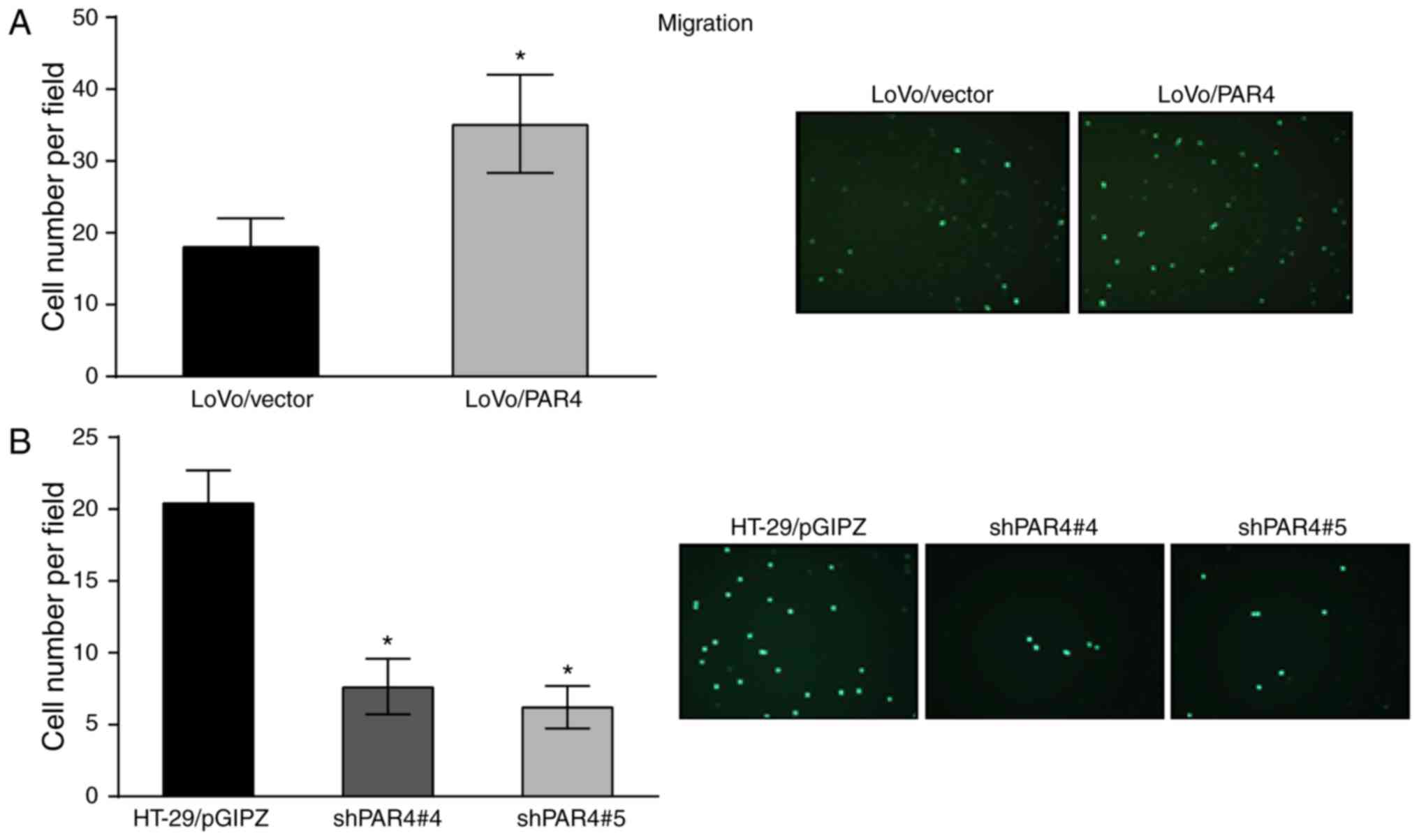
Effects of PAR4 on the migration of
CRC cells
To determine whether PAR4 is able to induce changes
in colon cancer cell motility, the migration effect of PAR4 on LoVo
cells and HT-29 cells was investigated. The migratory activity was
investigated using a Transwell migration assay. As presented in
Fig. 4A, LoVo/PAR4 cells exhibited
increased migratory activity compared with LoVo/vector cells after
16 h. Quantification of migratory cells indicated that expression
of PAR4 led to a >2-fold increase compared with the LoVo/vector
cells. Similarly, knockdown of PAR4 in HT-29 also decreased the
migratory activity compared to the relative control cells.
Quantification of migration of HT-29/shPAR4#4 and HT-29/shPAR4#5
cells demonstrated that knockdown of PAR4 (P<0.05) led to a
>2-fold decrease in migratory activity compared with the
HT-29/pGIPZ control (Fig. 4B). These
results indicated that the expression of PAR4 increased colon
cancer cell migration.
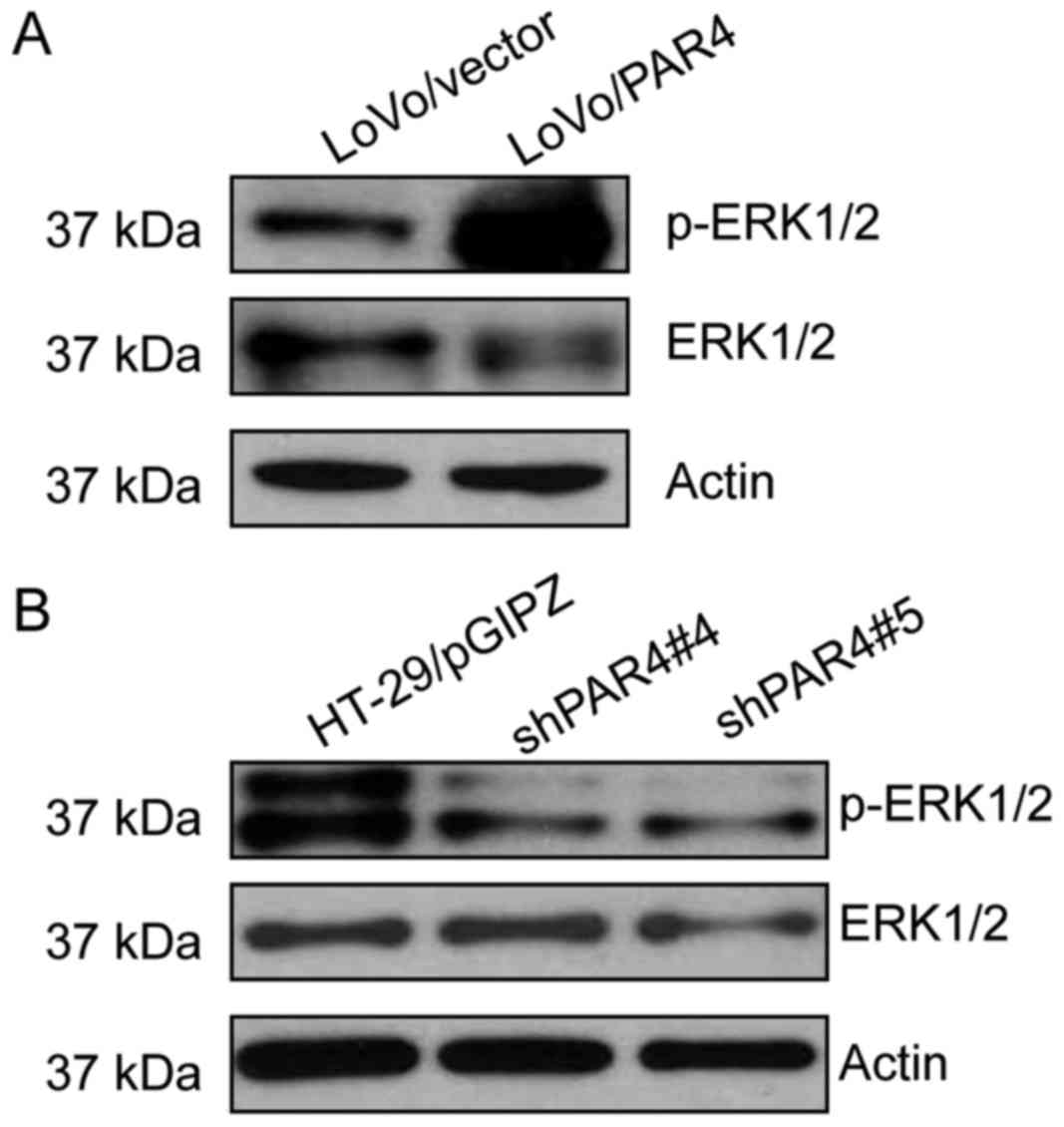
PAR4 activates ERK1/2 in CRC
cells
In order to explore the potential underlying
molecular mechanism for the effects of PAR4 in CRC cells, the
expression level of phospho-ERK1/2 in CRC cells was investigated.
As presented in Fig. 5, the level of
phospho-ERK1/2 was increased in LoVo/PAR4 cells compared with the
LoVo/vector cells. However, the level of phospho-ERK1/2 was
decreased markedly in HT-29/shPAR4#4 or HT-29/shPAR4#5 clones
compared with the HT-29/pGIPZ control (Fig. 5B). These results indicated that PAR4
overexpression in CRC cells increased the level of
phospho-ERK1/2.
Discussion
PAR4, the most recently identified member of the PAR
family, is a G-protein-coupled receptor, and serves physiological
functions in the presence of thrombin, trypsin and cathepsin G
(22). Besides platelet activation
and relaxation of esophageal smooth muscle, PAR4 is involved in the
progression of cancer (23,24). In human hepatocellular carcinoma
(HCC), PAR1 and PAR4 were identified to trigger HCC cell migration
through activating common pro-migratory signaling pathways
(14,25). In gastric cancer, PAR1 and PAR2
promote gastric carcinogenesis by trigging intracellular signals,
whereas PAR4 serves a negative function in the progression of
gastric cancer (26). In our previous
research, the expression of PAR4 was decreased in gastric, lung and
esophageal cancer compared with the normal tissues, and the
downregulation of expression was associated with poor cell
differentiation and lymph node invasion (7,9,10). However, the expression of PAR4 was
increased in CRC tissues, and the upregulation of expression partly
resulted from the hypomethylation of the promoter (6). In the present study, in order to
investigate further the function of PAR4 in the progression of CRC,
PAR4 was overexpressed in CRC LoVo cells, and it was identified
that PAR4 promotes LoVo cell proliferation and
anchorage-independent growth compared with the control. When PAR4
was knocked down in HT-29 cells, proliferation and colony formation
of HT-29 cells were decreased compared with the vector only. The
results suggested that PAR4 serves a function in the proliferation
and survival of CRC cells. When the cells were injected into the
nude mouse subcutaneously, it was identified that the tumor growth
of LoVo/PAR4 was more rapid compared with that of the LoVo/vector,
suggesting that PAR4 promoted colorectal tumorigenesis. The
tumorigenesis of PAR4 is possibly a result of the effect of PAR4
increasing cell proliferation and survival. However, no significant
difference in tumor growth was identified between HT-29/pGIPZ and
HT-29/shPAR4#4 or shPAR4#5 cells (data not shown), which may be
improved by increasing the number of animals in future research. In
the present study, it was also identified that overexpression of
PAR4 in LoVo cells promoted cell migration, and knockdown of PAR4
in HT-29 cells decreased cell migration. The results indicated that
PAR4 promoted CRC cell migration. In fact, in Hep3B hepatocellular
carcinoma cells, PAR4 and PAR1 have been identified to promote cell
migration depending on reactive oxygen species formation and
receptor tyrosine kinase transactivation (25). Similarly, the study of Zhang et
al (21) revealed that human
trefoil factor 2 (hTFF2) promoted the invasion of gastric cancer
cells that overexpressed PAR4, but there was no promotion effect
when the PAR4 was knocked down. In our previous study, it was
identified that the expression levels of PAR4 were markedly
increased in CRC tissues compared with the matched non-cancerous
tissues, particularly in cancer with positive lymph node metastasis
(6). These results are consistent
with the promotion effect on migration of PAR4, and suggested that
PAR4 serves a function in lymph mode metastasis of CRC. Finally,
the potential underlying molecular mechanism for the function of
PAR4 in the progress of CRC was investigated. It was investigated
that overexpression of PAR4 in LoVo cells increased the level of
phospho-ERK1/2, and that knockdown of PAR4 in HT-29 cells decreased
ERK1/2 expression. These results suggested that PAR4 increased the
level of phospho-ERK1/2. In fact, it has been identified previously
that the function of PAR4 was associated with the activation of the
ERK2 signaling pathway. A study by Gratio et al (19) identified that the increase in the
extracellular phosphorylation level of ERK1/2 and ErbB-2 was
associated with the promotion effect of PAR4 on colon cancer cell
proliferation. In recent research, Smith et al (27) identified that the internalization of
activated PAR4 is associated with proper ERK1/2 and protein kinase
B activation. Recombinant hTFF2 promoted gastrointestinal cancer
AGS and LoVo cell migration via phosphorylation of ERK1/2 when PAR4
was overexpressed in the cells (21).
In that study, it was identified that PAR4 activated the ERK1/2
signaling pathway, which is possibly involved in the promoted
effect of CRC cell proliferation, migration and tumorigenesis.
In conclusion, the results of the present study
indicated that PAR4 serves a function in proliferation and
migration of colon cancer cells, which possibly results in
tumorigenesis and invasion of CRC tumors. Further investigation of
the molecular mechanism of the involvement of PAR4 in tumorigenesis
and metastasis in CRC is warranted.
Acknowledgements
Not applicable.
Funding
This work was supported by the Chinese National
Natural Science Foundation (grant nos. 81160302 and 81260084), and
Yunnan Province Basic Research for Application Fund of Yunnan China
(grant nos. 2012FB050 and 2011FZ124).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ and WW were involved in the design of the present
study. HSZ, PJ, CZ and WW performed the cell experiments and
interpreted the cell data. HSZ and PJ conducted the animal assays.
HSZ and SL were involved in acquisition of data and performed the
western blot analysis. SL and WW analyzed and interpreted the data.
HSZ, PJ and HZ wrote, reviewed and/or revised the manuscript. HZ
was also involved in critically revising the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was reviewed and approved by The
Second Affiliated Hospital of Kunming Medical University. All
animals were raised according to the protocols approved by the
Kunming Medical Experimental Animal Care Commission.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cottrell S, Bicknell D, Kaklamanis L and
Bodmer WF: Molecular analysis of apc mutations in familial
adenomatous polyposis and sporadic colon carcinomas. Lancet.
340:626–630. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instabilities in human cancers. Nature. 396:643–649. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Macfarlane SR, Seatter MJ, Kanke T, Hunter
GD and Plevin R: Proteinase-activated receptors. Pharmacol Rev.
53:245–282. 2001.PubMed/NCBI
|
|
6
|
Yu G, Jiang P, Xiang Y and Zhang Y, Zhu Z,
Zhang C, Lee S, Lee W and Zhang Y: Increased expression of
protease-activated receptor 4 and trefoil factor 2 in human
colorectal cancer. PLoS One. 10:e01226782015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Yu G, Jiang P, Xiang Y, Li W, Lee
W and Zhang Y: Decreased expression of protease-activated receptor
4 in human gastric cancer. Int J Biochem Cell Biol. 43:1277–1283.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li SM, Jiang P, Xiang Y, Wang WW, Zhu YC,
Feng WY, Li SD and Yu GY: Protease-activated receptor (par)1, par2
and par4 expressions in esophageal squamous cell carcinoma.
Dongwuxue Yanjiu. 35:420–425. 2014.PubMed/NCBI
|
|
9
|
Jiang P, Yu GY and Zhang Y, Xiang Y, Hua
HR, Bian L, Wang CY, Lee WH and Zhang Y: Down-regulation of
protease-activated receptor 4 in lung adenocarcinoma is associated
with a more aggressive phenotype. Asian Pac J Cancer Prev.
14:3793–3798. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee S, Jiang P, Wang W, Feng W and Yu G:
The decreased expression of protease-activated receptor 4 in
esophageal squamous carcinoma. Neoplasma. 61:546–552. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Elste AP and Petersen I: Expression of
proteinase-activated receptor 1–4 (par 1–4) in human cancer. J Mol
Histol. 41:89–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Darmoul D, Gratio V, Devaud H, Lehy T and
Laburthe M: Aberrant expression and activation of the thrombin
receptor protease-activated receptor-1 induces cell proliferation
and motility in human colon cancer cells. Am J Pathol.
162:1503–1513. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Booden MA, Eckert LB, Der CJ and Trejo J:
Persistent signaling by dysregulated thrombin receptor trafficking
promotes breast carcinoma cell invasion. Mol Cell Boil.
24:1990–1999. 2004. View Article : Google Scholar
|
|
14
|
Kaufmann R, Rahn S, Pollrich K, Hertel J,
Dittmar Y, Hommann M, Henklein P, Biskup C, Westermann M,
Hollenberg MD and Settmacher U: Thrombin-mediated hepatocellular
carcinoma cell migration: Cooperative action via
proteinase-activated receptors 1 and 4. J Cell Physiol.
211:699–707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kaufmann R, Junker U, Nuske K, Westermann
M, Henklein P, Scheele J and Junker K: Par-1- and par-3-type
thrombin receptor expression in primary cultures of human renal
cell carcinoma cells. Int J Oncol. 20:177–180. 2002.PubMed/NCBI
|
|
16
|
Segal L, Katz LS, Shapira H, Sandbank J,
Geras-Raaka E, Gershengorn MC and Oron Y: Par-3 knockdown enhances
adhesion rate of panc-1 cells via increased expression of
integrinalphav and e-cadherin. PloS One. 9:e938792014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Greenberg DL, Mize GJ and Takayama TK:
Protease-activated receptor mediated rhoa signaling and
cytoskeletal reorganization in lncap cells. Biochemistry.
42:702–709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang YQ, Li JJ and Karpatkin S: Thrombin
inhibits tumor cell growth in association with up-regulation of
p21(waf/cip1) and caspases via a p53-independent, stat-1-dependent
pathway. J Boil Chem. 275:6462–6468. 2000. View Article : Google Scholar
|
|
19
|
Gratio V, Walker F, Lehy T, Laburthe M and
Darmoul D: Aberrant expression of proteinase-activated receptor 4
promotes colon cancer cell proliferation through a persistent
signaling that involves src and erbb-2 kinase. Int J Cancer.
124:1517–1525. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu G, Lee YC, Cheng CJ, Wu CF, Song JH,
Gallick GE, Yu-Lee LY, Kuang J and Lin SH: Rsk promotes prostate
cancer progression in bone through ing3, ckap2, and ptk6-mediated
cell survival. Mol Cancer Res. 13:348–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Yu G, Wang Y, Xiang Y, Gao Q,
Jiang P, Zhang J, Lee W and Zhang Y: Activation of
protease-activated receptor (par) 1 by frog trefoil factor (tff) 2
and par4 by human tff2. Cell Mol Life Sci. 68:3771–3780. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sambrano GR, Weiss EJ, Zheng YW, Huang W
and Coughlin SR: Role of thrombin signalling in platelets in
haemostasis and thrombosis. Nature. 413:74–78. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kataoka H, Hamilton JR, McKemy DD, Camerer
E, Zheng YW, Cheng A, Griffin C and Coughlin SR: Protease-activated
receptors 1 and 4 mediate thrombin signaling in endothelial cells.
Blood. 102:3224–3231. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bretschneider E, Kaufmann R, Braun M,
Nowak G, Glusa E and Schror K: Evidence for functionally active
protease-activated receptor-4 (par-4) in human vascular smooth
muscle cells. Br J Pharmacol. 132:1441–1446. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mußbach F, Henklein P, Westermann M,
Settmacher U, Bohmer FD and Kaufmann R: Proteinase-activated
receptor 1- and 4-promoted migration of hep3b hepatocellular
carcinoma cells depends on ros formation and rtk transactivation. J
Cancer Res Clin Oncol. 141:813–825. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sedda S, Marafini I, Caruso R, Pallone F
and Monteleone G: Proteinase activated-receptors-associated
signaling in the control of gastric cancer. World J Gastroenterol.
20:11977–11984. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smith TH, Coronel LJ, Li JG, Dores MR,
Nieman MT and Trejo J: Protease-activated receptor-4 signaling and
trafficking is regulated by the clathrin adaptor protein complex-2
independent of β-arrestins. J Boil Chem. 291:18453–18464. 2016.
View Article : Google Scholar
|