Introduction
Deregulated cell proliferation is one of the
critical events that propel tumor cells and their progeny into
uncontrolled expansion and invasion. Sigma-1 receptor (sigma-1R), a
25-kDa integral membrane protein, has been reported to play a role
in tumor cell proliferation (1). This
receptor was found to represent a unique, non-opioid,
non-phencyclidine, haloperiodol-sensitive receptor family. Based on
binding affinity, two subtypes of sigma receptors have been
identified, namely sigma-1R and sigma-2R (2). Sigma-1R has been cloned from several
species and various tissues, and the protein is 223 amino acids in
length, with a molecular weight of 25 kDa (3,4). Sigma-1R
is a distant homolog to the sterol isomerase in yeast and fungi,
with a single putative transmembrane domain and a cytoplasmic
reticulum retention sequence (3,5). Although
lacking enzymatic activity, sigma-1R shares some pharmacological
characteristics with this enzyme. The protein is considered to have
two transmembrane-spanning helices (amino acids 9–28 and 81–101),
with a 125-amino acid C-terminus (6,7). Sigma-1R
is a chaperone protein binding with the chaperone binding
immunoglobulin protein (BiP) in the lumen of the endoplasmic
reticulum (ER). Agonists of the receptor stimulate its chaperone
activity, which exerts a potent neuroprotective effect by
ameliorating ER stress and reducing the levels of reactive oxygen
radicals (5). Approaches using
site-directed mutagenesis and photo-affinity probes identify that
the ligand-binding region of sigma-1R is mainly located in the
N-terminus of the receptor; however, the C-terminus facilitates the
binding of the receptor with its ligands (8), has chaperone function and enhances
bradykinin-stimulated calcium release (5,9). The
sigma-1R chaperone function was found to be ligand-regulated in the
intact protein; however, the ligand-binding and chaperone functions
are separable (8).
Initial research on sigma-1R was focused on its role
in the nervous system. Mounting evidence indicates that sigma-1R is
involved in a number of nervous system diseases, such as
methamphetamine or cocaine addiction, amnesia, pain depression,
Alzheimer's disease, Parkinson's disease, stroke, Huntington's
disease and HIV infection (10).
Sigma-1R was later found to be expressed a thigh density in various
tumor cells, suggesting the importance of this receptor in tumor
cellular functions, as well as serving as a target binding site for
tumor-imaging agents (11,12). Accumulating evidence shows that
sigma-1R ligands affect tumor cell proliferation. Human mammary
adenocarcinoma, colon carcinoma and melanoma cell proliferation was
inhibited by sigma-1R ligands, such as haloperidol, DTG and
SKF-10047, in vitro cell culture experiments (13). Similarly, sigma-1R ligands, including
haloperidol, 2-IBP {N-[2-(piperidino) ethyl]-2-iodo-benzamide} and
IPAB [2-piperidinyl-(aminoethyl)-4-iodobenzamide] were found to
inhibit the growth of small-cell lung cancer cells (14). Some sigma-1R putative antagonists, but
not putative agonists, inhibited tumor cell proliferation in
vitro and in mouse tumor xenograft experiments (1,15).
However, whether sigma-1R overexpression in cancer cells affects
their proliferation has not yet been addressed.
Protein kinase C (PKC) is a family of
serine/threonine kinases associated with tumor promotion. PKC play
an important role in cell cycle regulation, cell survival,
malignant transformation and apoptosis (16). PKC enzymes are stimulated by signals
such as increase in the concentration of diacylglycerol (DAG) or
calcium ions. Activated PKC phosphorylate hydroxyl groups of serine
and threonine amino acid residues on various substrate proteins,
depending on cell type, regulate various functions, including
receptor desensitization, membrane structure events, gene
transcription, immune response and cell growth. To date, at least
12 isoforms of PKC have been cloned, each displaying different
enzymatic properties, tissue expression and intracellular
localization (17,18). The PKC family comprises three groups:
Classic PKCs (PKCα, PKCβ1, PKCβ2 and PKCγ), novel PKCs (PKCδ, PKCε,
PKCη, PKCθ and PKCµ) and atypical PKCs (PKCζ, PKCλ and PKCι), which
require different stimulators for their full activation.
It has been well-documented that the PKC family
regulates cell proliferation and migration in breast tumor cell
lines (19). PKCα plays a key role in
controlling the proliferation of breast cancer cells through the
activation of extracellular signal-regulated kinase (ERK) (20) and telomerase (21). Stimulating PKCβI and βII in MCF-7
cells enhance their growth by upregulating the expression of cyclin
D1 (22). PKCη upregulation in
malignant breast cells was found to be associated with cancer cell
growth and survival via hormone-dependent cell growth pathways
(23,24). PKCζ stimulates estrogen-mediated
breast cancer cell growth by stabilizing steroid receptor
coactivator-3 (25,26). Finally, PKCδ is a potential
pro-proliferative factor in breast cancer cells, as TPA-mediated
PKCδ activation leads to the activation and nuclear translocation
of estrogen receptor (ER)α and enhances ER-dependent reporter gene
expression (27). Although both
sigma-1R and PKC are involved in cell proliferation, the
possibility of a connection between them has not yet been
addressed.
While evaluating the role of sigma-1R C-terminus in
inositol 1,4,5-trisphosphate (IP3) receptor activation (9), we found that the sigma-1R-overexpressing
cell line, MCF-41, proliferated significantly faster compared with
the sigma-1R-defective line, MCF-7 (9,11). Based
on these observations and the roles of sigma receptors and PKC
isoenzymes in cell proliferation, the present study aimed to
investigate how sigma-1R overexpression affects the proliferation
rate of MCF-7 cells, and establish the connection via a signaling
pathway between sigma-1R and PKC subtype enzymes in the
pro-proliferative receptor function, which may provide potential
molecular targets for anticancer treatment.
Materials and methods
Production of stably transfected cell
lines and cell culture
Breast tumor cell lines stably transfected with
pcDNA3.1 expression vectors harboring intact sigma-1R (MCF-41line)
or its C-terminus (aa 102–223, sg101 line) were developed as
previously described (9). MCF-7
(ATCC, Manassas, VA, USA), MCF-41 and sg101 cells were cultured in
Dulbecco's minimal essential medium (DMEM) containing 1.5 g/l
NaHCO3, 10% fetal bovine serum, insulin (10 mg/l) and
penicillin/streptomycin (100 U/100 µg/ml). All cell cultures
were performed in a humidified atmosphere of 5% CO2 and
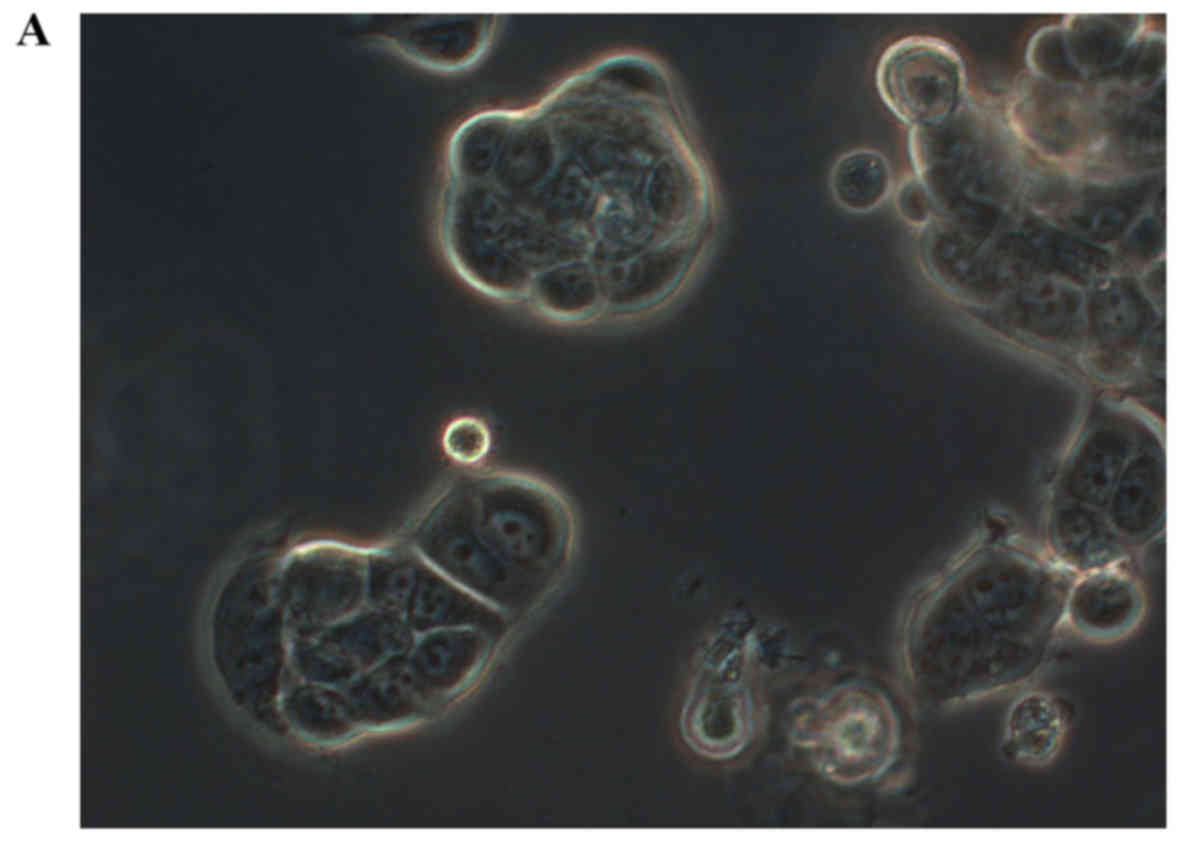
95% air at 37°C. On the third day of the cell cultures, cells were
observed from the culture flask under a phase-contrast inverted
microscope. Images of the cells were captured using a Carl Zeiss
Axiovert 200 microscope. No specific staining was performed.
Cell proliferation assays
Cells were seeded into 96-well plates at a density
of 1×104 cells per well, and cultured in 5%
CO2 at 37°C overnight. On the second day, the medium was
replaced by fresh medium, with or without various concentrations of
fetal bovine serum, and six parallel wells were arranged for one
experiment. Media were refreshed every 3 days. CCK-8 reagent (10
µl) was added to each well at the indicated time points (days 0, 1,
2, 3, 4, 5, 6 and 7) and incubated for 1 h at 37°C. Optical
absorbance was read by a Victor V plate reader at a wavelength of
450 nm. The readings were normalized to blank medium. Cell
proliferation was calculated as the ratio of the readings of the
selected day to the readings of day 0.
Proliferation of enzyme inhibiting
assays
The protocol was similar to that described above for
the proliferation assay. The main difference was that the cell
medium was changed after 24 h and replaced by different
concentrations of enzyme inhibitors, as described in the text, with
complete serum or serum without insulin and fetal bovine serum.
Measurements were performed as described in the proliferation
assays and the readings were normalized to blank medium, either
with or without serum and insulin.
Statistical analysis
Values are presented as mean ± standard deviation.
Data were analyzed by the Student's t-test or one-way analysis of
variance with post hoc comparisons using Student-Newman-Keuls test.
The data were analyzed using SPSS software v20.0 (IBM Corp.,
Armonk, NY, USA). P<0.05 was considered to indicate
statistically significant differences.
Results
Sigma-1R overexpression in MCF-7 cells
was associated with higher proliferation rate and changed cell
morphology
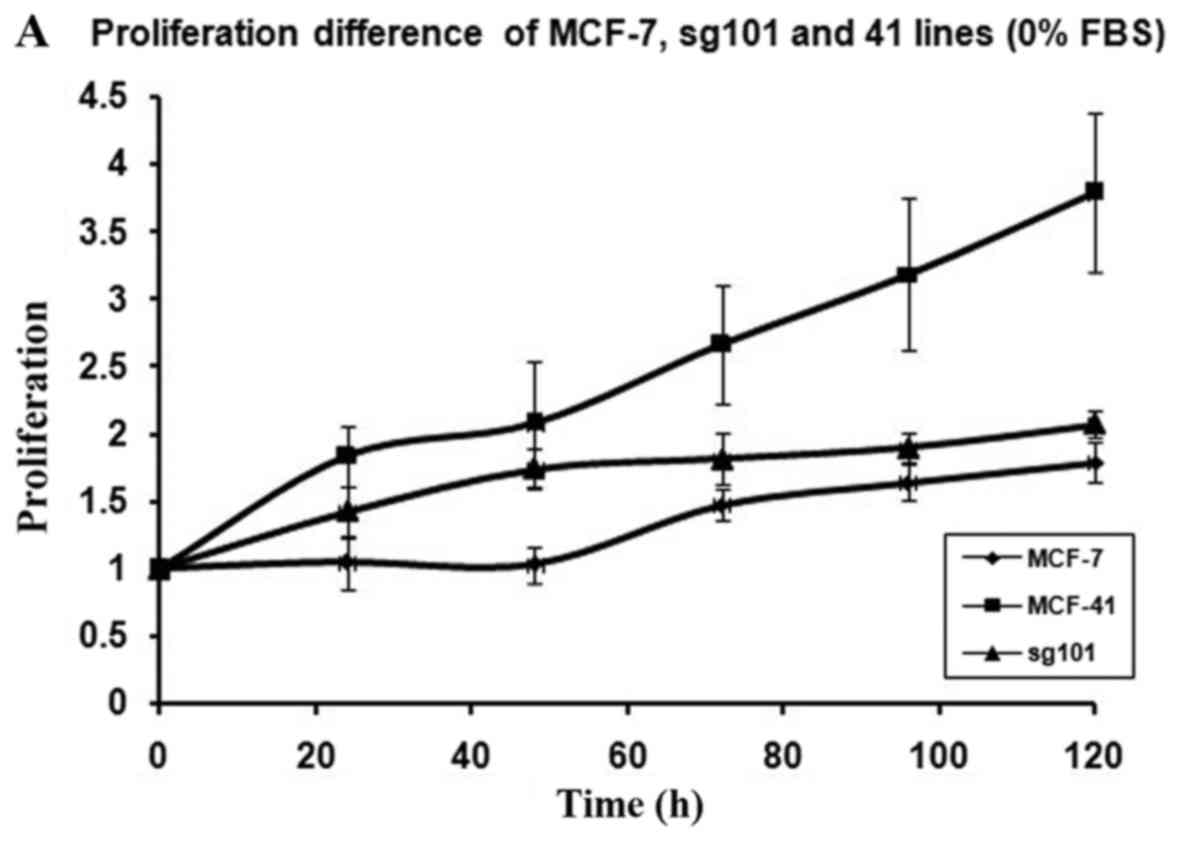
In order to investigate how serum differently
affected the growth rates in the MCF-7, sg101 and MCF-41 cell
lines, the cells were cultured in media containing various serum
concentrations (0, 0.3 and 10%) and the proliferation rates were
measured at indicated time points, as shown in Fig. 1A-C. As illustrated in Table I, MCF-41 cell proliferation was 4.5-,
3.5- and 3.4-fold during a 96-h cell culture in media containing
10, 0.3 and 0% serum, respectively. However, the proliferation of
wild-type MCF-7 cells was only 1.8-, 1.5- and 1.3-fold during same
period. Thus, MCF-41 cells exhibited a significantly higher
proliferation rate compared with wild-type MCF-7 cells, even at low
serum concentrations. Of note, sigma-1R overexpression enhanced
cell proliferation in the absence of serum, i.e., absence of
exogenous growth signals. The C-terminal segment of sigma-1R has
been shown to enhance bradykinin-stimulated calcium signaling
(9). We also determined the
proliferation rate of the sg101 cell line in media with different
serum concentrations. As shown in Fig.
1 and Table I, the proliferation
rate of sg101 cells was 2.9-, 2.4- and 1.9-fold during a 96-h
culture in media with varied concentration of serum, as mentioned
above. The proliferation rates of sg101 cells were significantly
higher compared with those of MCF-7 cells, but slightly lower
compared with the MCF-41 cell line. The results demonstrated that
sigma-1R played a key role in promoting the proliferation in breast
tumor cells, and identified the C-terminus of sigma-1R as the
functional domain driving cell proliferation. These results were
consistent with previous findings supporting that the C-terminus of
sigma-1R is the cellular function domain of the receptor (9). In addition to enhancing cell
proliferation rate, overexpression of sigma-1R affected cell
morphology. As shown in Fig. 2, the
shape of MCF-7 cells was round-like, MCF-41 cells appeared more
expanded and displayed pseudo-flagella, and the sg101 cell line
displayed more prominent expansion compared with MCF-7 cells, but
less prominent expansion compared with MCF-41 cells. Thus, sigma-1R
expression was found to be associated with cell morphology,
providing additional evidence supporting the cellular function of
sigma-1R.
 | Table I.Summary proliferation rates of
MCF-41, sg101 and MCF-7 after 96 h of cell culture in different
media which contain 10, 0.3 and 0% fetal bovine serum. |
Table I.
Summary proliferation rates of
MCF-41, sg101 and MCF-7 after 96 h of cell culture in different
media which contain 10, 0.3 and 0% fetal bovine serum.
|
| Cell lines |
|---|
|
|
|
|---|
| Serum concentration
(%) | MCF-7 | MCF-41 | Sg101 |
|---|
| 10.0 | 1.8 | 4.5 | 2.9 |
|
0.3 | 1.5 | 3.5 | 2.4 |
|
0.0 | 1.3 | 3.4 | 1.9 |
Sigma-1R overexpression increased
MCF-7 cell proliferation rate via the PKC pathway
Tumors often progress from benign to malignant due
to continuous mutations that enhance the sensitivity of cells to
growth factor signals (28). The
present study revealed that sigma-1R overexpression promotes tumor
cell proliferation, which may be a part of this process. In order
to identify the key molecules involved in the mechanism underlying
the role of sigma-1R in proliferation, various enzyme inhibitors
for 3 well-documented entities located upstream and downstream in
the PKC signaling cascade, or PKC subclass isoenzymes associated
with cell proliferation, were used in the CCK-8 assay and data were
analyzed.
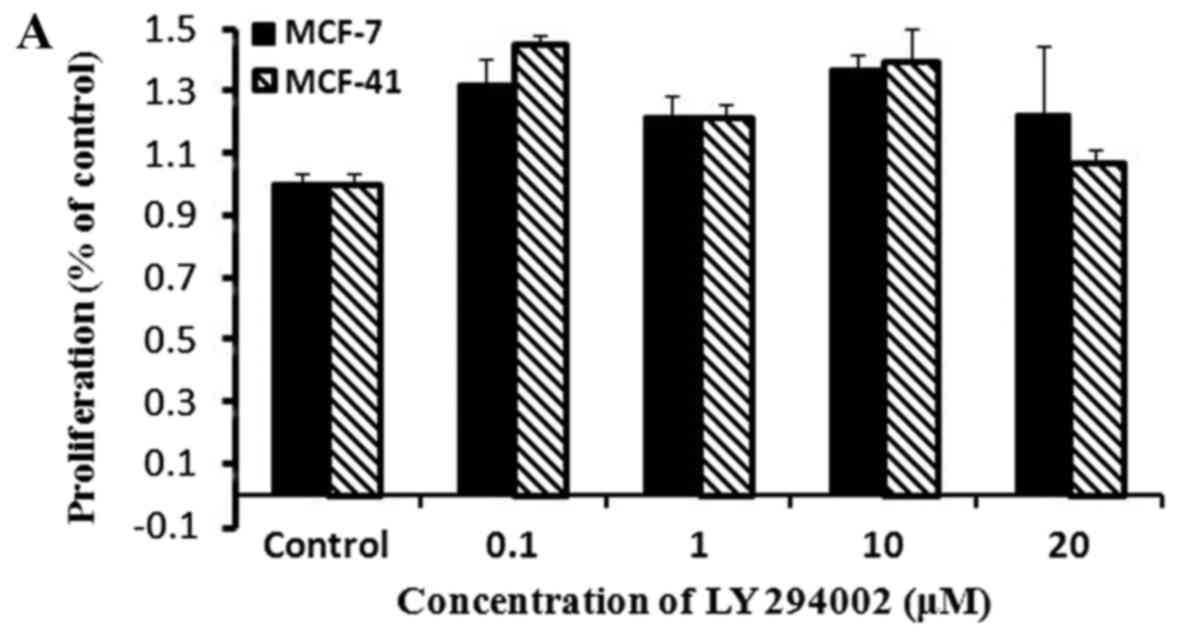
PI3K inhibition did not affect cell
proliferation
To examine whether sigma-1R-overexpressing cells
exhibited enhanced proliferation through the PI3K signaling
pathway, the CCK-8 assay was performed with the PI3K inhibitor,
LY294002. Cells were cultured in media containing various
concentrations of LY294002, and proliferation rates were measured
by CCK-8 as described in Materials and methods. The results are
presented in Fig. 3A, and show that
the PI3K inhibitor did not affect the proliferation rate of either
MCF-7 or MCF-41 cells over a period of 48 h. These results
indicated that the PI3K signaling pathway was not associated with
sigma-1R overexpression enhancing cell proliferation. LY294002 and
all inhibitors used in following studies were examined to confirm
their effects and specification meet the research goals according
to manufacturer's handbook when they were delivered from chemical
companies (data not shown).
MEK1/2 inhibition did not affect cell
proliferation
We investigated whether the MEK1/2 signaling pathway
was involved in sigma-1R enhancing cell proliferation. An
experiment similar to that performed with the PI3K inhibitor was
conducted using a MEK1/2 inhibitor, PD98059. Cells were cultured in
media with various concentrations of PD98059, and CCK-8 assays were
performed at the indicated time points, as described in Fig. 3B. The data revealed that PD98059 did
not affect the proliferation of MCF-7 or MCF-41 cells within a
period of 48 h. Thus, MEK1/2 is not involved in the signaling
pathway of sigma-1R driving cell proliferation.
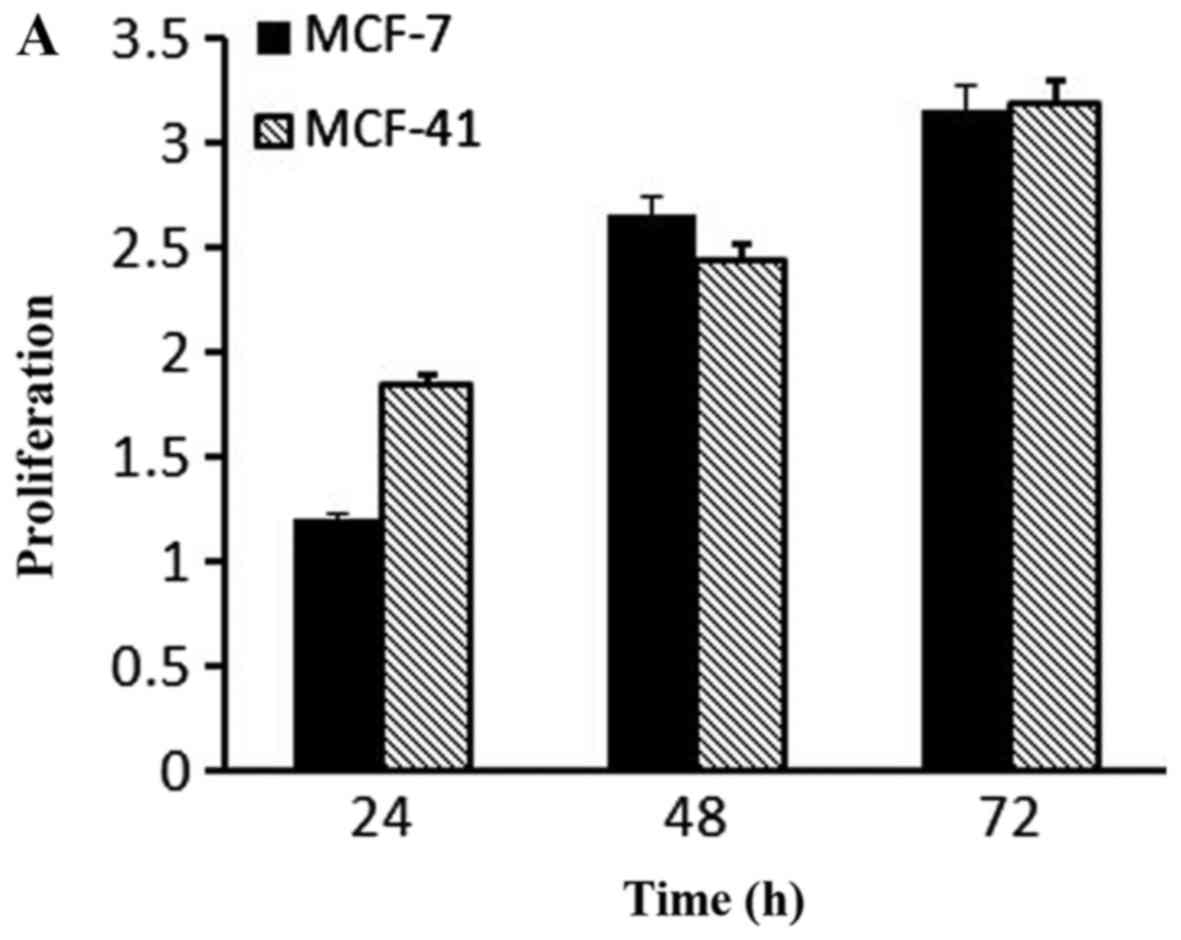
PKC inhibition affected cell
proliferation
Based on the fact that PKC isozymes play critical
roles in cell proliferation, we examined whether PKC were
associated with sigma-1R overexpression driving cell growth. The
effect of PKC inhibition on cell growth was examined by the CCK-8
assay with 10 µM sotrastaurin, a universal PKC inhibitor, at 24, 48
and 72 h. As shown in Fig. 4A,
sotrastaurin inhibited MCF-41 cell proliferation more potently
compared with the MCF-7 cell line. We investigated further to
determine which PKC subtype was involved in this function. LY333531
and dimethoxy-substituted phenyl-thiophene (DSPT), inhibitors of
nPKC and aPKC, respectively, were used in the CCK-8 assay, and the
results revealed that neither inhibitor affected proliferation in
the two cell lines (Fig. 4B and C).
However, GF109203×, a classic PKC subtype enzyme inhibitor,
inhibited cell proliferation (Fig.
4D). The results are shown in Fig.
4D: MCF-7 cell proliferation rates decreased slightly when
GF109203× concentration was increased from 0 to 1 µM, but the
proliferation rate mildly increased when the inhibitor
concentration was further increased to 10 µM. In the MCF-41 cell
line, however, the proliferation rates decreased significantly
following an inhibitor concentration increase from 0 to 0.01 µM,
and the rates decreased even more when the inhibitor concentration
was increased from 1 to 10 µM. This experiment clearly indicated
that sigma-1R overexpression driving MCF-41cell proliferation was
associated with classic PKC subtype isoenzymes.
Discussion
Overexpression of sigma receptors has been found in
various types of human cancer cells (11,12).
Previous studies revealed that sigma-1R ligands affected tumor cell
proliferation (13–15). Overexpression of some receptors, such
as TRIM29, was recently reported to facilitate cancer cell
proliferation (29,30). However, little was known on whether
the overexpression of sigma-1R in cancer cells regulated cell
proliferation. In the present study, we demonstrated that
overexpression of this receptor constitutively enhanced MCF-7 cell
proliferation, even in absence of any of the receptor ligand,
strongly indicating that sigma-1R is an oncoprotein. This result
was consistent with a previous study reporting that
sigma-1R-overexpressing cell lines exhibited constitutively
enhanced bradykinin-induced calcium release, driving sigma
agonist-independent dissociation of ANK 220 from IP3R-3, resulting
in its activation (9). Thus, it is
possible that the positive effect of sigma-1R on cell proliferation
may be due to the receptor sensitizing cells to various growth
signals associated with the IP3R-3/Ca2+ signaling
pathway. In the present study, sigma-1R overexpression driving cell
proliferation was inhibited by GF109203×, suggesting that the
pro-proliferative effects of sigma-1R are closely associated with
classic PKC subtype enzymes.
PKC isozymes are known to be involved in cell
proliferation, survival, invasion and migration. Activation of PKC
isozymes may be triggered by changes in intracellular cofactors of
PKCs, which are activated by stimulation from receptor tyrosine
kinases (RTKs), G-protein-coupled receptors (GPCR) and integrins
(31). RTKs include receptors for
epidermal growth factor (EGF), fibroblast growth factor (FGF),
platelet-derived growth factor (PDGF), vascular endothelial growth
factor (VEGF), hepatocyte growth factor (Met), and insulin receptor
family (IR) (32–34). Thus, various growth factors in the
serum can stimulate RTKs on the cell surface. GPCRs recruit and
activate PKC by releasing factor Gαq, which stimulates
phospholipase C (PLC)-β to hydrolyze phosphatidylinositol
biphosphate (PIP2) and produce IP3 and DAG, the two major secondary
messengers required for stimulation of PKC (35). Integrins and other extracellular
matrix protein clusters may also activate PKC via the PLC signaling
cascade, which only acts in the formation of cellular focal
adhesions (36). Thus, overexpression
of sigma-1R regulating PKC activation to promote cell proliferation
may be mediated only through the RTK or GPCR cascade rather than
through the integrins pathway. The proliferation data revealed that
sigma-1R overexpression drove cell proliferation in the absence of
serum in the culture medium, indicating that sigma-1R regulates
PKCs even when RTKs are silent. Therefore, our data indicated that
sigma-1R regulating PKCs may be through the GPCR signaling pathway.
A recent study confirmed that sigma-1R directly interacts with
Rac1-GTPase in the brain mitochondria, providing more evidence
supporting our hypothesis (37).
In breast cancer cells, several PKC isozymes, such
as PKCα, β, δ, ε, ζ, η and θ, are involved in cell proliferation,
differentiation, survival and apoptosis (19). In the present study, we first tested
whether the Ras/Raf/MEK/ERK proliferation signaling pathway was
associated with sigma-1R overexpression driving cell proliferation.
The MEK1/2 inhibitor, PD98059, was employed in the cell
proliferation assay and the data demonstrated it did not affect
MCF-7 or MCF-41 cell proliferation, indicating that this signaling
pathway is not involved in sigma-1R-mediated cell proliferation.
Second, we tested whether the PI3K/Akt/mTOR proliferation signaling
pathway was involved in sigma-1R overexpression driving cell
proliferation. The PI3K inhibitor, LY294002, was used in the
proliferation assay and it was found to exert no effect on cell
proliferation. Therefore, this pathway was not associated with the
effects of sigma-1R driving cell proliferation. Finally, a
universal PKC inhibitor, sotrastaurin, was used in the cell
proliferation assay, and the data revealed that the compound
inhibited both MCF-7 and MCF-41 cell proliferation, suggesting that
the PKC isoenzymes contribute to sigma-1R-induced cell
proliferation. These findings were consistent with previous
research reporting that PKC isozymes stimulate survival- or
proliferation-associated signaling pathways in cancer (19). The PKC family can be classified into
three subfamilies: Classic, novel and atypical PKCs. We
investigated further to determine which PKC isoenzymes are involved
in the pro-proliferative function of sigma-1R. The atypical PKC
subtype inhibitor DSPT, the novel PKC subtype inhibitor LY333531,
and the classic PKC subtype inhibitor GF109203× were used in MCF-41
and MCF-7 cell proliferation assays, and GF109203× inhibited cell
proliferation to a significantly greater extent in the
sigma-1R-overexpressing MCF-41 cell line compared with the
sigma-1R-defective MCF-7 cell line. These results demonstrated that
only classic PKC subtype enzymes are associated with
sigma-1R-mediated cell proliferation. One signaling pathway for
cell proliferation is the PKCζ-MEK-ERK (38), but our data demonstrated that MEK1/2
inhibition did not affect MCF-41 or MCF-7 cell proliferation,
providing further evidence that atypical PKCs, such as PKCζ, are
not involved insigma-1R-induced cell proliferation. It should be
noted that in the experiments, the PI3K and MEK/ERK inhibitors
exist in cell culture media up to 48 h, the cells still survive.
The main reason may be that multiple signaling pathways are
associated with cell growth and survival, and these signaling
pathways usually serve as backup each other, when one or more
pathways are blocked, the others can compensate them and this keep
cells still survive.
GF109203× is a potent PKC inhibitor with an
IC50 of 20, 17, 16 and 20 nM for PKCα, PKCβI, PKCβII and
PKCγ, respectively, in cell-free assays. In the present study, the
data revealed that GF109203× inhibited MCF-41 and MCF-7 cell
proliferation, consistently with previous studies reporting that
overexpression of PKCα may contribute to increased
anchorage-independent growth, tumorigenicity and metastasis
(39). The PKCβ isoform has been
implicated in mammary tumorigenesis inhuman and rodent models, and
is generally considered to be a growth-promoting kinase; in
addition, the PKCβ-specific inhibitor LY379196 significantly
reduces the growth of MCF-7, MDA-MB-231and BT-474 breast cancer
cells (23). All these previous
studies support that classic PKCs are key enzymes in tumor cell
proliferation. Our data clearly indicated that sigma-1R-induced
MCF-7 cell proliferation is mediated through the classic PKC
subtype pathway, at least in part. However, the accurate PKC of
classic PKC subtype enzymes is not identified due to short of
specific chemical currently which inhibit one of classic PKC
enzymes without affecting the other member in the group of PKC, and
details on the mechanism underlying sigma-1R activation of classic
PKC isoenzymes are unknown at present, and further investigation is
required focus on these points in the future study.
In conclusion, our data demonstrated that the
sigma-1R-overexpressing cell line, MCF-41, grew significantly
faster compared with MCF-7, whereas this enhancement of cell
proliferation was completely eliminated by the addition of PKC
inhibitor to the culture media. Among PKC isoenzyme inhibitors,
only the classic PKC subtype inhibitor, GF109203×, inhibited MCF-41
cell proliferation significantly compared with MCF-7 cells. In
conclusion, sigma-1R overexpression driving MCF-7 cell
proliferationis, at least in part, mediated through the classic PKC
subtype signaling pathway. Due to sigma-1R highly expressing in
various tumor cells (11,12), our finding provides new critical
molecule targets for anti-tumor drug design in cancer therapy.
Acknowledgements
The authors would like to thank Dr Wayne D. Bowen at
Brown University for his reviewing and valuable suggestions.
Funding
The present study was supported by the Scientific
and Technological Development Funding of Zhejiang Province (grant
no. 2016C33018).
Availability of data and materials
The datasets used in the present study are available
from corresponding author on reasonable request.
Authors' contributions
ZW conceived and designed the experiments. YW, XB,
XL and CZ performed the experiments. YW and ZW wrote the
manuscript. All authors reviewed and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Aydar E, Palmer CP and Djamgoz MB: Sigma
receptors and cancer: Possible involvement of ion channels. Cancer
Res. 64:5029–5035. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quirion R, Bowen WD, Itzhak Y, Junien JL,
Musacchio JM, Rothman RB, Su TP, Tam SW and Taylor DP: A proposal
for the classification of sigma binding sites. Trends Pharmacol
Sci. 13:85–86. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanner M, Moebius FF, Flandorfer A, Knaus
HG, Striessnig J, Kempner E and Glossmann H: Purification,
molecular cloning, and expression of the mammalian sigma1-binding
site. Proc Natl Acad Sci USA. 93:8072–8077. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hellewell SB and Bowen WD: A sigma-like
binding site in rat pheochromocytoma (PC12) cells: Decreased
affinity for (+)-benzomorphans and lower molecular weight suggest a
different sigma receptor form from that of guinea pig brain. Brain
Res. 527:244–253. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayashi T and Su TP: Sigma-1 receptor
chaperones at the ER-mitochondrion interface regulate Ca(2+)
signaling and cell survival. Cell. 131:596–610. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aydar E, Palmer CP, Klyachko VA and
Jackson MB: The receptor as a ligand-regulated auxiliary potassium
channel subunit. Neuron. 34:399–410. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Palmer CP, Mahen R, Schnell E, Djamgoz BA
and Aydar E: Sigma-1 receptors bind cholesterol and remodel lipid
rafts in breast cancer cell lines. Cancer Res. 67:11166–11175.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chu UB and Ruoho AE: Biochemical
pharmacology of the sigma-1 receptor. Mol Pharmacol. 89:142–153.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu Z and Bowen WD: Role of sigma-1
receptor C-terminal segment in inositol 1,4,5-trisphosphate
receptor activation: Constitutive enhancement of calcium signaling
in MCF-7 tumor cells. J Biol Chem. 283:28198–28215. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maurice T and Su TP: The pharmacology of
sigma-1 receptors. Pharmacol Ther. 124:195–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vilner BJ, John CS and Bowen WD: Sigma-1
and sigma-2 receptors are expressed in a wide variety of human and
rodent tumor cell lines. Cancer Res. 55:408–413. 1995.PubMed/NCBI
|
|
12
|
van Waarde A, Rybczynska AA, Ramakrishnan
NK, Ishiwata K, Elsinga PH and Dierckx RA: Potential applications
for sigma receptor ligands in cancer diagnosis and therapy. Biochim
Biophys Acta. 1848:2703–2714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brent PJ and Pang GT: Sigma binding site
ligands inhibit cell proliferation in mammary and colon carcinoma
cell lines and melanoma cells in culture. Eur J Pharmacol.
278:151–160. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moody TW, Leyton J and John C: Sigma
ligands inhibit the growth of small cell lung cancer cells. Life
Sci. 66:1979–1986. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim FJ, Schrock JM, Spino CM, Marino JC
and Pasternak GW: Inhibition of tumor cell growth by Sigma1 ligand
mediated translational repression. Biochem Biophys Res Commun.
426:177–182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Erin M and Kazanietz GG: Protein kinase C
and other diacylglycerol effector in cancer. Nat Rev Cancer.
7:281–294. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nishizuka Y: The molecular heterogeneity
of protein kinase C and its implications for cellular regulation.
Nature. 334:661–665. 1988. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ono Y, Fujii T, Ogita K, Kikkawa U,
Igarashi K and Nishizuka Y: The structure, expression, and
properties of additional members of the protein kinase C family. J
Biol Chem. 263:6927–6932. 1988.PubMed/NCBI
|
|
19
|
Kang JH: Protein kinase C (PKC) isozymes
and cancer. New J Sci. 2014:2314182014. View Article : Google Scholar
|
|
20
|
Gupta AK, Galoforo SS, Berns CM, Martinez
AA, Corry PM, Guan KL and Lee YJ: Elevated levels of ERK2 in human
breast carcinoma MCF-7 cells transfected with protein kinase C
alpha. Cell Prolif. 29:655–663. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H, Zhao L, Yang Z, Funder JW and Liu
JP: Telomerase is controlled by protein kinase Calpha in human
breast cancer cells. J Biol Chem. 273:33436–33442. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li H and Weinstein IB: Protein kinase C
beta enhances growth and expression of cyclin D1 in human breast
cancer cells. Cancer Res. 66:11399–11408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pal D, Outram SP and Basu A: Upregulation
of PKCη by PKCε and PDK1 involves two distinct mechanisms and
promotes breast cancer cell survival. Biochim Biophy Acta.
1830:4040–4045. 2013. View Article : Google Scholar
|
|
24
|
Karp G, Maissel A and Livneh E: Hormonal
regulation of PKC: Estrogen up-regulates PKCeta expression in
estrogen responsive breast cancer cells. Cancer Lett. 246:173–181.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yi P, Feng Q, Amazit L, Lonard DM, Tsai
SY, Tsai MJ and O'Malley BW: Atypical protein kinase C regulates
dual pathways for degradation of the oncogenic coactivator
SRC-3/AIB1. Mol Cell. 29:465–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Allen-Petersen BL, Carter CJ, Ohm AM and
Reyland ME: Protein kinase Cδ is required for ErbB2-driven mammary
gland tumorigenesis and negatively correlates with prognosis in
human breast cancer. Oncogene. 33:1306–1315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Servi B, Hermani A, Medunjanin S and
Mayer D: Impact of PKCdelta on estrogen receptor localization and
activity in breast cancer cells. Oncogene. 24:4946–4955. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gomperts B, Kramer IM and Tatham P: Signal
transduction. 2nd edition. Elsevier Academic Press; Fourth
printing: pp. 246–251. 2004
|
|
29
|
Tan ST, Liu SY and Wu B: TRIM29
overexpression promotes proliferation and survival of bladder
cancer cells through NF-KB signaling. Cancer Res Treat.
48:1302–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu J, Welm B, Boucher KM, Ebbert MT and
Bernard PS: TRIM29 functions as a tumor suppressor in
nontumorigenic breast cells and invasive ER+ breast cancer. Am J
Pathol. 180:839–847. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oliva JL, Griner EM and Kazanietz MG: PKC
isozymes and diacylglycerol-regulated proteins as effectors of
growth factor receptors. Growth Factors. 23:245–252. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lemmon MA and Schlessinger J: Cell
signaling by receptor tyrosine kinases. Cell. 141:1117–1134. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schlessinger J: Cell signaling by receptor
tyrosine kinases. Cell. 103:211–225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gschwind A, Fischer OM and Ullrich A: The
discovery of receptor tyrosine kinases: Targets for cancer therapy.
Nat Rev Cancer. 4:361–370. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dorsam RT and Gutkind JS:
G-protein-coupled receptors and cancer. Nat Rev Cancer. 7:79–94.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fogh BS, Multhaupt HA and Couchman JR:
Protein kinase C, focal adhesions and the regulation of cell
migration. J Histochem Cytochem. 62:172–184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Natsvlishvili N, Goguadze N, Zhuravliova E
and Mikeladze D: Sigma-1 receptor directly interacts with
Rac1-GTPase in the brain mitochondria. BMC Biochem. 16:112015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Naranatt PP, Akula SM, Zien CA, Krishnan
HH and Chandran B: Kaposi's sarcoma-associated herpesvirus induces
the phosphatidylinositol 3-kinase-PKC-zeta-MEK-ERK signaling
pathway in target cells early during infection: Implications for
infectivity. J Virol. 77:1524–1539. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ways DK, Kukoly CA, deVente J, Hooker JL,
Bryant WO, Posekany KJ, Fletcher DJ, Cook PP and Parker PJ: MCF-7
breast cancer cells transfected with protein kinase C-alpha exhibit
altered expression of other protein kinase C isoforms and display a
more aggressive neoplastic phenotype. J Clin Invest. 95:1906–1915.
1995. View Article : Google Scholar : PubMed/NCBI
|