Introduction
The risk factors for lung cancer are usually
identified by comparing the studied feature between healthy
subjects and lung cancer patients. This approach has determined a
series of risk factors, both genetic and epigenetic (1–7). Since the
association between lung cancer and chronic obstructive pulmonary
disease (COPD) has been reported in numerous studies (8–12), the
risk factors that influence the morbidity of lung cancer in
subjects with COPD constitute a separate problem.
A common initial pathogenic event in COPD and lung
cancer is oxidative stress, caused by smoking or environmental
factors. This initiates several pathways depending on genetic and
epigenetic factors, which to some extent, overlap in COPD and lung
cancer (13,14). The common feature is initial
activation of transcription factors and subsequent trans-activation
of inflammation-related genes. In addition, oxidative DNA damage is
inflicted, which may lead to genomic instability and the initiation
of cancer, if insufficiently or incorrectly repaired (15). Thus, when searching for genetic
factors that increase the risk of lung cancer in COPD subjects, it
seems logical to examine polymorphisms in DNA repair genes, and in
genes that code for proteins involved in inflammation and tissue
remodeling, since these have been associated with COPD and cancer
progression (8–12).
Previously, our group has compared the genotype
distribution of single nucleotide polymorphisms (SNPs) in the genes
encoding several matrix metalloproteinases (MMPs), DNA repair
proteins, tumor necrosis factor α (TNFα) and haptoglobin
(HP) in two groups of male Caucasian subjects: The first
group consisted of patients with COPD and lung cancer, whereas the
second group consisted of patients with COPD only. It was observed
that distributions of genotypes of MMP3 and HP
polymorphisms were significantly different between the studied
groups (16,17). The HP gene is located on
chromosome 16q22 and has two major alleles in humans (18). Allele 1 contains five exons whereas
allele 2 has a duplication of exons 3 and 4 of allele 1 potentially
resulting from unequal crossing over between two HP1 alleles
(18). As a result, there are three
common haptoglobin phenotypes: The homodimer HP1/1, the
linear polymer HP1/2, and the large circular polymer
HP2/2 (18). In order to
search for other factors that may affect the extent of oxidative
stress, the present study focused on proteins involved in iron
metabolism. Iron is an established determinant of oxidative stress,
being responsible for the induction of DNA damage and activation of
many signaling pathways (19). Here,
results are reported on blood serum levels of several proteins
involved in iron metabolism, inflammation and the oxidative stress
response compared between the same groups of subjects as in our
previous work.
Materials and methods
Subjects
The patients were diagnosed and treated at the 2nd
Department of Respiratory Medicine and 3rd Department of
Respiratory Medicine and Oncology of the Institute of Tuberculosis
and Lung Diseases in Warsaw, Poland from January 2007 to December
2009. The study protocol conformed to the Declaration of Helsinki,
and was approved by the Institutional Ethics Committee of the
Institute of Tuberculosis and Lung Diseases (Warsaw, Poland). Each
participating patient had provided signed informed consent
following a detailed explanation of the study protocols. The
subjects were examined in two groups, selected according to gender,
age, smoking habits and diagnosed disease. There were 53 male COPD
patients with lung cancer and 54 male patients with COPD only. The
detailed characteristics of the subject groups have been presented
in our previous paper (17).
ELISA measurements
A total of 10 ml of blood was collected from each
patient into a tube containing no anticoagulant. Blood samples were
incubated in an upright position for 30 min at room temperature to
allow clotting and centrifuged in a horizontal rotor for 10 min at
1,500 × g at room temperature. Serum was transferred to sterile
eppendorf tubes and stored at −70°C in 0.5 ml aliquots. ELISA kits
for the following proteins were used according to the
manufacturer's instructions: Ferritin (Alpha Diagnostic
International, San Antonio, TX, USA, Catalog no. 1810); hepcidin
prohormone (DRG Instruments GmbH, Marburg, Germany; catalog no.
EIA-4644); soluble transferrin receptor (sTfR) (BioVendor, Brno,
Czech Republic; catalog no. RD194011100, TNFα and transferrin
(Assaypro St. Charles, MO, USA; catalog no. ET2010-1 and ET3105-1,
respectively); and 8-oxo-2′-deoxyguanosine (HT (High throughput)
8-oxo-dG ELISA kit; R&D Systems, Inc., Minneapolis, MN, USA;
catalog no. 4380-096-K). Thawed serum was not reused for testing.
Each ELISA test included a standard curve, the shape of which had
been compared with the data provided by the manufacturer as well as
a positive control (sample with known antigen concentration) and
negative control (sample containing no antigen). All samples and
standards were run in triplicate and coefficients of variation were
compared with the data provided by the manufacturer.
Total iron-binding capacity
TIBC values of serum were determined using the
Randox TIBC colorimetric assay (Randox Laboratories Ltd., Crumlin,
UK) according to the manufacturer's recommendations. A total of 0.5
ml of serum from each subject was used for the measurement.
Ceruloplasmin ferroxidase activity
(CFA)
The CFA of serum samples was measured according to
the method described by Erel (20).
The chromogen, (3-(2-pyridyl)-5,6-bis(2-[5-furylsulfonic
acid])-1,2,4-triazine), forms a coloured complex with ferrous ions,
but not with ferric ions. The difference in the ferrous ion
concentration prior to and following the enzymatic reaction
indicates the quantity of the oxidized ferrous ion. The quantity of
enzyme that converted 1 µmol of substrate into product per minute
was defined as 1 unit.
Statistical analysis
Data are presented as the mean with standard error
of the mean and/or standard deviation as indicated. Differences
between the studied groups in blood levels of proteins and 8-oxodG
as well as in TIBC and CFA were analysed by Mann-Whitney U-tests.
Analysis of transferrin blood level between groups with different
HP genotypes determined previously (17) was performed using one-way analysis of
variance (ANOVA) followed by post-hoc Tukey's tests. To estimate
correlations between the parameters under study, Pearson's
correlation coefficients (r) were computed. In all tests,
significance was accepted at P<0.05. All statistical analyses
were performed using Statistica 7 software (StatSoft, Inc., Tulsa,
OK, USA).
Results
Comparison of oxidative stress, iron
metabolism, and inflammation markers between COPD patients with and
without lung cancer
The blood serum levels of the oxidative stress
marker, 8-oxodG, and several proteins involved in iron metabolism
and inflammation were analyzed in two groups of patients: The first
group consisted of patients with COPD and lung cancer, whereas the
second group consisted of patients with COPD only.
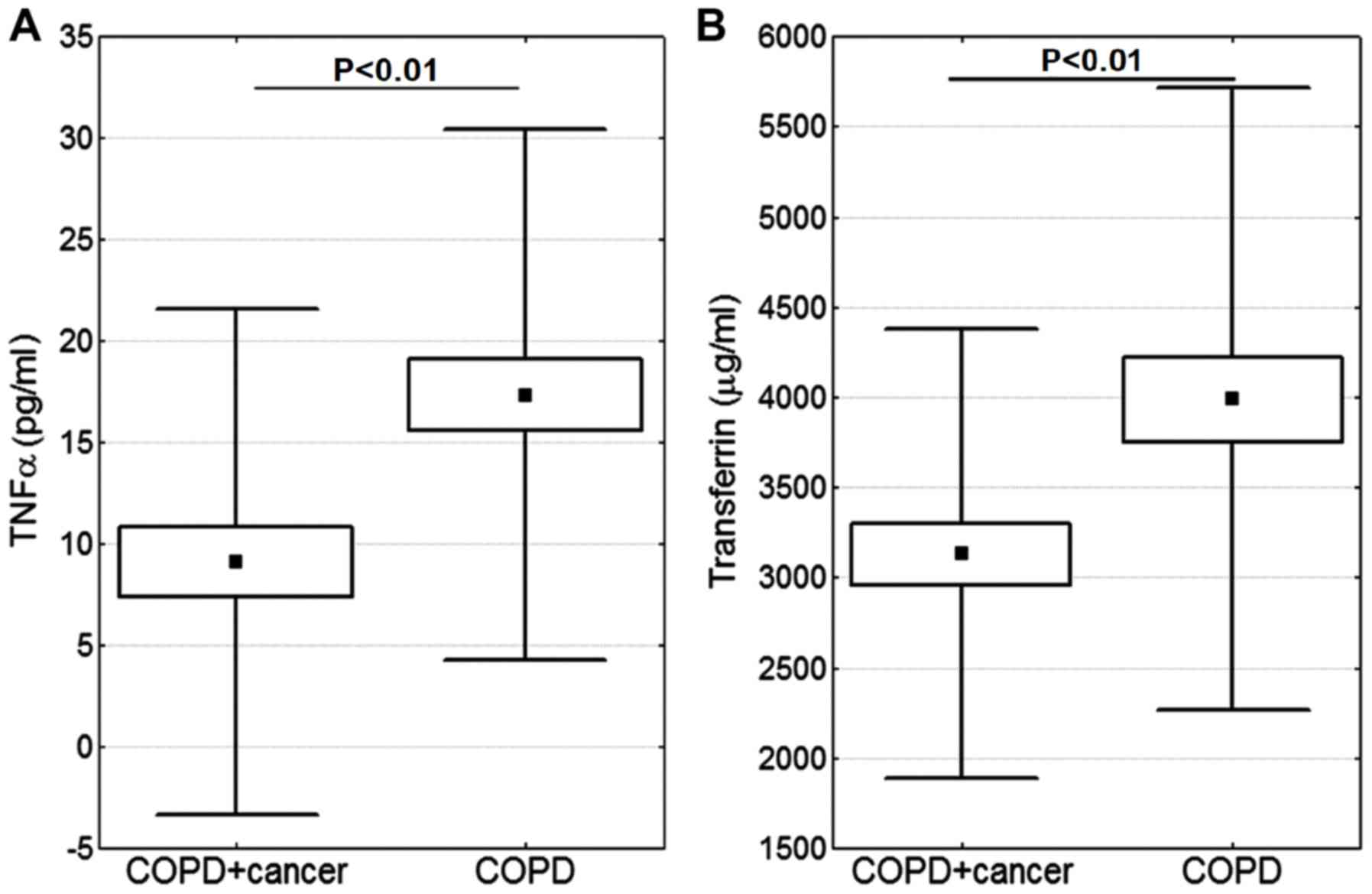
The blood serum level of TNFα was lower in the COPD
+ cancer group compared with in the COPD group (P<0.01; Fig. 1A). Additionally, the difference in
transferrin level was significant, and its level was lower in the
COPD + cancer subjects (P<0.01; Fig.
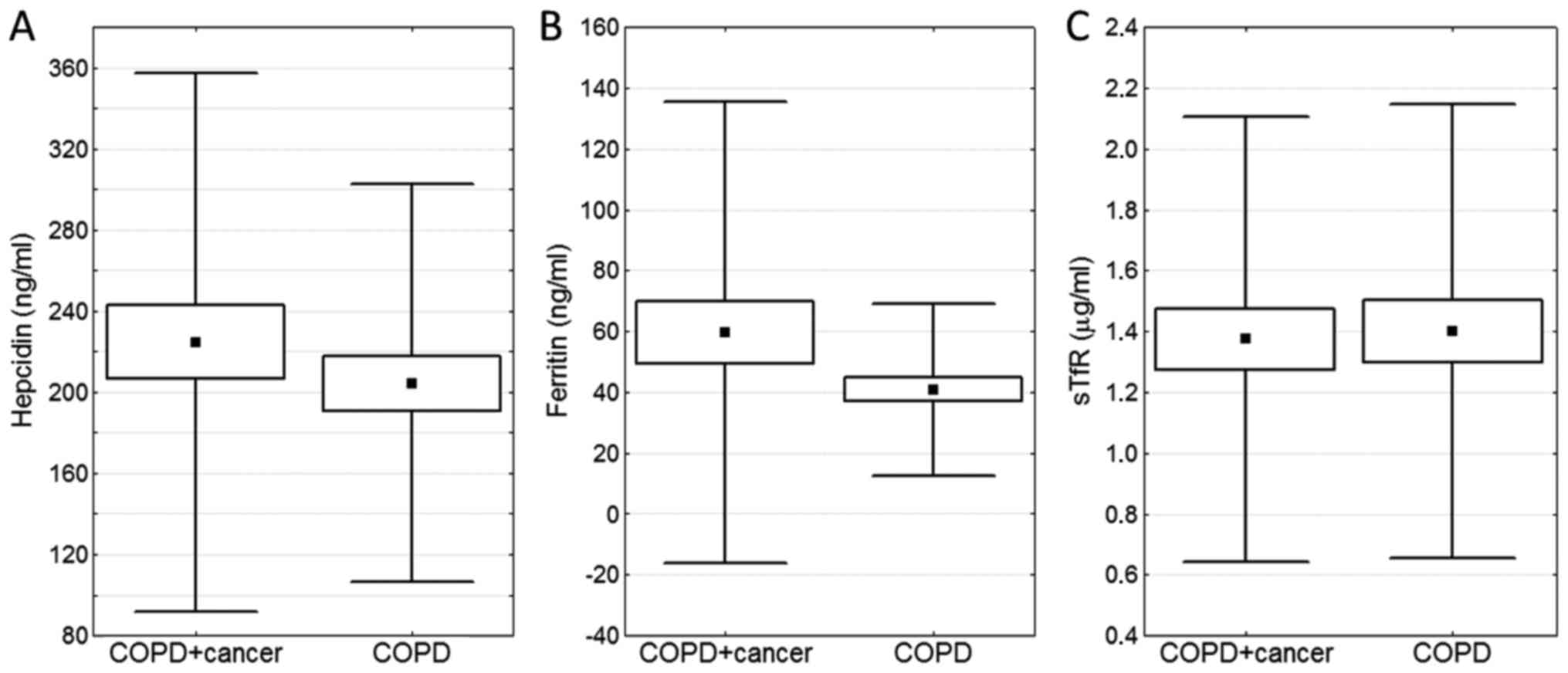
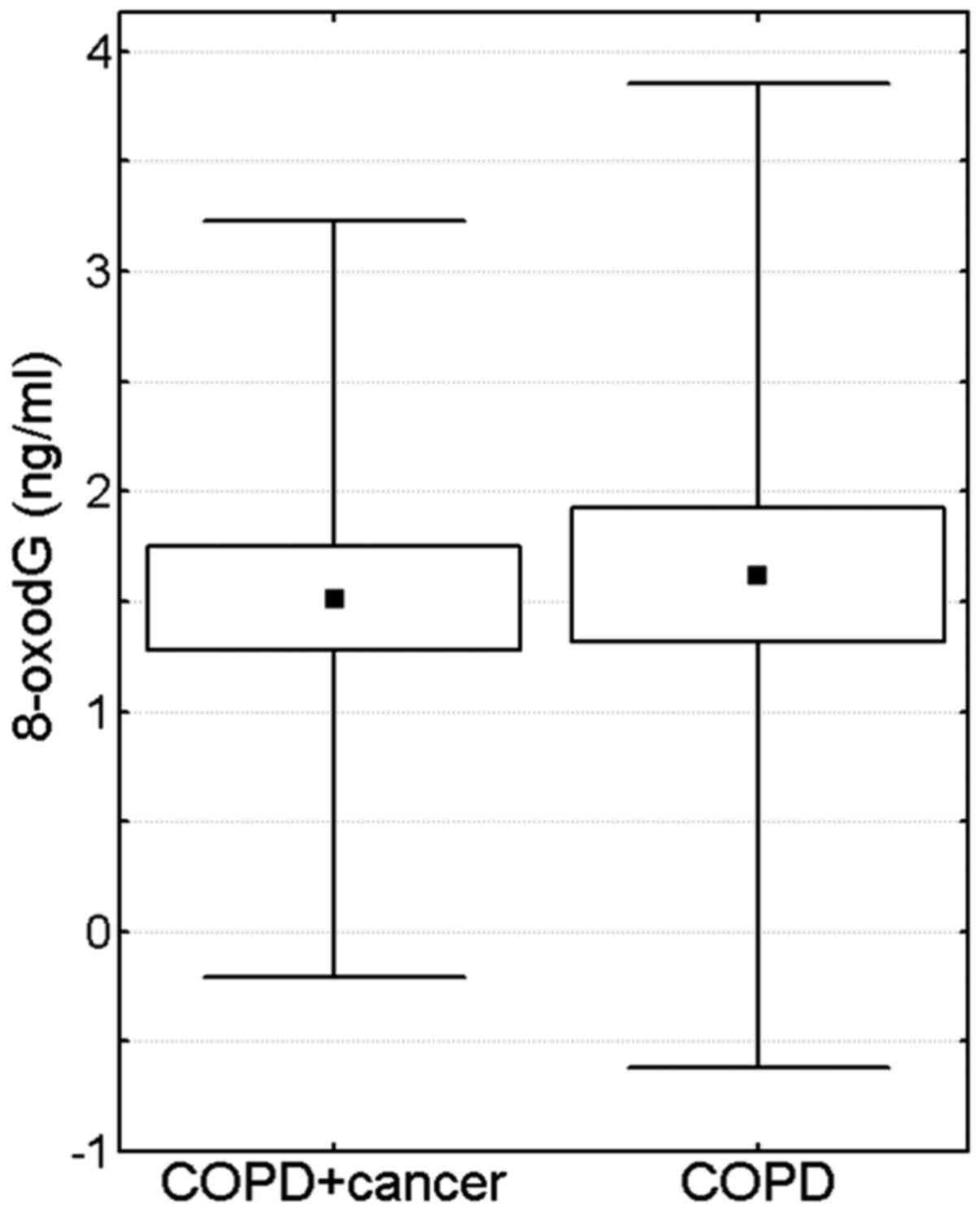
1B). No statistically significant differences were observed in
the blood serum levels of other proteins involved in iron
metabolism (hepcidin, ferritin and sTfR) (Fig. 2) or 8-oxodG (Fig. 3).
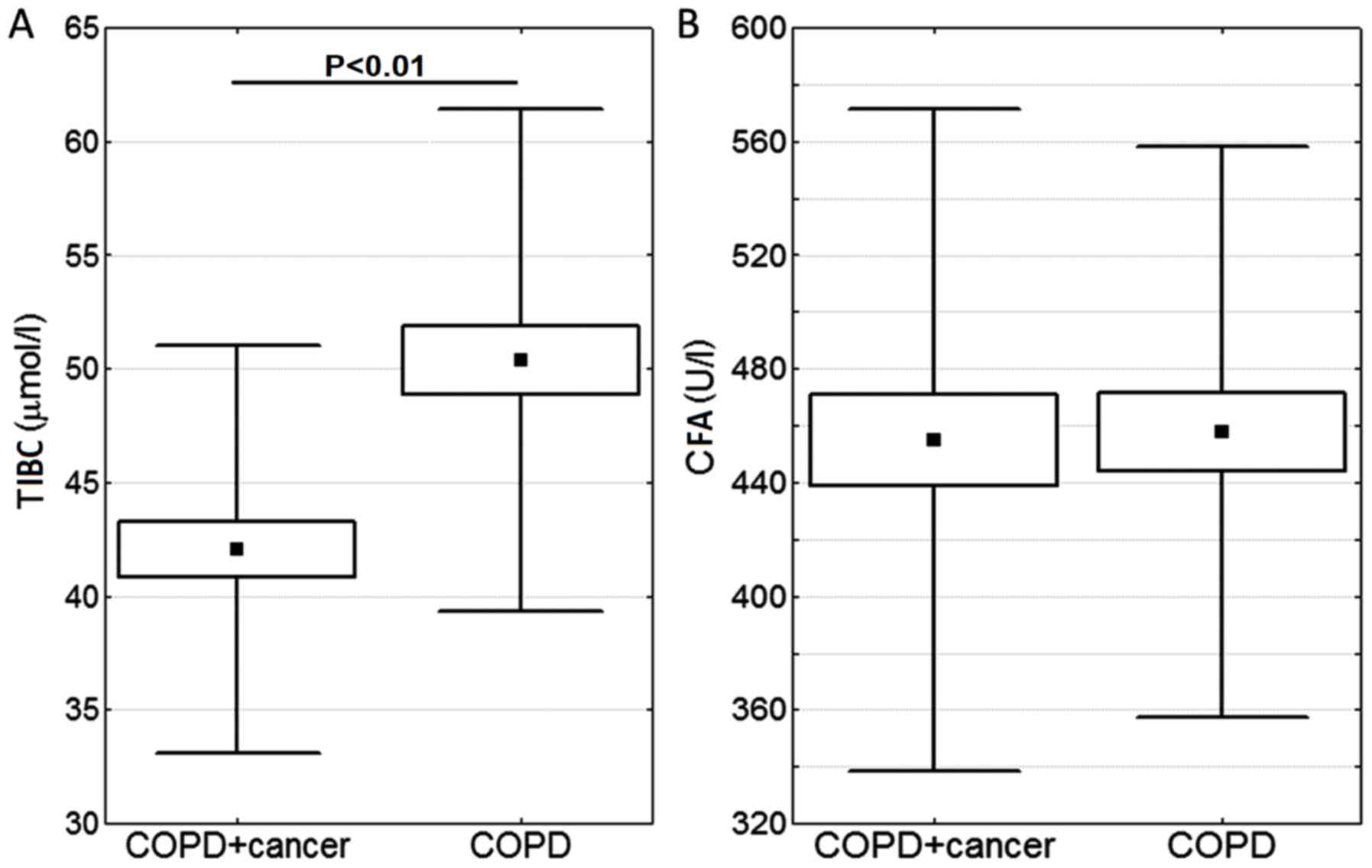
Furthermore, TIBC and CFA were compared (Fig. 4). The lower TIBC index in COPD +
cancer subjects was statistically significantly different to that
of the COPD group (P<0.01).
Correlation analysis
Table I sumarizes the
analysis of correlations between measured parameters. Notable
differences between the groups under study were observed. Namely,
in COPD-only patients, hepcidin level was positively correlated
with CFA and 8-oxodG level (r=0.31 and 0.60; P<0.05); whereas in
patients with COPD and lung cancer, hepcidin level also showed
positive correlation with ferritin level (r=0.45; P<0.05), as
well as negative correlation with transferrin level and TIBC
(r=−0.27 and −0.29; P<0.05). In the case of ferritin level, in
COPD-only patients it was negatively correlated with tranferrin
level and TIBC (r=−0.33 and −0.40; P<0.05), and positively
correlated with 8-oxodG level (r=0.39; P<0.05); whereas in
patients with COPD and lung cancer, ferritin level was not
correlated with the levels of transferrin or 8-oxodG, but instead
correlation with hepcidin and CFA (r=0.45 and 0.29; P<0.05) was
observed.
 | Table I.Pearson's correlation coefficients
(r) for correlations between the measured parameters in COPD and
COPD + lung cancer subjects. |
Table I.
Pearson's correlation coefficients
(r) for correlations between the measured parameters in COPD and
COPD + lung cancer subjects.
| A, All subjects
(n=107) |
|---|
|
| TNFα | sTfR | Ferritin | Transferrin | TIBC | CFA | Hepcidin | 8-oxodG |
|---|
| TNFα | 1.00 | – | – | – | – | – | – | – |
| sTfR | −0.09 | 1.00 | – | – | – | – | – | – |
| Ferritin | −0.08 | 0.03 | 1.00 | – | – | – | – | – |
| Transferrin | 0.47 | 0.02 | −0.26 | 1.00 | – | – | – | – |
| TIBC | 0.21 | 0.13 | −0.34 | 0.42 | 1.00 | – | – | – |
| CFA | 0.11 | 0.17 | 0.26 | −0.06 | −0.05 | 1.00 | – | – |
| Hepcidin | −0.11 | 0.13 | 0.38 | −0.24 | −0.11 | 0.35 | 1.00 | – |
| 8-oxodG | −0.18 | 0.08 | 0.18 | −0.31 | −0.11 | 0.29 | 0.51 | 1.00 |
|
| B, COPD-only
subjects (n=54) |
|
|
| TNFα | sTfR |
Ferritin |
Transferrin | TIBC | CFA |
Hepcidin | 8-oxodG |
|
| TNFα | 1.00 | – | – | – | – | – | – | – |
| sTfR | −0.05 | 1.00 | – | – | – | – | – | – |
| Ferritin | −0.05 | −0.07 | 1.00 | – | – | – | – | – |
| Transferrin | 0.40 | 0.03 | −0.33 | 1.00 | – | – | – | – |
| TIBC | 0.01 | 0.22 | −0.40 | 0.30 | 1.00 | – | – | – |
| CFA | 0.02 | 0.14 | 0.24 | −0.18 | 0.02 | 1.00 | – | – |
| Hepcidin | −0.15 | 0.24 | 0.17 | −0.19 | 0.13 | 0.31 | 1.00 | – |
| 8-oxodG | −0.23 | 0.09 | 0.39 | −0.32 | −0.07 | 0.48 | 0.60 | 1.00 |
|
| C, COPD + lung
cancer subjects (n=53) |
|
|
| TNFα | sTfR |
Ferritin |
Transferrin | TIBC | CFA |
Hepcidin | 8-oxodG |
|
| TNFα | 1.00 | – | – | – | – | – | – | – |
| sTfR | −0.17 | 1.00 | – | – | – | – | – | – |
| Ferritin | −0.04 | 0.08 | 1.00 | – | – | – | – | – |
| Transferrin | 0.46 | −0.02 | −0.24 | 1.00 | – | – | – | – |
| TIBC | 0.23 | 0.03 | −0.33 | 0.44 | 1.00 | – | – | – |
| CFA | 0.18 | 0.20 | 0.29 | 0.08 | −0.15 | 1.00 | – | – |
| Hepcidin | −0.05 | 0.05 | 0.45 | −0.27 | −0.29 | 0.38 | 1.00 | – |
| 8-oxodG | −0.15 | 0.07 | 0.15 | −0.33 | −0.25 | 0.08 | 0.48 | 1.00 |
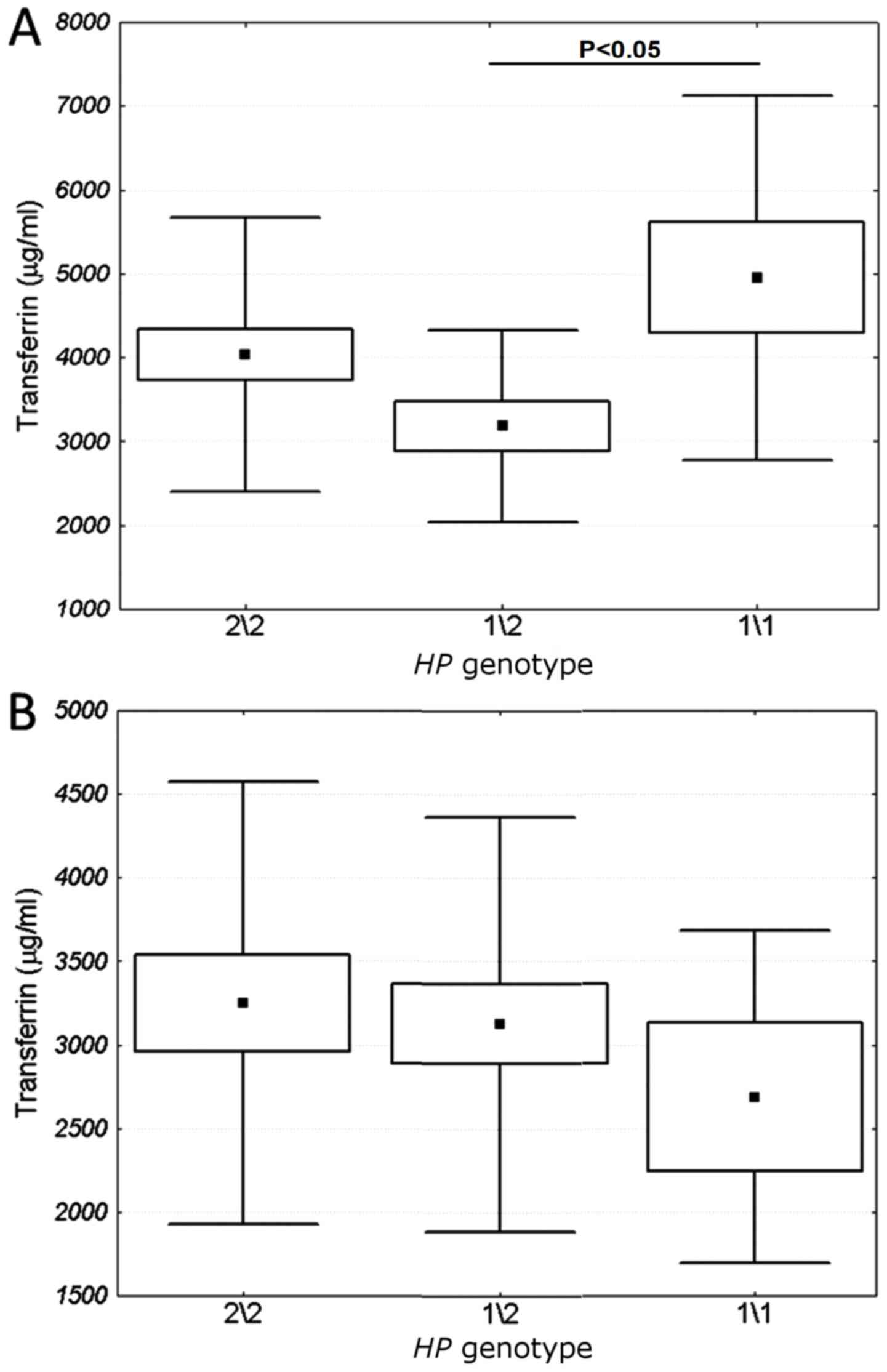
Impact of HP polymorphism on iron
metabolism, and inflammation markers
The influence of HP polymorphism on TIBC, as
well as on the blood serum levels of TNFα and transferrin was
examined. From one-way ANOVA, it was determined that HP
genotype 1/2 was concomitant with a low transferrin blood level in
the subjects with COPD only (Fig.
5A), but not in the subjects with COPD and lung cancer
(Fig. 5B).
Discussion
The oxidant and noxious stress responses that occur
in the lungs of cigarette smokers cause damage to epithelial cells,
leading to their apoptosis and the development of emphysema, as a
main characteristic of COPD (21). In
addition, reactive oxygen species are genotoxic agents and their
interaction with DNA results in the formation of oxidized DNA
bases, DNA strand breaks and chromosomal damage (22). These lesions, if unrepaired or
misrepaired, may lead to cell death or genomic instability and,
consequently, result in a number of diseases, including those
associated with neoplastic changes (23,24).
There are several reports of increased lung cancer
risk being associated with polymorphisms in the DNA repair genes
(25–29). Nevertheless, an analysis of pooled
data from 14 studies on the significance of polymorphisms in the
DNA repair genes for cancer risk, published in 2008 by the
International Lung Cancer Consortium (30), indicated that mostly weak associations
could be found for single polymorphisms. It should be stressed that
all these analyses (25–30) were performed by comparing healthy
subjects with those with lung cancer. In our previous work, COPD
patients were compared with patients with both COPD and lung
cancer. No significant differences in the distribution of
polymorphisms in DNA repair genes were identified between the two
groups. Similarly, the frequency of polymorphisms in the tumor
protein p53 and cyclin dependent kinase inhibitor 1 genes did not
differ between COPD and COPD + cancer subjects (17). This result is consistent with the
similar blood level of 8-oxodG, a marker of oxidative DNA damage,
in the same two groups of subjects, as reported in the present
study.
During a previous study on the same groups of
subjects, no statistically significant difference was found in the
frequency of SNPs in the gene encoding TNFα. By contrast, a
significant difference between the compared groups was identified
in the distribution of genotypes of the HP gene (17). HP belongs to the acute phase
proteins that are present in serum; their increase and altered
glycosylation accompany inflammation and tumorigenesis (31). The biological function of HP is
to bind free hemoglobin in blood, thus preventing iron loss and
allowing the recycling of heme iron in the reticuloendothelial
system in the liver (32–34). Therefore, acting as a hemoglobin
scavenger, HP lowers iron concentration, and facilitates the
anti-inflammatory response (34).
Among the known HP phenotypes, HP1/1 is the most
effective in binding free hemoglobin, thus suppressing the
oxidative stress and inflammatory response associated with free
hemoglobin. HP2/2 is the least effective, whereas
HP1/2 has an intermediate anti-oxidative activity (35–37).
The formation of reactive oxygen radicals frequently
occurs in Haber-Weiss and Fenton-type reactions with the
participation of ferrous ions (Fe2+), therefore,
homeostasis disorders of this element may be important in the
development of cancer (19). In fact,
such has been observed in patients with various cancers: Iron was
identified as a potential carcinogen approximately 25 years ago
(38–41) also in lung cancer (42). Recent reviews (43,44)
summarize the research on iron metabolism in the cancer cell and
the possible applications of iron targeted therapy. In the present
study, it was revealed that HP1/2 genotype was associated
with low transferrin blood level in patients with COPD. This
relationship was absent in patients with COPD and lung cancer,
likely reflecting carcinogenesis-related changes in iron
metabolism. Such changes were also manifested in the significantly
lower blood levels of TNFα, transferrin and TIBC in the patients
with COPD and lung cancer.
In conclusion, the results of the present study
indicate the importance of iron metabolism in the development and
progression of lung cancer in COPD-affected subjects. In future
studies by our group, the aim will be to further investigate the
mechanisms of COPD and lung cancer development and the relationship
between these two diseases, with focus on the roles of the immune
system, iron metabolism and the oxidative stress response in their
pathogenesis. Specifically, analysis of the expression of genes
related to these processes is intended on the transcriptomic level,
in circulating immune cells isolated from COPD and lung cancer
patients as well as from tumor samples. Further aims will be to
verify if epigenetic factors including microRNA expression and DNA
methylation may be responsible for the effects observed in the
present study (for example, the association between transferrin
level and haptoglobin genotype), and to investigate their role in
COPD and lung cancer pathogenesis.
Acknowledgements
The authors thank Professor Irena Szumiel (Institute
of Nuclear Chemistry and Technology, Centre for Radiobiology and
Biological Dosimetry, Warsaw, Poland; Retired) for her advice and
assistance in writing the manuscript.
Funding
The present study was supported by the Ministry of
Science and Higher Education, Warsaw, Poland (grant no. N402 109
32/3503).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MK, PŚ, KRŚ and LKS designed the study. KB, TB, BS,
MC, JG and JK collected the data. KB, PŚ, KRŚ and LKS analysed the
data. KB and MK performed statistical analysis. KB and LKS wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol conformed to the Declaration of
Helsinki, and was approved by the Institutional Ethics Committee of
the Institute of Tuberculosis and Lung Diseases (Warsaw, Poland).
Each participating patient had provided signed informed consent
following a detailed explanation of the study protocols.
Patient consent for publication
Study participants provided their consent for the
publication of this data and any associated images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dik S, Scheepers PT and Godderis L:
Effects of environmental stressors on histone modifications and
their relevance to carcinogenesis: A systematic review. Crit Rev
Toxicol. 42:491–500. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Edwards TM and Myers JP: Environmental
exposures and gene regulation in disease etiology. Environ Health
Perspect. 115:1264–1270. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hursting SD and Berger NA: Energy balance,
host-related factors, and cancer progression. J Clin Oncol.
28:4058–4065. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hursting SD and Hursting MJ: Growth
signals, inflammation, and vascular perturbations: Mechanistic
links between obesity, metabolic syndrome, and cancer. Arterioscler
Thromb Vasc Biol. 32:1766–1770. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maynard S, Schurman SH, Harboe C, de
Souza-Pinto NC and Bohr VA: Base excision repair of oxidative DNA
damage and association with cancer and aging. Carcinogenesis.
30:2–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weidman JR, Dolinoy DC, Murphy SK and
Jirtle RL: Cancer susceptibility: Epigenetic manifestation of
environmental exposures. Cancer J. 13:9–16. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adcock IM, Caramori G and Barnes PJ:
Chronic obstructive pulmonary disease and lung cancer: New
molecular insights. Respiration. 81:265–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Decramer M, Janssens W and Miravitlles M:
Chronic obstructive pulmonary disease. Lancet. 379:1341–1351. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Decramer M, Rennard S, Troosters T, Mapel
DW, Giardino N, Mannino D, Wouters E, Sethi S and Cooper CB: COPD
as a lung disease with systemic consequences-clinical impact,
mechanisms, and potential for early intervention. COPD. 5:235–256.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang IA, Relan V, Wright CM, Davidson MR,
Sriram KB, Francis Savarimuthu SM, Clarke BE, Duhig EE, Bowman RV
and Fong KM: Common pathogenic mechanisms and pathways in the
development of COPD and lung cancer. Expert Opin Ther Targets.
15:439–456. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Young RP and Hopkins RJ: How the genetics
of lung cancer may overlap with COPD. Respirology. 16:1047–1055.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bozinovski S, Vlahos R, Anthony D,
McQualter J, Anderson G, Irving L and Steinfort D: COPD and
squamous cell lung cancer: Aberrant inflammation and immunity is
the common link. Br J Pharmacol. 173:635–648. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Durham AL and Adcock IM: The relationship
between COPD and lung cancer. Lung Cancer. 90:121–127. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aoshiba K, Zhou F, Tsuji T and Nagai A:
DNA damage as a molecular link in the pathogenesis of COPD in
smokers. Eur Respir J. 39:1368–1376. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brzóska K, Bartlomiejczyk T, Sochanowicz
B, Cymerman M, Grudny J, Kołakowski J, Kapka-Skrzypczak L,
Kruszewski M, Sliwiński P and Roszkowski-Śliż K: Matrix
metalloproteinase 3 polymorphisms as a potential marker of enhanced
susceptibility to lung cancer in chronic obstructive pulmonary
disease subjects. Ann Agric Environ Med. 21:546–551. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grudny J, Kolakowski J, Kruszewski M,
Szopiński J, Sliwiński P, Wiatr E, Winek J, Załęska J, Zych J and
Roszkowski-Śliż K: Association of genetic dependences between lung
cancer and chronic obstructive pulmonary disease. Pneumonol Alergol
Pol. 81:308–318. 2013.PubMed/NCBI
|
|
18
|
Goldenstein H, Levy NS and Levy AP:
Haptoglobin genotype and its role in determining heme-iron mediated
vascular disease. Pharmacol Res. 66:1–6. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kruszewski M: Labile iron pool: The main
determinant of cellular response to oxidative stress. Mutat Res.
531:81–92. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Erel O: Automated measurement of serum
ferroxidase activity. Clin Chem. 44:2313–2319. 1998.PubMed/NCBI
|
|
21
|
Boukhenouna S, Wilson MA, Bahmed K and
Kosmider B: Reactive oxygen species in chronic obstructive
pulmonary disease. Oxid Med Cell Longev. 2018:57303952018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Markkanen E: Not breathing is not an
option: How to deal with oxidative DNA damage. DNA Repair (Amst).
59:82–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewandowska H, Kalinowska M, Lewandowski
W, Stepkowski TM and Brzóska K: The role of natural polyphenols in
cell signaling and cytoprotection against cancer development. J
Nutr Biochem. 32:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liguori I, Russo G, Curcio F, Bulli G,
Aran L, Della-Morte D, Gargiulo G, Testa G, Cacciatore F, Bonaduce
D and Abete P: Oxidative stress, aging, and diseases. Clin Interv
Aging. 13:757–772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hung RJ, Hall J, Brennan P and Boffetta P:
Genetic polymorphisms in the base excision repair pathway and
cancer risk: A HuGE review. Am J Epidemiol. 162:925–942. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
López-Cima MF, Gonzalez-Arriaga P,
Garcia-Castro L, Pascual T, Marrón MG, Puente XS and Tardón A:
Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and
lung cancer risk in a population of northern Spain. BMC Cancer.
7:1622007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Manuguerra M, Saletta F, Karagas MR,
Berwick M, Veglia F, Vineis P and Matullo G: XRCC3 and XPD/ERCC2
single nucleotide polymorphisms and the risk of cancer: A HuGE
review. Am J Epidemiol. 164:297–302. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park JY, Lee SY, Jeon HS, Bae NC, Chae SC,
Joo S, Kim CH, Park JH, Kam S, Kim IS and Jung TH: Polymorphism of
the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer
Epidemiol Biomarkers Prev. 11:23–27. 2002.PubMed/NCBI
|
|
29
|
Zhou W, Liu G, Miller DP, Thurston SW, Xu
LL, Wain JC, Lynch TJ, Su L and Christiani DC: Polymorphisms in the
DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk.
Cancer Epidemiol Biomarkers Prev. 12:359–365. 2003.PubMed/NCBI
|
|
30
|
Hung RJ, Christiani DC, Risch A, Popanda
O, Haugen A, Zienolddiny S, Benhamou S, Bouchardy C, Lan Q, Spitz
MR, et al: International lung cancer consortium: Pooled analysis of
sequence variants in DNA repair and cell cycle pathways. Cancer
Epidemiol Biomarkers Prev. 17:3081–3089. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dempsey E and Rudd PM: Acute phase
glycoproteins: Bystanders or participants in carcinogenesis? Ann N
Y Acad Sci. 1253:122–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Delanghe JR and Langlois MR: Haptoglobin
polymorphism and body iron stores. Clin Chem Lab Med. 40:212–216.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dobryszycka W: Biological functions of
haptoglobin-new pieces to an old puzzle. Eur J Clin Chem Clin
Biochem. 35:647–654. 1997.PubMed/NCBI
|
|
34
|
Nielsen MJ and Moestrup SK: Receptor
targeting of hemoglobin mediated by the haptoglobins: Roles beyond
heme scavenging. Blood. 114:764–771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Levy AP, Asleh R, Blum S, Levy NS,
Miller-Lotan R, Kalet-Litman S, Anbinder Y, Lache O, Nakhoul FM,
Asaf R, et al: Haptoglobin: Basic and clinical aspects. Antioxid
Redox Signal. 12:293–304. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sadrzadeh SM and Bozorgmehr J: Haptoglobin
phenotypes in health and disorders. Am J Clin Pathol. 121
Suppl:S97–S104. 2004.PubMed/NCBI
|
|
37
|
Wassell J: Haptoglobin: Function and
polymorphism. Clin Lab. 46:547–552. 2000.PubMed/NCBI
|
|
38
|
Huang X: Iron overload and its association
with cancer risk in humans: Evidence for iron as a carcinogenic
metal. Mutat Res. 533:153–171. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Knekt P, Reunanen A, Takkunen H, Aromaa A,
Heliövaara M and Hakulinen T: Body iron stores and risk of cancer.
Int J Cancer. 56:379–382. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Selby JV and Friedman GD: Epidemiologic
evidence of an association between body iron stores and risk of
cancer. Int J Cancer. 41:677–682. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Toyokuni S and Sagripanti JL: Association
between 8-hydroxy-2′-deoxyguanosine formation and DNA strand breaks
mediated by copper and iron. Free Radic Biol Med. 20:859–864. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou W, Park S, Liu G, Miller DP, Wang LI,
Pothier L, Wain JC, Lynch TJ, Giovannucci E and Christiani DC:
Dietary iron, zinc, and calcium and the risk of lung cancer.
Epidemiology. 16:772–779. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Richardson DR, Kalinowski DS, Lau S,
Jansson PJ and Lovejoy DB: Cancer cell iron metabolism and the
development of potent iron chelators as anti-tumour agents. Biochim
Biophys Acta. 1790:702–717. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang SJ, Gao C and Chen BA: Advancement of
the study on iron metabolism and regulation in tumor cells. Chin J
Cancer. 29:451–455. 2010. View Article : Google Scholar : PubMed/NCBI
|