Introduction
The incidence of metastatic ovarian cancer varies
greatly across different studies (it was reported to occur in 2.3
to 23.7% of cases of malignant ovarian cancer). Such tumors mainly
arise from the colon (10.9 to 33.2%), breast (1.8 to 33.3%), or
stomach (4.5 to 30.4%) (1).
Metastatic ovarian cancer occurs in 5 to 9.7% of women with
colorectal cancer, who exhibit a median survival period of only 19
to 27 months after detection (2,3). Surgical
treatment has been recommended in several retrospective studies
because it results in a better prognosis (4–6), and
chemotherapy exhibits poor sensitivity, although recurrence and
distant metastasis often occur during the follow-up period.
Furthermore, the preoperative clinical diagnosis of metastatic
ovarian cancer is difficult, and pathological examinations of
surgical samples are required to definitively differentiate it from
primary ovarian cancer. In previous studies of its utility for
distinguishing primary ovarian cancer from metastatic ovarian
cancer, imaging analysis was shown to only produce non-specific
findings (7,8).
Despite recent progress in colorectal cancer
treatment, including the development of molecular-targeting
therapy, there are little available data about the best way to
treat patients affected by metastatic colorectal cancer to the
ovary. Furthermore, there is insufficient information about the
clinicopathological and genomic predictors of the outcomes of such
patients, and the optimal clinical management strategy remains
unclear (9,10). Ganesh et al (9), examined the genetic aberrations found in
metastatic colorectal cancer to the ovary, including primary
colorectal cancer, ovarian metastasis, and extra-ovarian
metastasis. The frequencies of somatic mutations in oncogenes and
tumor suppressor genes demonstrated a high concordance rate between
matched primary and metastatic (from sites other than the ovary)
colorectal tumors. Increased frequencies of mutations were seen in
the Kirsten rat sarcoma viral oncogene homolog (KRAS), Smad
family member 4 (SMAD4), and neurotrophic receptor tyrosine
kinase 1 (NTRK1) genes.
Recent studies have shown that genomic alterations
in solid cancers can be characterized by massive parallel
sequencing of the circulating cell-free tumor DNA released from
cancer cells in plasma, which is known as circulating tumor DNA
(ctDNA). This technique is effectively a non-invasive ‘liquid
biopsy’ examination (11,12). Liquid biopsy examinations of ctDNA can
be used to detect molecular alterations, including tumor-specific
mutations, and have recently been used as a diagnostic, prognostic,
and predictive tool, especially in colorectal cancer (to detect
RAS mutations) and lung cancer (to detect epidermal growth
factor receptor [EGFR] mutations) (13–15).
However, to the best of our knowledge, there have not been any
reports about the use of liquid biopsy examinations in patients
with metastatic colorectal cancer to the ovary. In this study, we
subjected plasma ctDNA samples (liquid biopsy samples) to multiple
gene mutation analysis using CAncer Personalized Profiling by deep
Sequencing (CAPP-Seq), a novel next-generation sequencing-based
approach to ultrasensitive ctDNA detection (16,17), in
patients with ovarian metastases from colorectal cancer. Then, we
compared the findings with the results obtained for primary and
metastatic tumor samples.
Materials and methods
Patients and samples
Two patients whose metastatic ovarian tumors had
been surgically resected at Wakayama Medical University Hospital
between May 2017 and June 2017 were included in this study. The
tumor staging was carried out according to the TNM classification.
The present study was approved by the ethics committee of Wakayama
Medical University Faculty of Medicine (authorization no: 2025) and
Kindai University Faculty of Medicine (authorization no: 29-066).
All of the patients in this study provided written informed consent
for the use of their plasma and tissue samples.
Tumor DNA extraction
Formalin-fixed paraffin-embedded (FFPE) specimens
were subjected to a histological review, and only those containing
sufficient tumor cells (at least 75% tumor cells), as determined by
hematoxylin and eosin staining, were used for DNA extraction. The
collected DNA was purified with the use of an AllPrep DNA/RNA FFPE
kit (Qiagen, Inc., Valencia, CA, USA), according to the
manufacturer's instructions. The quality and quantity of the DNA
were verified using the NanoDrop 2000 device (Thermo Fisher
Scientific, Inc., Wilmington, DE, USA) and PicoGreen dsDNA assay
kit (Thermo Fisher Scientific, Inc.). The extracted DNA was stored
at −80°C until the analysis.
Tumor DNA sequencing
We used a 40 ng of DNA for the QIAseq Human
Comprehensive Cancer Panel (275 genes; Qiagen, Inc.). Library
preparation was performed according to the manufacturer's
instructions. The purified libraries were pooled and then sequenced
with a NextSeq 500 instrument (Illumina, Inc., San Diego, CA, USA).
Reads were aligned with the hg19 human reference genome, and
variant detection was performed according to the manufacturer's
pipeline (18). Germline mutations
were excluded with the use of the Human Genetic Variation Database
(http://www.genome.med.kyoto-u.ac.jp/SnpDB) and the
Exome Aggregation Consortium database (19).
ctDNA extraction
Samples (8.5 ml) of peripheral blood were collected
from the patients in cell-free DNA collection tubes (Roche
Diagnostics, Indianapolis, IN, USA). Plasma ctDNA was purified
using an AVENIO cfDNA isolation kit (Roche Diagnostics), according
to the manufacturer's instructions. The quality and quantity of the
DNA were verified using the NanoDrop 2000 device (Thermo Fisher
Scientific, Inc.) and PicoGreen dsDNA assay kit (Thermo Fisher
Scientific, Inc.). The extracted ctDNA was stored at −80°C until
the analysis.
ctDNA sequencing
We used a maximum of 50 ng of DNA for the CAPP-Seq
ctDNA analyses using the AVENIO ctDNA surveillance kit (197 genes;
Roche Diagnostics), according to the manufacturer's instructions.
The purified libraries were pooled and sequenced on an Illumina
NextSeq 500 (Illumina, Inc.) using the 300-cycle high output kit.
Variants were called with the AVENIO ctDNA analysis software (Roche
Diagnostics), which includes bioinformatics methods from CAPP-Seq
(16) and integrated digital error
suppression (17). Germline mutations
were excluded with the use of the Human Genetic Variation Database
(http://www.genome.med.kyoto-u.ac.jp/SnpDB) and the
Exome Aggregation Consortium database (19).
Results
Patient 1
A 61-year-old female was referred to our institution
with suspected ovarian cancer. She had been diagnosed with sigmoid
colon cancer 2 years ago, which was completely surgically resected
in other institution. The tumor had a moderately differentiated
histology, and pathologically, it extended into the subserosa
(pT3). There were no lymph node or distant metastases (pN0, M0).
The patient received adjuvant chemotherapy (8 courses of
capecitabine). Two years later, a tumor appeared in the right
ovary. Magnetic resonance imaging (MRI) revealed a solid 4.3-cm
mass with an irregular surface, and positron emission
tomography-computed tomography (PET-CT) detected high radiotracer
uptake by the tumor [maximum standardized uptake value (SUVmax):
10.66]. The patient's levels of carcinoembryonic antigen (CEA) and
cancer antigen (CA) 19-9 were elevated (to 8.8 ng/ml and 58.4 U/ml,
respectively), whereas her CA125 level was normal. The patient
underwent a total abdominal hysterectomy and bilateral
salpingo-oophorectomy, and the pathological diagnosis was
metastatic ovarian cancer. We obtained an FFPE sample from the
metastatic ovarian tumor (O-1) and a preoperative plasma ctDNA
sample (L-1). The patient's clinical course and clinicopathological
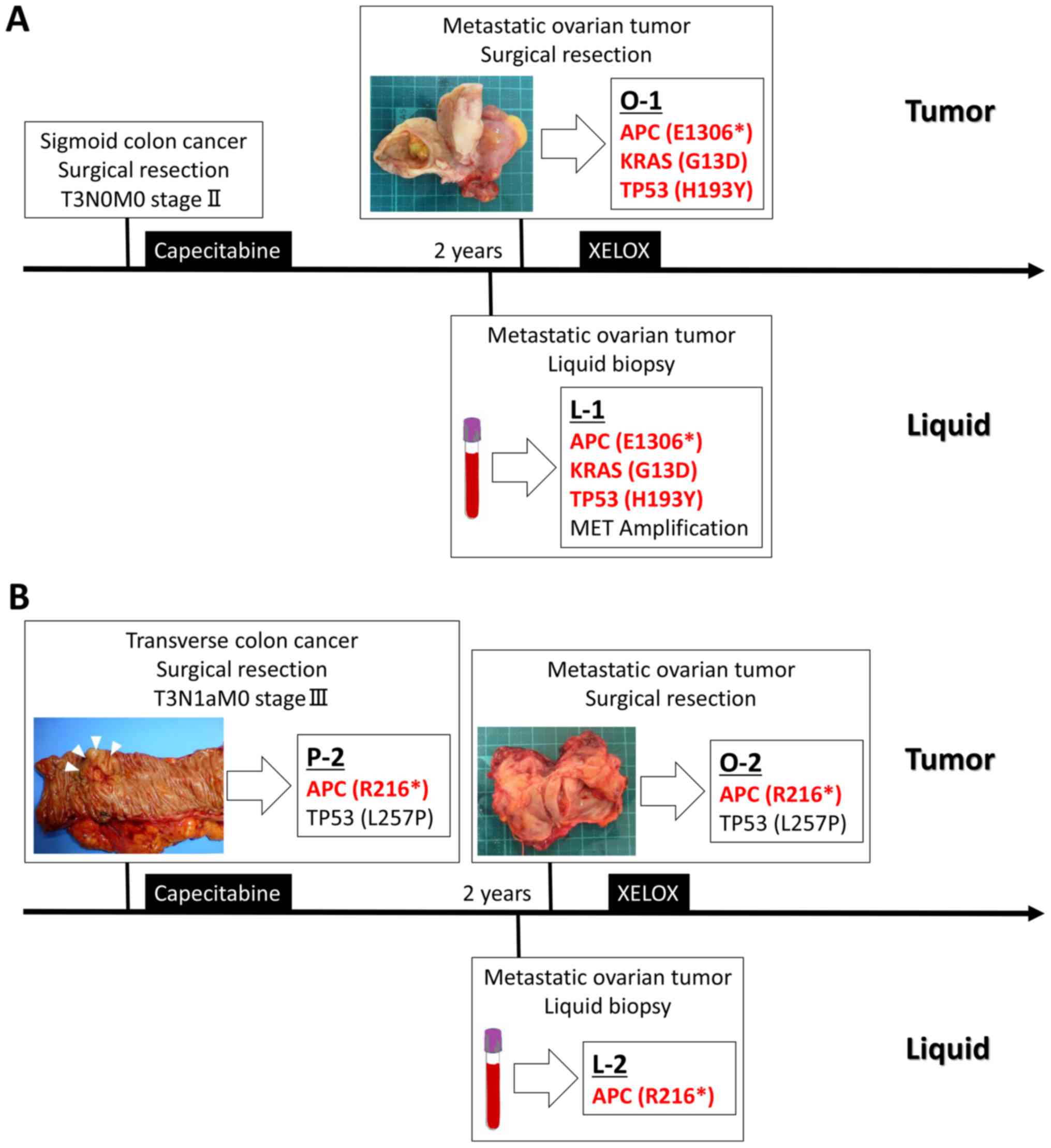
features are summarized in Fig. 1A.
Targeted re-sequencing was performed using the QIAseq Human
Comprehensive Cancer Panel for O-1 and the AVENIO ctDNA
surveillance kit for L-1. Intronic changes and exonic SNP listed in
the HGVD database were excluded from our analysis, as described in
Materials and Methods. Non-synonymous somatic point mutations that
resulted in changes in protein amino acid sequences are summarized
in Table I. In O-1, mutations were
identified in three regions, the KRAS G13D, adenomatous
polyposis coli (APC) E1306*, and tumor protein p53 (TP53) H193Y
genes. Sequencing of the plasma ctDNA detected three mutations and
one copy number variation; i.e., KRAS G13D, APC
E1306*, and TP53 H193Y mutations and MET gene amplification.
A comparison of the genetic profiles of O-1 and L-1 revealed that
the KRAS G13D, APC E1306*, and TP53 H193Y gene
mutations, which are well-known genetic signatures of colorectal
cancer were present in both O-1 and L-1. These results suggest that
liquid biopsy-based gene mutation profiling could facilitate the
preoperative diagnosis of metastatic colorectal cancer to the
ovary. The patient received postoperative adjuvant chemotherapy (8
courses of capecitabine and oxaliplatin; XELOX).
 | Table I.Non-synonymous mutations in Patient
1. |
Table I.
Non-synonymous mutations in Patient
1.
|
|
|
|
|
| Variant frequency
(%) |
|---|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
| Liquid | Ovary |
|---|
|
|
|
|
|
|
|
|---|
| Chromosome | Position | Gene | Variant effect | Amino acid
change | L-1 | O-1 |
|---|
| Chr5 | 112839510 | APC | Nonsense | E1306* | 11.85 | 41.2 |
| Chr12 | 25245347 | KRAS | Missense | G13D | 7.58 | 26.0 |
| Chr17 | 7674954 | TP53 | Missense | H193Y | 9.48 | 41.0 |
Patient 2
A 66-year-old female was referred to our institution
with suspected ovarian cancer. She had been diagnosed with
transverse colon cancer 2 years earlier, which was completely
surgically resected. The tumor had a well-differentiated histology,
and pathologically, it extended into the subserosa (pT3). There was
a single lymph node metastasis, but no distant metastasis (pN1a,
M0). The patient was treated with adjuvant chemotherapy (8 courses
of capecitabine). Two years later, a tumor appeared in the right
ovary. MRI revealed a multilocular, partially solid, 5.0-cm mass
with an irregular surface, and PET-CT detected high radiotracer
uptake by the tumor (SUVmax: 6.39). The patient's CEA level was
elevated (11.1 ng/ml), while her CA19-9 and CA125 levels were
normal. She underwent a total abdominal hysterectomy and bilateral
salpingo-oophorectomy, and the pathological diagnosis was
metastatic ovarian cancer. We obtained an FFPE sample of the
primary transverse colon cancer (P-2), an FFPE sample of the
metastatic ovarian tumor (O-2), and a plasma ctDNA sample before
the second operation (L-2). The patient's clinical course and
clinicopathological features are summarized in Fig. 1B. Targeted re-sequencing was performed
using the QIAseq Human Comprehensive Cancer Panel for the FFPE
samples and the AVENIO ctDNA surveillance kit for the plasma ctDNA.
The intronic changes and exonic SNP listed in the HGVD database
were excluded from our analysis, as described in Materials and
Methods. The non-synonymous somatic point mutations that resulted
in changes in protein amino acid sequences are summarized in
Table II. Mutations were identified
in the same region; i.e., APC R216* and TP53 L257P,
in both the primary (P-2) and metastatic (O-2) tumors. Sequencing
of the plasma ctDNA sample (L-2) detected a single mutation:
APC R216*. In a comparison of the genetic profiles of P-2,
O-2, and L-2, it was found that APC R216* was present in all
P-2, O-2, and L-2, whereas it was not detected in L-2. Conversely
APC R216* was detected in L-2, which is well-known genetic
signature of colorectal cancer. These findings also support the
usefulness of liquid biopsy-based gene mutation profiling in the
preoperative diagnosis of metastatic colorectal cancer to the
ovary. The patient received postoperative adjuvant chemotherapy (8
courses of capecitabine and oxaliplatin; XELOX).
 | Table II.Non-synonymous mutations in Patient
2. |
Table II.
Non-synonymous mutations in Patient
2.
|
|
|
|
|
| Variant frequency
(%) |
|---|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
| Colon | Liquid | Ovary |
|---|
|
|
|
|
|
|
|
|---|
| Chromosome | Position | Gene | Variant effect | Amino acid
change | P-2 | L-2 | O-2 |
|---|
| Chr5 | 112792446 | APC | Nonsense | R216* | 17.2 | 0.11 | 20.3 |
| Chr17 | 7577511 | TP53 | Missense | L257P | 23.6 | 0 | 28.5 |
Discussion
To the best of our knowledge, the current study is
the first to characterize the genetic profile of metastatic
colorectal cancer to the ovary, by subjecting liquid biopsy samples
to a novel comprehensive gene mutation analysis technique; i.e.,
CAPP-Seq. We investigated the mutation profiles of tumor and liquid
samples of metastatic colorectal cancer to the ovary to elucidate
the potential of liquid biopsy examinations as a tool for
preoperative diagnosis. In comparisons of the mutation profiles of
the patients' tumor and liquid samples, matching colorectal cancer
mutation signatures were observed in both patients, suggesting that
liquid biopsy has potential as a tool for aiding the preoperative
diagnosis and treatment of metastatic ovarian cancer from
colorectal cancer.
Liquid biopsy examinations are non-invasive and have
the potential to improve cancer (including recurrence) detection,
non-invasive tumor genotyping, and disease monitoring. However,
most early-stage and many advanced-stage solid tumors exhibit very
low levels of ctDNA, which complicates ctDNA detection and
analysis. In this study, we used CAPP-Seq, a novel approach which
combines the hybrid affinity capture of hundreds of genomic regions
with deep sequencing and a specialized bioinformatics workflow, for
the ultrasensitive quantitation of ctDNA during comprehensive gene
expression analysis (16). CAPP-Seq
is able to detect ctDNA in patients with early and advanced stages
of various human malignancies, including lung cancer and lymphoma
(17,20). However, in the gynecological field
there have not been any reports about the comprehensive gene
mutation analysis of liquid biopsy samples, which might be a
powerful tool for advancing personalized medicine for gynecological
malignancies.
We revealed that liquid biopsy examinations have
potential to facilitate the preoperative diagnosis of metastatic
ovarian cancer from colorectal cancer. After examining FFPE tumor
samples, Crobach et al reported that the gene mutation
profiles of primary and metastatic ovarian cancer differ (10). Inactivating APC mutations were
found in 4.7% of primary endometrioid or mucinous ovarian tumors,
whereas they were identified in 71% of colorectal cancer
metastases; thus, APC mutation analysis can be used to
differentiate between primary endometrioid/mucinous ovarian tumors
and colorectal cancer metastases to the ovary. In patient 1,
KRAS G13D, APC E1306*, and TP53 H193Y
mutations were detected in both O-1 and L-1, with MET gene
amplification in L-1, which has been reported to be a common gene
mutation pattern in colorectal cancer. In patient 2, APC
R216* was detected in all P-2, O-2, and L-2, which is also
well-known genetic signature of colorectal cancer. These findings
suggest that it might be possible to predict the development of
metastatic ovarian cancer from colorectal cancer based on the
detection of APC gene mutations in liquid biopsy
samples.
The detection of KRAS mutations in liquid
biopsy samples also plays an important role in selecting the
optimal treatment for metastatic ovarian cancer from colorectal
cancer. Anti-EGFR antibodies combined with cytotoxic agents are one
of the standard treatments for advanced colorectal cancer. Although
the addition of anti-EGFR antibodies was found to be related to a
significantly reduced risk of disease progression and improved
overall survival in patients with KRAS wild-type tumors,
this treatment had no benefit in patients whose tumors carried
KRAS gene mutations in codons 12 and 13 (21,22)
Furthermore, less favorable clinical outcomes, particularly shorter
survival, have been reported to be associated with the presence of
KRAS mutations in codon 13 (23). Thus, KRAS mutation status was
confirmed to be a powerful biomarker for predicting the efficacy of
anti-EGFR antibody treatment (24).
In patient 1, a KRAS G13D mutation, which confers resistance
to anti-EGFR antibodies and is associated with greater progression,
was detected in the liquid biopsy sample.
Tumor heterogeneity might also hinder personalized
molecular-targeted treatment, which depends on patients' somatic
mutation profiles. In a previous study, we detected intra- and
inter-tumor multi-clonality during mutational profiling of
multi-regional colon cancer using next-generation sequencing of
FFPE tumor samples (25). The
KRAS-NRAS status of a tumor can vary between regions, making
it difficult to decide whether anti-EGFR antibodies should be used.
Examining tumor heterogeneity is challenging because we can not use
whole surgically resected tumor samples for gene mutation analysis,
and collecting tumor samples can be difficult in advanced cases
involving repeated biopsies. In another previous study, by using
liquid biopsy, we also detected newly appearing KRAS-NRAS
mutation after first-line chemotherapy in metastatic colorectal
cancer with wild-type of KRAS exon 2 in tumor tissue at the
diagnosis, which could be a negative predictive marker for
panitumumab (anti-EGFR antibody) (26). Non-invasive liquid biopsy
examinations, whose results are not influenced by tumor
heterogeneity, might be useful for determining the optimal
chemotherapy regimen, including for novel molecular-targeted
agents, immediately just before the treatment.
In conclusion, this is the first study to examine
the tumor gene mutation profile of metastatic colorectal cancer to
the ovary using both tumor and liquid samples. The difficulty of
obtaining a preoperative diagnosis and tumor heterogeneity can both
influence the effectiveness of molecular-targeted agents, such as
anti-EGFR antibodies. Thus, in the clinical setting liquid biopsy
examinations, which are non-invasive and easy to perform
repeatedly, might be useful. The characterization of the genetic
profiles of tumors based on liquid biopsy examinations might lead
to the development of novel personalized treatment strategies. To
confirm the findings of this study, a further investigation should
be conducted.
Acknowledgements
Not applicable.
Funding
The present study was supported by Ministry of
Education, Culture, Sports, Science and Technology of Japan
Grants-in-Aid for Scientific Research Grant (grant no.
JP18K16776).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
NI, KS, KN and KI designed the research. NI, KS, TN,
TY and ST conducted the experiments. NI and KS analyzed the data.
The manuscript was drafted by KS, KN and KI. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committee of
Wakayama Medical University Faculty of Medicine (authorization
number: 2025) and Kindai University Faculty of Medicine
(authorization number: 29-066). All of the patients in this study
provided written informed consent for the use of their plasma and
tissue samples.
Patient consent for publication
All participants provided written informed consent
for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kubeček O, Laco J, Špaček J, Petera J,
Kopecký J, Kubečková A and Filip S: The pathogenesis, diagnosis,
and management of metastatic tumors to the ovary: A comprehensive
review. Clin Exp Metastasis. 34:295–307. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanna NN and Cohen AM: Ovarian neoplasms
in patients with colorectal cancer: Understanding the role of
prophylactic oophorectomy. Clin Colorectal Cancer. 3:215–222. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan KL, Tan WS, Lim JF and Eu KW:
Krukenberg tumors of colorectal origin: A dismal outcome-experience
of a tertiary center. Int J Colorectal Dis. 25:233–238. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fujiwara A, Noura S, Ohue M, Shingai T,
Yamada T, Miyashiro I, Ohigashi H, Yano M, Ishikawa O, Kamiura S
and Tomita Y: Significance of the resection of ovarian metastasis
from colorectal cancers. J Surg Oncol. 102:582–587. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McCormick CC, Giuntoli RL II, Gardner GJ,
Schulick RD, Judson K, Ronnett BM, Vang R and Bristow RE: The role
of cytoreductive surgery for colon cancer metastatic to the ovary.
Gynecol Oncol. 105:791–795. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rayson D, Bouttell E, Whiston F and Stitt
L: Outcome after ovarian/adnexal metastectomy in metastatic
colorectal carcinoma. J Surg Oncol. 75:186–192. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ojo J, De Silva S, Han E, Lin P,
Wakabayashi M, Nelson R and Lai LL: Krukenberg tumors from
colorectal cancer: presentation, treatment and outcomes. Am Surg.
77:1381–1385. 2011.PubMed/NCBI
|
|
8
|
Willmott F, Allouni KA and Rockall A:
Radiological manifestations of metastasis to the ovary. J Clin
Pathol. 65:585–590. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ganesh K, Shah RH, Vakiani E, Nash GM,
Skottowe HP, Yaeger R, Cercek A, Lincoln A, Tran C, Segal NH, et
al: Clinical and genetic determinants of ovarian metastases from
colorectal cancer. Cancer. 123:1134–1143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Crobach S, Ruano D, van Eijk R, Fleuren
GJ, Minderhout I, Snowdowne R, Tops C, van Wezel T and Morreau H:
Target-enriched next-generation sequencing reveals differences
between primary and secondary ovarian tumors in formalin-fixed,
paraffin-embedded tissue. J Mol Diagn. 17:193–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Misale S, Yaeger R, Hobor S, Scala E,
Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M,
Siravegna G, et al: Emergence of KRAS mutations and acquired
resistance to anti-EGFR therapy in colorectal cancer. Nature.
486:532–536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Murtaza M, Dawson SJ, Tsui DW, Gale D,
Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS,
et al: DNon-invasive analysis of acquired resistance to cancer
therapy by sequencing of plasma DNA. Nature. 497:108–112. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peeters M, Oliner KS, Parker A, Siena S,
Van Cutsem E, Huang J, Humblet Y, Van Laethem JL, André T, Wiezorek
J, et al: Massively parallel tumor multigene sequencing to evaluate
response to panitumumab in a randomized phase III study of
metastatic colorectal cancer. Clin Cancer Res. 19:1902–1912. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang
F, Xu L and Yin R: Circulating tumor DNA is effective for the
detection of EGFR mutation in non-small cell lung cancer: A
meta-analysis. Cancer Epidemiol Biomarkers Prev. 24:206–212. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oxnard GR, Thress KS, Alden RS, Lawrance
R, Paweletz CP, Cantarini M, Yang JC, Barrett JC and Jänne PA:
Association between plasma genotyping and outcomes of treatment
with osimertinib (AZD9291) in advanced non-small-cell lung cancer.
J Clin Oncol. 34:3375–3382. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Newman AM, Bratman SV, To J, Wynne JF,
Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et
al: An ultrasensitive method for quantitating circulating tumor DNA
with broad patient coverage. Nat Med. 20:548–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Newman AM, Lovejoy AF, Klass DM, Kurtz DM,
Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C, et al:
Integrated digital error suppression for improved detection of
circulating tumor DNA. Nat Biotechnol. 34:547–555. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu C, Ranjbar Nezami MR, Wu Z, DiCarlo J
and Wang Y: Detecting very low allele fraction variants using
targeted DNA sequencing and a novel molecular barcode-aware variant
caller. BMC Genomics. 18:52017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Narahara M, Higasa K, Nakamura S, Tabara
Y, Kawaguchi T, Ishii M, Matsubara K, Matsuda F and Yamada R:
Large-scale East-Asian eQTL mapping reveals novel candidate genes
for LD mapping and the genomic landscape of transcriptional effects
of sequence variants. PLoS One. 9:e1009242014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scherer F, Kurtz DM, Newman AM, Stehr H,
Craig AF, Esfahani MS, Lovejoy AF, Chabon JJ, Klass DM, Liu CL, et
al: Distinct biological subtypes and patterns of genome evolution
in lymphoma revealed by circulating tumor DNA. Sci Transl Med.
8:364ra1552016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD,
Robitaille S, et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bokemeyer C, Bondarenko I, Makhson A,
Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G,
Stroh C, et al: Fluorouracil, leucovorin, and oxaliplatin with and
without cetuximab in the first-line treatment of metastatic
colorectal cancer. J Clin Oncol. 27:663–671. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bazan V, Agnese V, Corsale S, Calò V,
Valerio MR, Latteri MA, Vieni S, Grassi N, Cicero G, Dardanoni G,
et al: Specific TP53 and/or Ki-ras mutations as independent
predictors of clinical outcome in sporadic colorectal
adenocarcinomas: Results of a 5-year Gruppo Oncologico dell'Italia
Meridionale (GOIM) prospective study. Ann Oncol. 16 Suppl
4:iv50–iv55. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Van Cutsem E, Köhne CH, Láng I, Folprecht
G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D,
Tejpar S, et al: Cetuximab plus irinotecan, fluorouracil, and
leucovorin as first-line treatment for metastatic colorectal
cancer: Updated analysis of overall survival according to tumor
KRAS and BRAF mutation status. J Clin Oncol. 29:2011–2019. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kogita A, Yoshioka Y, Sakai K, Togashi Y,
Sogabe S, Nakai T, Okuno K and Nishio K: Inter- and intra-tumor
profiling of multi-regional colon cancer and metastasis. Biochem
Biophys Res Commun. 458:52–56. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shitara K, Yonesaka K, Denda T, Yamazaki
K, Moriwaki T, Tsuda M, Takano T, Okuda H, Nishina T, Sakai K, et
al: Randomized study of FOLFIRI plus either panitumumab or
bevacizumab for wild-type KRAS colorectal cancer-WJOG 6210G. Cancer
Sci. 107:1843–1850. 2016. View Article : Google Scholar : PubMed/NCBI
|