Introduction
As one of the most common gynecologic cancers in
China, endometrial carcinoma (EC) has become a major threat to
women's health, with an incidence rate of 8.56/100,000 and
mortality rate of 1.94/100,000 (1).
Endometrial cancer is a complex disease with various risk factors,
the most common being unopposed estrogen exposure and obesity
(2,3).
Genetic factors also play crucial roles in the development of
endometrial cancer. While some of the low-frequency,
high-penetrance mutations in genes such as MLH1, MSH2, EPCAM,
MSH6 or PMS2 contribute to Lynch syndrome, a hereditary
cancer syndrome that increases one's risk of developing endometrial
cancer and colorectal cancer (4),
high-frequency, low-penetrance genetic variants such as single
nucleotide polymorphisms (SNPs) are more often associated with
sporadic endometrial cancers. There are two common methods to
identify disease-related SNPs, genome-wide association studies
(GWAS) (5) and candidate genes
studies (6). The identification of
potentially relevant SNPs could help us further study the
occurrence, progression and prognosis of ECs on a populational
basis.
Epithelial-mesenchymal transition (EMT) is an
important event in tumor cell metastasis, whereby epithelial cells
lose their polarity and cell-cell contacts, and shift to
mesenchymal cells with a more dispersed morphology as well as an
increased motility for migration and invasion (7). Endometrial cancer typically arises from
the glandular epithelium, and relies heavily on the EMT process for
invasion and metastasis (8).
Hallmarks of EMT in EC have been widely reported, such as the
levels of E-cadherin, N-cadherin, β-catenin, matrix
metalloproteinases and several transcription factors (9). Thus, the cadherin 1 (CDH1) gene,
encoding E-cadherin, is an important factor regulating the EMT
process. We therefore hypothesize that SNPs in CDH1 are
possibly associated with EC susceptibility among the Chinese Han
population.
E-cadherin is a type of cell adhesion molecule
within the surface of the epithelium. As a calcium ion-dependent
glycoprotein, E-cadherin contributes to maintaining cell-cell and
cell-matrix adherent junctions (10).
Loss of E-cadherin expression can therefor lead to increased cell
motility, thus imparting the EMT process, which accelerates cell
invasion during tumor progression. In fact, downregulation of
E-cadherin has been found to correlate with EC via CDH1
mutation (11). However, germline
mutations are relatively rare and tend to be associated with
familiar cancers. SNPs on the other hand, being much more common
genetic variants, could serve as better indicators of sporadic
cancers in future genetic screening and disease prediction. So far
in CDH1, SNPs such as rs13689, rs2059254 and rs12919719 have
been found to be associated with breast cancer susceptibility in a
Chinese population (12), while other
SNPs have been associated with endometriosis susceptibility; could
potentially affect the clinical outcome of epithelial ovarian
cancer, etc (13,14). However, no studies have yet been
reported concerning the association studies of CDH1 SNPs
with EC risk among Chinese Han women.
We conducted candidate genes studies, but analyzing
all known SNPs in the target gene would be too costly and
time-consuming. A more common strategy, based on linkage
disequilibrium (LD), is to find haplotype-tagging SNPs (htSNPs),
which by definition are a small subset of SNPs capable of capturing
the full information of haplotypes (15). This strategy can greatly reduce the
expense and scale of the genotyping process, and has been widely
used in population-based association studies. In this study, we
picked out the htSNPs in CDH1 and comprehensively analyzed
the associations between their genetic polymorphisms and EC
susceptibility in a Chinese Han population, followed by genotype
imputation to fine-map more SNPs that may also be relevant.
Functional annotation was also conducted using various
bioinformatic tools to predict the functional characteristics of
potential causal variants. In the end, we demonstrated that several
SNPs in CDH1 may modulate endometrial cancer
susceptibility.
Materials and methods
Study population
This case-control study included 516 cases of
endometrial adenocarcinoma from Peking University Third Hospital,
Beijing Cancer Hospital and Beijing Hospital between 1999 and 2011,
all of whom were Chinese Han women with definite pathological
diagnoses. Patients who had previous histories of cancer,
metastasized cancer originated from other organs, and those who had
been treated with radiotherapy or chemotherapy were excluded from
the study. A total of 706 controls were from Chinese Han women who
participated in a community-based screening program for
non-infectious diseases in Beijing, with no history of cancer. The
case and control groups were age-matched, and epidemiological
information was collected for both groups from clinical records or
questionnaires, including: Age, body mass index (BMI), age at
menarche, age at menopause, age at first full-term pregnancy
(FFTP), number of child birth, smoking history, and family history
of cancer in first-degree relatives. The study was approved by the
Ethics Committee of Peking University Health Science Center.
SNP selection
Haplotype-tagging SNPs were selected according to
the HapMap database (2009-02-06: HapMap Data Release no. 27; CHB
(Chinese Beijing) population) using Haploview v.4.2 software.
Specific methods and selection criteria have been described in
previous research (15–19). For CDH1, we identified 10
htSNPs (rs7200690, rs12185157, rs7198799, rs17715799, rs2011779,
rs10431923, rs7186053, rs6499199, rs4783689 and rs13689) in the
CDH1 locus (2 kb upstream to 2 kb downstream). As rs2011779
was failed to be directly genotyped in our lab, only the other 9
htSNPs remained to be analyzed in the following study.
DNA isolation and genotyping
assay
For the EC cases, genomic DNAs were extracted from
archived formalin-fixed paraffin-embedded (FFPE) blocks of
non-tumor tissues. For the control group, genomic DNAs were
extracted from blood leukocytes. Conventional proteinase K
digestion, phenol-chloroform extraction and ethanol precipitation
were performed to prepare genomic DNA. Genotyping of all htSNPs
were conducted using the ABI 7900HT® Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). TaqMan® assay was performed in
compliance with the manufacturer's instructions. Primer and probes
were supplied by Applied Biosystems and PCR conditions were the
same as described by Ruan et al (20). For quality control, positive and
negative controls were included in each genotyping plate, and 3% of
the samples were repeatedly genotyped, with a concordance rate of
more than 99% between the duplicates.
LD block determination and haplotype
construction
Lewontin coefficient (D') and squared correlation
coefficient (r2) between the genotyped SNPs in
CDH1 (in cases and controls respectively) were calculated
using the Haploview v4.2 software, along with the construction of
LD plots and haplotype blocks. The most probable haplotypes for
each individual were estimated according to
expectation-maximization (EM) algorithm using the SAS 9.1 PROC
HAPLOTYPE procedure (21).
Genotype imputation
Genotype imputation serves as an in-silico method to
predict missing genotypes of variants that are not directly assayed
in existing case-control studies (22,23). Thus,
to find out more variants potentially related to EC risk, we
performed genotype imputation using the MACH software (24), with reference haplotypes in
CDH1 (spanning from 5 kb upstream to 5 kb downstream)
obtained from the CHB population in the 1000 Genomes Project. The
imputation helper module in GenGen software tools (http://gengen.openbioinformatics.org/en/latest/) was
later applied to convert MACH output files to appropriate formats
that could be directly used in subsequent association analyses.
Finally, the well-imputed variants were analyzed for their allelic
associations with EC risk using the PLINK v1.07 software (25).
Statistical analysis
The epidemiological characteristics between cases
and controls were compared using Pearson's χ2 test or
Student's t-test. For each htSNP, Hardy-Weinberg equilibrium was
assessed using one-degree of freedom goodness-of-fit test based on
genotypes of the controls. Two-sided χ2 test was
conducted to compare the distributions of alleles and genotypes
between two groups, with each genotype categorized according to
three models: Codominant, dominant and recessive. Cochran-Armitage
trend test was also conducted in order to predict the effect of
allele dose in each SNP on its association with EC risk. Univariate
and multivariate logistic regression models were carried out to
estimate the effect of genotypes on EC susceptibility through
calculating odd ratios (ORs) and 95% confidence intervals (95%
CIs), including unadjusted models as well as models adjusted for
BMI, age at menarche, menopause status, age at FFTP, number of
birth, and family history of cancer in first-degree relatives.
In order to identify higher-order interactions
associated with EC risk, generalized multifactor dimensionality
reduction (GMDR) method (GMDR software Beta 0.9) was applied to
analyze SNP-SNP and SNP-environment interactions. Originated from
the MDR method, GMDR has several advantages: permitting adjustment
for covariates, being able to handle both dichotomous and
quantitative phenotypes, and applicable to multiple types of
population-based study designs (26).
During the analysis, cross-validation and/or permutation testing
could be applied to evaluate the significance of the models, and
the best candidate model with the maximum testing accuracy and/or
cross-validation consistency (CVC) could be selected (27). In our study, SNP-SNP interactions were
analyzed for 1 to 8-factor models, including 9 htSNPs and 14 newly
imputed SNPs that were found to be associated with EC risk during
genotype imputation analysis. SNP-environment interactions were
also examined for 1-to-8-factor models, including the above 23 SNPs
along with age, BMI, family history of cancer in first-degree
relatives, number of birth. Both analyses were adjusted for
covariates including age, BMI, family history of cancer in
first-degree relatives, age at menarche, menopause status, and
number of birth.
Functional annotation
In order to further predict the potential functional
characteristics of the susceptible SNPs, and to explore the roles
they might play in the development of EC, each variant was being
functionally annotated via publicly available bioinformatic
databases or annotation software. To analyze the correlation
between selected variants and mRNA expression levels of
corresponding genes, expression Quantitative Trait Loci (eQTL)
information was extracted from GTEx Portal (28) (https://gtexportal.org/home/), which could present
eQTL results of certain SNPs in various tissues or cell lines. By
using HaploReg v4.1 (29) (http://www.broadinstitute.org/mammals/haploreg/haploreg.php)
and rSNPBase (30) (http://rsnp.psych.ac.cn/), SNPs were comprehensively
and reliably annotated, focusing on DNase I hypersensitivity,
histone modification, transcriptional factor binding and motif
alteration, as well as eQTL results. UCSC Genome Browser (31) (http://genome.ucsc.edu) was applied to analyze the
potential functions of SNPs (especially transcriptional factor
ChIP-seq data) in mixed EC cells (ECC-1) (32), which may indicate the particular roles
of these variants in endometrial cancer.
Results
Characteristics of the study
population
The demographic and epidemiological characteristics
of the 516 EC cases and 706 cancer-free controls are presented in
Table I. The two groups were
adequately matched in age (P=0.7810). There was no significant
difference in smoking history (P=0.6177) between cases and
controls, but cases had higher BMIs (P<0.0001) than controls.
Moreover, cases more likely had an earlier menarche (P<0.0001),
later menopause (P=0.0002), with a lower proportion of individuals
in postmenopausal status (P<0.0001), and a younger age at first
full-term pregnancy (FFTP) (P=0.0493). In addition, there were also
statistical significant differences in the number of childbirth
(P<0.0001). The above variables with significant difference were
taken into account in the subsequent multivariate logistic
regression models to adjust for any possible confounding bias.
 | Table I.Characteristics of endometrial cancer
patients and cancer-free controls. |
Table I.
Characteristics of endometrial cancer
patients and cancer-free controls.
| Variables | Case, n=516 | Control, n=706 | P-value |
|---|
| Age, years (mean ±
SD) | 56.09±10.47 | 55.95±6.56 | 0.7810 |
| Age, years, n
(%) |
|
| 0.1327 |
|
<55 | 229 (44.38) | 344 (48.73) |
|
|
≥55 | 287 (55.62) | 362 (51.27) |
|
| BMI (mean ±
SD) | 25.85±4.11 | 24.92±3.17 |
<0.0001a |
| Age at menarche,
years (mean ± SD) | 14.76±1.84 | 15.58±1.94 |
<0.0001a |
| Age at menopause,
years (mean ± SD) | 50.51±3.52 | 49.61±3.53 | 0.0002a |
| Age at FFTP, years
(mean ± SD) | 25.17±3.36 | 25.54±2.95 | 0.0493a |
| Menopause status, n
(%) |
|
|
<0.0001a |
|
Premenopause | 185 (35.85) | 122 (17.28) |
|
|
Postmenopause | 331 (64.15) | 584 (82.72) |
|
| No. of childbirth,
n (%) |
|
|
<0.0001a |
| 0 | 60 (11.63) | 8 (1.13) |
|
| 1 | 186 (36.05) | 364 (51.56) |
|
| ≥2 | 270 (52.33) | 334 (47.31) |
|
| Family history of
cancer in first-degree relatives, n (%) |
|
| 0.0451a |
|
Yes | 82 (15.89) | 144 (20.40) |
|
| No | 434 (84.11) | 562 (79.60) |
|
| Smoking history, n
(%) |
|
| 0.6177 |
|
Yes | 19 (3.68) | 30 (4.25) |
|
| No | 497 (96.32) | 676 (95.75) |
|
Associations between htSNP genotypes
and BC susceptibility
All 9 htSNPs were in conformation with
Hardy-Weinburg Equilibrium (P>0.05) in the control group (data
not shown). Allele and genotype distributions of the 9 htSNPs are
shown in Table II. Two-sided
χ2 test revealed significant differences in allele
frequencies between cases and controls for rs17715799, rs10431923,
rs6499199, rs4783689 and rs13689. Both univariate and multivariate
unconditional logistic regression analyses indicated that the
genotypes of 6 htSNPs were related with endometrial cancer
susceptibility. The genotypes of SNPs rs17715799 (A>T),
rs6499199 (C>T) and rs13689 (T>C) were significantly
associated with increased endometrial cancer susceptibility
(Table I). On the other hand, the
genotypes of SNPs rs12185157 (A>G), rs10431923 (T>G) and
rs4783689 (C>T) were significantly associated with decreased EC
susceptibility (Table II).
 | Table II.Genotype and allele frequencies of
the htSNPs in CDH1 and their associations with EC
susceptibility. |
Table II.
Genotype and allele frequencies of
the htSNPs in CDH1 and their associations with EC
susceptibility.
| SNPs | Genotype | Cases (%) | Controls (%) |
P-valuea |
P-valueb | Ptrend | OR (95% CI) | P-value | OR (95%
CI)c |
P-valuec |
|---|
| rs7200690 | CC | 344 | 435 | 0.1924 |
| 0.0862 | Reference |
| Reference |
|
|
|
| (66.67) | (61.61) |
|
|
|
|
|
|
|
|
| CT | 147 | 231 |
|
|
| 0.81 | 0.0891 | 0.83 | 0.1707 |
|
|
| (28.49) | (32.72) |
|
|
| (0.63–1.03) |
| (0.63–1.09) |
|
|
| TT | 25 | 40 |
|
|
| 0.79 | 0.3745 | 0.79 | 0.4322 |
|
|
| (4.84) | (5.67) |
|
|
| (0.47–1.33) |
| (0.44–1.42) |
|
|
| T allele | 197 | 311 |
| 0.0772 |
|
|
|
|
|
|
| frequency | (19.09) | (22.03) |
|
|
|
|
|
|
|
|
| CT/TT vs. |
|
|
|
|
| 0.80 | 0.0698 | 0.82 | 0.1373 |
|
| CC (D) |
|
|
|
|
| (0.63–1.02) |
| (0.64–1.06) |
|
|
| TT vs. CT/ |
|
|
|
|
| 0.85 | 0.5281 | 0.84 | 0.5546 |
|
| CC (R) |
|
|
|
|
| (0.51–1.42) |
| (0.47–1.50) |
|
| rs12185157 | AA | 150 | 185 | 0.1495 |
| 0.0433d | Reference |
| Reference |
|
|
|
| (29.07) | (26.20) |
|
|
|
|
|
|
|
|
| AG | 257 | 339 |
|
|
| 0.92 | 0.5444 | 0.91 | 0.5267 |
|
|
| (49.81) | (48.02) |
|
|
| (0.70–1.21) |
| (0.68–1.22) |
|
|
| GG | 109 | 182 |
|
|
| 0.72 | 0.0401d | 0.67 | 0.0209d |
|
|
| (21.12) | (25.78) |
|
|
| (0.52–0.99) |
| (0.47–0.94) |
|
|
| G allele | 475 | 703 |
| 0.0661 |
|
|
|
|
|
|
| frequency | (46.03) | (49.79) |
|
|
|
|
|
|
|
|
| AG/GG vs. |
|
|
|
|
| 0.85 | 0.2017 | 0.82 | 0.162 |
|
| AA (D) |
|
|
|
|
| (0.66–1.09) |
| (0.63–1.08) |
|
|
| GG vs. |
|
|
|
|
| 0.75 | 0.0405d | 0.71 | 0.0192d |
|
| AG/AA (R) |
|
|
|
|
| (0.58–0.99) |
| (0.53–0.95) |
|
| rs7198799 | CC | 368 | 508 | 0.9659 |
| 0.8385 | Reference |
| Reference |
|
|
|
| (71.32) | (71.95) |
|
|
|
|
|
|
|
|
| CT | 135 | 180 |
|
|
| 1.04 | 0.7936 | 1.13 | 0.3898 |
|
|
| (26.16) | (25.50) |
|
|
| (0.80–1.34) |
| (0.85–1.50) |
|
|
| TT | 13 | 18 |
|
|
| 1.00 | 0.9935 | 0.94 | 0.8791 |
|
|
| (2.52) | (2.55) |
|
|
| (0.48–2.06) |
| (0.42–2.12) |
|
|
| T allele | 161 | 216 |
| 0.8375 |
|
|
|
|
|
|
| frequency | (15.60) | (15.30) |
|
|
|
|
|
|
|
|
| CT/TT vs. |
|
|
|
|
| 1.03 | 0.807 | 1.11 | 0.438 |
|
| CC (D) |
|
|
|
|
| (0.80–1.33) |
| (0.85–1.46) |
|
|
| TT vs. CT/ |
|
|
|
|
| 0.99 | 0.9736 | 0.91 | 0.8175 |
|
| CC (R) |
|
|
|
|
| (0.48–2.04) |
| (0.40–2.05) |
|
| rs17715799 | AA | 300 | 457 | 0.0074d |
| 0.0031d | Reference |
| Reference |
|
|
|
| (58.14) | (64.73) |
|
|
|
|
|
|
|
|
| AT | 172 | 216 |
|
|
| 1.21 | 0.1264 | 1.25 | 0.0993 |
|
|
| (33.33) | (30.59) |
|
|
| (0.95–1.55) |
| (0.96–1.63) |
|
|
| TT | 44 | 33 |
|
|
| 2.03 | 0.0034d | 2.06 | 0.005d |
|
|
| (8.53) | (4.67) |
|
|
| (1.26–3.26) |
| (1.24–3.41) |
|
|
| T allele | 260 | 282 |
| 0.0021d |
|
|
|
|
|
|
| frequency | (25.19) | (19.97) |
|
|
|
|
|
|
|
|
| AT/TT vs. |
|
|
|
|
| 1.32 | 0.0192d | 1.36 | 0.0163d |
|
| AA (D) |
|
|
|
|
| (1.05–1.67) |
| (1.06–1.75) |
|
|
| TT vs. |
|
|
|
|
| 1.90 | 0.0069d | 1.91 | 0.0107d |
|
| AT/AA (R) |
|
|
|
|
| (1.19–3.03) |
| (1.16–3.13) |
|
| rs10431923 | TT | 217 | 214 | 0.0001d |
|
<0.0001d | Reference |
| Reference |
|
|
|
| (42.05) | (30.31) |
|
|
|
|
|
|
|
|
| GT | 217 | 348 |
|
|
| 0.41 |
<0.0001d | 0.42 |
<0.0001d |
|
|
| (42.05) | (49.29) |
|
|
| (0.32–0.54) |
| (0.31–0.56) |
|
|
| GG | 82 | 144 |
|
|
| 0.25 |
<0.0001d | 0.24 |
<0.0001d |
|
|
| (15.89) | (20.40) |
|
|
| (0.18–0.35) |
| (0.17–0.35) |
|
|
| G allele | 381 | 636 |
|
<0.0001d |
|
|
|
|
|
|
| frequency | (36.92) | (45.04) |
|
|
|
|
|
|
|
|
| GT/GG vs. |
|
|
|
|
| 0.35 |
<0.0001d | 0.35 |
<0.0001d |
|
| TT (D) |
|
|
|
|
| (0.27–0.46) |
| (0.27–0.46) |
|
|
| GG vs. |
|
|
|
|
| 0.43 |
<0.0001d | 0.41 |
<0.0001d |
|
| GT/TT (R) |
|
|
|
|
| (0.33–0.58) |
| (0.30–0.56) |
|
| rs7186053 | GG | 261 | 332 | 0.4634 |
| 0.236 | Reference |
| Reference |
|
|
|
| (50.58) | (47.03) |
|
|
|
|
|
|
|
|
| AG | 210 | 306 |
|
|
| 0.87 | 0.2653 | 0.90 | 0.416 |
|
|
| (40.70) | (43.34) |
|
|
| (0.69–1.11) |
| (0.70–1.16) |
|
|
| AA | 45 | 68 |
|
|
| 0.84 | 0.4104 | 0.77 | 0.255 |
|
|
| (8.72) | (9.63) |
|
|
| (0.56–1.27) |
| (0.49–1.21) |
|
|
| A allele | 300 | 442 |
| 0.2356 |
|
|
|
|
|
|
| frequency | (29.07) | (31.30) |
|
|
|
|
|
|
|
|
| AG/AA vs. |
|
|
|
|
| 0.87 | 0.2194 | 0.88 | 0.286 |
|
| GG (D) |
|
|
|
|
| (0.69–1.09) |
| (0.69–1.12) |
|
|
| AA vs. |
|
|
|
|
| 0.90 | 0.5874 | 0.81 | 0.3396 |
|
| AG/GG (R) |
|
|
|
|
| (0.60–1.33) |
| (0.53–1.25) |
|
| rs6499199 | CC | 367 | 535 | <0.0001 |
| 0.001 | Reference |
| Reference |
|
|
|
| (71.12) | (75.78) |
|
|
|
|
|
|
|
|
| CT | 112 | 160 |
|
|
| 1.02 | 0.8856 | 0.96 | 0.8008 |
|
|
| (21.71) | (22.66) |
|
|
| (0.77–1.34) |
| (0.72–1.29) |
|
|
| TT | 37 | 11 |
|
|
| 4.90 | <0.0001 | 4.50 | <0.0001 |
|
|
| (7.17) | (1.56) |
|
|
| (2.47–9.74) |
| (2.21–9.18) |
|
|
| T allele | 186 | 182 |
| 0.0005 |
|
|
|
|
|
|
| frequency | (18.02) | (12.89) |
|
|
|
|
|
|
|
|
| CT/TT vs. |
|
|
|
|
| 1.27 | 0.0678 | 1.20 | 0.197 |
|
| CC (D) |
|
|
|
|
| (0.98–1.64) |
| (0.91–1.58) |
|
|
| TT vs. |
|
|
|
|
| 4.88 | <0.0001 | 4.54 | <0.0001 |
|
| CT/CC (R) |
|
|
|
|
| (2.47–9.66) |
| (2.23–9.23) |
|
| rs4783689 | CC | 283 | 321 | 0.0041 |
| 0.0012 | Reference |
| Reference |
|
|
|
| (54.84) | (45.47) |
|
|
|
|
|
|
|
|
| CT | 189 | 303 |
|
|
| 0.71 | 0.0051 | 0.72 | 0.0117 |
|
|
| (36.63) | (42.92) |
|
|
| (0.56–0.90) |
| (0.55–0.93) |
|
|
| TT | 44 | 82 |
|
|
| 0.61 | 0.0149 | 0.63 | 0.0329 |
|
|
| (8.53) | (11.61) |
|
|
| (0.41–0.91) |
| (0.41–0.96) |
|
|
| T allele | 277 | 467 |
| 0.0009 |
|
|
|
|
|
|
| frequency | (26.84) | (33.07) |
|
|
|
|
|
|
|
|
| CT/TT vs. |
|
|
|
|
| 0.69 | 0.0012 | 0.70 | 0.0038 |
|
| CC (D) |
|
|
|
|
| (0.55–0.86) |
| (0.55–0.89) |
|
|
| TT vs. |
|
|
|
|
| 0.71 | 0.0807 | 0.73 | 0.1292 |
|
| CT/CC (R) |
|
|
|
|
| (0.48–1.04) |
| (0.48–1.10) |
|
| rs13689 | TT | 247 | 467 | <0.0001 |
| <0.0001 | Reference |
| Reference |
|
|
|
| (47.87) | (66.15) |
|
|
|
|
|
|
|
|
| CT | 207 | 218 |
|
|
| 1.80 | <0.0001 | 1.71 | <0.0001 |
|
|
| (40.12) | (30.88) |
|
|
| (1.41–2.29) |
| (1.32–2.27) |
|
|
| CC | 62 | 21 |
|
|
| 5.58 | <0.0001 | 5.16 | <0.0001 |
|
|
| (12.02) | (2.97) |
|
|
| (3.32–9.37) |
| (2.97–8.98) |
|
|
| C allele | 331 | 260 |
| <0.0001 |
|
|
|
|
|
|
| frequency | (32.07) | (18.41) |
|
|
|
|
|
|
|
|
| CT/CC vs. |
|
|
|
|
| 2.13 | <0.0001 | 2.01 | <0.0001 |
|
| TT (D) |
|
|
|
|
| (1.69–2.69) |
| (1.57–2.58) |
|
|
| CC vs. |
|
|
|
|
| 4.46 | <0.0001 | 4.20 | <0.0001 |
|
| CT/TT (R) |
|
|
|
|
| (2.68–7.41) |
| (2.44–7.23) |
|
Fine-scale genetic mapping of CDH1 by
genotype imputation
Based on the 1000 Genomes dataset (Chinese Han
Beijing population), there are 381 SNPs with a minor allele
frequency (MAF) >1% in CDH1 gene. To identify more SNPs
potentially related to EC risk, we performed genotype imputation
using the MACH software. By using our directly genotyped data of
the 9 htSNPs as well as reference haplotypes of CDH1
obtained from the 1000 Genomes Project (CHB population), the
genotypes of 96 SNPs were well-imputed in cases and controls. Using
allelic association tests in PLINK software, the minor alleles of
19 SNPs in CDH1 were identified to be significantly
associated with endometrial cancer susceptibility (P<0.05;
Table III), including htSNPs
rs17715799, rs10431923, rs6499199, rs4783689 and rs13689, which are
consistent with our above htSNP analysis (Table II).
 | Table III.Well-imputed SNPs associated with EC
susceptibility in CDH1 (P<0.05) by genotype imputation. |
Table III.
Well-imputed SNPs associated with EC
susceptibility in CDH1 (P<0.05) by genotype imputation.
| SNP | Position |
P-valuea | OR (95% CI) | Rsqb |
|---|
| rs12599393 | Chr16:
68829021 | 0.02208 | 1.26
(1.03–1.54) | 0.9573 |
| rs6499197 | Chr16:
68830473 | 0.01723 | 1.28
(1.05–1.58) | 0.8524 |
|
rs17715799c | Chr16:
68830511 | 0.002255 | 1.35
(1.11–1.64) | 0.9853 |
| rs8063605 | Chr16:
68836665 | 0.03434 | 0.64
(0.43–0.97) | 0.7150 |
|
rs10431923c | Chr16:
68839263 | 0.003279 | 0.78
(0.66–0.92) | 0.9816 |
| rs10431924 | Chr16:
68839302 | 0.006005 | 0.78
(0.66–0.93) | 0.9296 |
|
rs6499199c | Chr16:
68849837 | 0.0005845 | 1.48
(1.18–1.85) | 0.9903 |
| rs8057342 | Chr16:
68849904 | 0.0007468 | 1.47
(1.17–1.84) | 0.9753 |
| rs34022452 | Chr16:
68850384 | 0.0007224 | 1.48
(1.18–1.85) | 0.9606 |
| rs36029373 | Chr16:
68850406 | 0.004227 | 1.40
(1.11–1.76) | 0.9114 |
| rs8050039 | Chr16:
68852074 | 0.03827 | 1.42
(1.02–1.97) | 0.7898 |
| rs138957735 | Chr16:
68852748 | 0.02279 | 1.48
(1.05–2.07) | 0.7578 |
|
rs4783689c | Chr16:
68853671 | 0.0009422 | 0.74
(0.62–0.89) | 0.9961 |
| rs76685922 | Chr16:
68854135 | 0.009579 | 1.95
(1.17–3.26) | 0.7033 |
| rs34635465 | Chr16:
68854703 | 0.001848 | 2.33
(1.35–4.01) | 0.6781 |
| rs10500544 | Chr16:
68855064 | 0.0102 | 2.63
(1.22–5.66) | 0.6625 |
| rs1801026 | Chr16:
68867456 |
6.71×10−15 | 2.09
(1.73–2.52) | 0.9622 |
|
rs13689c | Chr16:
68868522 |
6.71×10−15 | 2.09
(1.73–2.52) | 0.9945 |
| rs17690554 | Chr16:
68869510 |
6.71×10−15 | 2.09
(1.73–2.52) | 0.9425 |
Multivariate logistic analysis to
identify independently associated SNPs
In order to seek out the SNPs independently
associated with EC susceptibility, we performed multiple logistic
regression analysis, which included the 6 susceptible htSNPs and 14
newly imputed SNPs that affected EC risk during genotype imputation
analysis. However, only 9 SNPs remained in the multiple logistic
regression model, including the 6 susceptible htSNPs (rs12185157,
rs17715799, rs10431923, 6499199, rs4783689, rs13689) and 3
susceptible imputed-SNPs (rs6499197, rs10431924, rs1801026), while
the other 11 susceptible imputed-SNPs dropped out of the model due
to their linear relationships with the other SNPs. We then narrowed
our analysis down on the 9 SNPs to further look for SNPs that
affected EC susceptibility independently. After adjusting for the
other SNPs and confounding factors, rs13689 became much more
significant in increasing EC risk (aOR=2.87, 95% CI=1.53–5.37,
P=0.0010), and rs10431923 as well as rs10431924 became much more
significant in reducing EC risk (rs10431923: aOR=0.04, 95%
CI=0.02–0.10, P<0.0001; rs10431924: aOR=0.14, 95% CI=0.07–0.26,
P<0.0001), whereas the statistical significance for the other
SNPs disappeared.
Association between high-order
interactions and endometrial cancer risk by GMDR analysis
GMDR is a nonparametric and genetic model-free
substitute for linear or logistic regression, serving to capture
and depict nonlinear interactions among genetic and environmental
factors (33). The interactions
between SNP-SNP and SNP-environment are analyzed by GMDR and
presented in Table IV. For SNP-SNP
interaction, we analyzed all 9 htSNPs and 14 imputed susceptible
SNPs for 1-to-8-factors models. After adjusting for covariates, it
was indicated that the best one-factor model for predicting EC risk
was rs10431923. It carried a testing balanced accuracy of 0.6088
and CVC of 10/10, suggesting that this htSNP may be the primary
factor contributing to EC risk among the total 23 SNPs. Among
multi-factor models, the one with the highest cross-validation
consistency (CVC) was a 7-factor model harboring rs7200690,
rs12185157, rs10431923, rs7186053, rs13689, rs6499197 and
rs10431924 (testing balanced accuracy=0.8372, CVC=10/10), showing
strong synergetic interactions among the 7 SNPs.
 | Table IV.Comparison of the models identified
by GMDR for SNP-SNP and SNP-environment interactions. |
Table IV.
Comparison of the models identified
by GMDR for SNP-SNP and SNP-environment interactions.
| Best
modelsa | Training balanced
accuracyb | Testing balanced
accuracyb | Sign test
(P)b | Cross-validation
consistencyb |
|---|
| SNP-SNP |
|
|
|
|
| X5 | 0.6083 | 0.6088 | 0.0107 | 10/10 |
| X5
X13 | 0.7688 | 0.7698 | 0.0010 | 10/10 |
| X5 X9
X13 | 0.7959 | 0.7887 | 0.0010 | 10/10 |
| X2 X5
X9 X13 | 0.8188 | 0.8020 | 0.0010 | 10/10 |
| X1 X2
X5 X9 X13 | 0.8412 | 0.8182 | 0.0010 | 10/10 |
| X1 X2
X5 X9 X11 X13 | 0.8599 | 0.8147 | 0.0010 | 6/10 |
| X1 X2
X5 X6 X9 X11 X13c | 0.8788 | 0.8372 | 0.0010 | 10/10 |
| X1 X2
X5 X6 X7 X8 X9 X11 | 0.8927 | 0.8159 | 0.0010 | 10/10 |
|
SNP-environment |
|
|
|
|
| X5 | 0.6083 | 0.6088 | 0.0107 | 10/10 |
| X5
X13 | 0.7688 | 0.7698 | 0.0010 | 10/10 |
| X5 X9
X13 | 0.7964 | 0.7796 | 0.0010 | 8/10 |
| X5 X9
X13 Nbirth | 0.8230 | 0.8077 | 0.0010 | 9/10 |
| X2 X5
X9 X13 Nbirth | 0.8447 | 0.8152 | 0.0010 | 8/10 |
| X1 X2
X5 X9 X13 Nbirthc | 0.8685 | 0.8299 | 0.0010 | 10/10 |
| X1 X2
X5 X9 X11 X13 Nbirth | 0.8851 | 0.8201 | 0.0010 | 5/10 |
| X1 X2
X5 X6 X9 X11 X13 Nbirth | 0.9010 | 0.8061 | 0.0010 | 5/10 |
As for SNP-environment interaction, after adjusting
for covariates, the best six-factor model (including rs7200690,
rs12185157, rs10431923, rs13689, rs10431924 and number of
childbirth) came to our notice as the model with the highest CVC
(10/10) and highest testing balanced accuracy (0.8299; Table IV). Together with two other
multi-factor models which also included number of childbirth as a
factor, it suggested that in accordance with our knowledge that
endometrial cancer is the comprehensive result of both genetic and
environmental factors. Genetic variants aside, an individual's
number of birth also serves as an important epidemiological factor
affecting her chance of developing endometrial cancer.
Functional annotation
According to our aforementioned results, 5 htSNPs,
namely rs12185157, rs10431923, rs6499199, rs4783689 and rs13689,
were found to be associated with EC susceptibility both in single
SNP association analysis and GMDR models. Among the 14 newly
imputed SNPs associated with EC risk, rs6499197 and rs10431924 were
considered to be relatively important since they were included in
some GMDR best models. To identify the possible effects of these 7
SNPs on relevant gene expression, expression Quantitative Trait
Loci (eQTL) information of the SNPs was extracted from GTEx Portal
and HaploReg, and their summarization can be seen in Table V. Among the 7 SNPs, 6 were found to be
associated with the RNA expression level of CDH1 or other
nearby genes in various tissues or cell lines (P<0.05). Notably,
individuals with minor allele homozygotes of one important
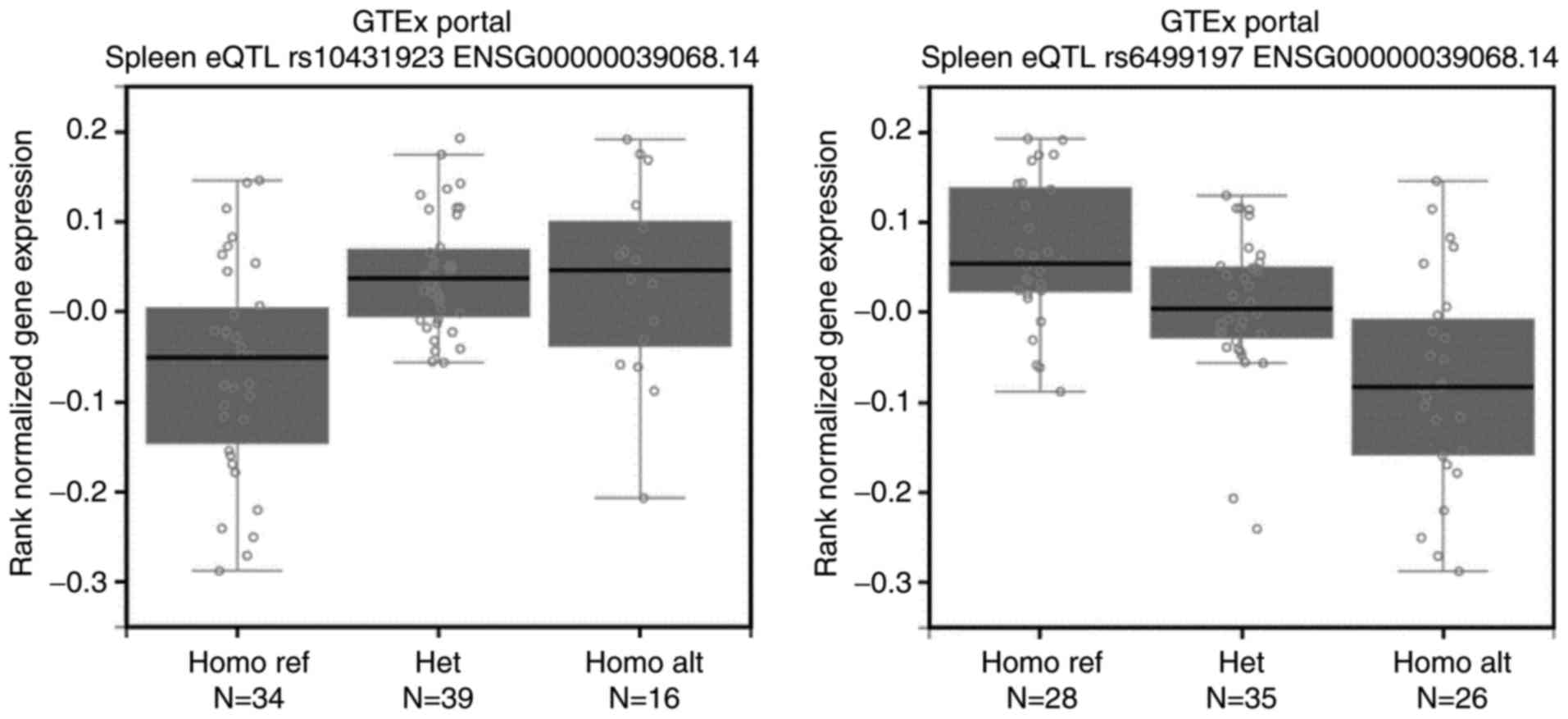
protective locus, rs10431923 (T>G), presented a higher mRNA
level of CDH1 than other genotype carriers
(P=1.73×10−6, Fig. 1).
Likewise, people carrying minor allele homozygotes of one newly
imputed risk locus, rs6499197 (A>G), showed a lower mRNA level
of CDH1 than other genotype carriers
(P=1.49×10−8; Fig. 1).
These results indicate that the above 6 SNP may influence
endometrial cancer susceptibility through regulating gene
expression, and thus may carry potential biological functions.
 | Table V.Functional annotation of selected
susceptible SNPs. |
Table V.
Functional annotation of selected
susceptible SNPs.
|
|
|
|
| eQTLc information |
|---|
|
|
|
|
|
|
|---|
| SNP | Type of
SNPa | MAF | TF binding in EC
cellsb | Tissue | Correlated
gene | P-value |
|---|
| rs12185157 | htSNP | 0.46 | CREB1, FOXM1,
p300 | EBV-transformed
lymphocytes | CDH1 |
6.99×10−6 |
| rs10431923 | htSNP | 0.41 | CEBPG, Egr-1, Max,
p300, TAF1, YY1, ZBTB7A | Spleen | CDH1 |
1.73×10−6 |
| rs4783689 | htSNP | 0.31 | CREB1, Egr-1, Max,
RAD21, TAF1 | Esophagus
Mucosa | FTLP14 |
2.87×10−14 |
| rs6499199 | htSNP | 0.17 | FOXM1, SRF,
TEAD4 | Testis | CTD-2033A16.2 |
1.92×10−5 |
| rs13689 | htSNP | 0.15 | CREB1, YY1, SRF,
USF-1 | – | – | – |
| rs6499197 | Imputed SNP | 0.45 | CEBPB, Egr-1,
FOXM1, Max, TAF1, TCF12, ZBTB7A | Spleen | CDH1 |
1.49×10−8 |
| rs10431924 | Imputed SNP | 0.42 | Egr-1, Max, NFIC,
p300, SRF, TEAD-4, USF-1, YY1, ZBTB7A | Spleen | CDH1 |
5.3×10−5 |
To further predict the potential functions of these
SNPs, multiple publicly available bioinformatic databases or
annotation software were used. The majority of the SNPs could
affect gene regulatory elements like DNase I hypersensitivity,
histone modifications including methylation and acetylation, as
well as transcriptional factor binding sites. In the EC cell line
ECC-1, which was identified as mixed EC cells (32), all selected SNPs were found to locate
in transcriptional factor binding regions (Table V). The functional annotations suggest
that these SNPs may affect its binding with transcriptional
factors, and affect gene's expression.
Discussion
To our knowledge, this is the first gene-wide
association study of CDH1 to comprehensively illustrate the
relation between SNPs and endometrial cancer susceptibility in
Chinese Han women. We first identified 6 single htSNPs (rs12185157,
rs10431923, rs4783689, rs17715799, rs6499199 and rs13689) in
CDH1 as susceptible SNPs associated with EC risk, followed
by genotype imputation analysis with fine-mapping of the
high-density SNPs within CDH1's target region, where 14 SNPs
were also identified as possibly associated with EC risk. Among all
susceptible SNPs, CDH1 rs10431923, rs10431924 and rs13689
remained to affect EC susceptibility after multiple logistic
regression analyses. To identify higher-order interactions
associated with EC risk, GMDR analyses were conducted, showing that
7 htSNPs and 2 newly imputed SNPs may be involved in SNP-SNP
interaction or SNP-environment interaction in endometrial cancer.
Subsequent functional annotations for these susceptible SNPs were
carried out, suggesting their considerable biological functions
underlying the associations that await future studies.
Our study results show that rs10431923 (G>T) was
the single most important independent protective factor for
endometrial cancer susceptibility in Chinese Han Women. It yielded
an aOR of 0.35 (dominant model) in the logistic regression, and
still served as a strong protective factor after adjusting for the
other susceptible SNPs in the multiple logistic regression.
Moreover, in our GMDR analysis, rs10431923 (G>T) was shown to be
the best one-factor model for predicting EC susceptibility, and was
involved in almost all other best fitting multi-factor models.
Consistent to our findings of a protective effect with GG genotype,
rs10431923 (T>G) was previously found to be related to Crohn's
disease in a North American population, with its TT genotype
related to abnormal aggregation of E-cadherin in epithelial cells
resulting in its impaired plasma membrane localization (34). Another study found no association
between rs10431923 with colorectal cancer risk (35), but due to small sample size and
Italian population, their results might not be generalizable to
Chinese Han Women. rs10431923 also showed considerable functional
potentials, where it may alter the mRNA expression of CDH1,
and was located in TF-binding regions (such as Egr-1, Max, p300,
YY1) with significant binding signals in the endometrial cancer
cells. This result indicates that the variation of rs10431923 may
influence the expression of CDH1 through modifications of
the transcription process, thus affecting E-cadherin levels and the
EMT process in endometrial cancer. Therefore, rs10431923 was
considered the most significant protective locus of EC
susceptibility, and its biological functions in the occurrence and
progression of EC call for intensive future studies.
Other protective htSNPs in CDH1, such as
rs12185157 (G>A) and rs4783689 (C>T), were also significantly
associated with EC risk in various statistical models. In
accordance with previous discoveries, rs12185157 was a part of a
three-SNPs diplotype associated with breast cancer susceptibility
in Chinese Han women (16). It was
also involved in 6 best-fitting gene-gene models and 5 best-fitting
gene-environment models in our GMDR analysis, suggesting its
interaction with various susceptible SNPs and environmental risk
factors for EC. The other protective htSNP, rs4783689, was found to
be associated with a higher risk of endometriosis for C allele
carriers in a Japanese population (36). No existing studies have yet revealed
the associations between these SNPs and endometrial cancer
susceptibility. Functional annotations also suggested their
functional potentials, with rs12185157 located in histone
modifications regions in cervical carcinoma cells, and was able to
change the TF binding motif USF2. It was also located in the
TF-binding site of ZEB1, a key activator for the EMT process and
metastasis (37), suggesting its
potential role in regulating CDH1 expression in endometrial
cancer.
Several risk htSNPs were also discovered to be
associated with increased EC risk, among which rs13689 (T>C)
stood out as the most important risk SNP. With an aOR of 4.20
(recessive model) and an aOR of 2.87 after adjusting for the other
susceptible SNPs, rs13689 was considered a strong independent risk
locus for EC risk. It was presented in almost all the best fitting
multi-factor models in our GMDR analyses, indicating its vital role
in the interaction among SNPs and (or) environmental factors.
rs13689 was previously identified as a significant risk factor for
breast cancer susceptibility in Chinese Han Population (16), echoing our findings in EC. Functional
annotation indicated its location in the 3′-UTR of CDH1,
suggesting potentials in stabilizing mRNA through miRNAs. It also
showed functional potential in regulating some TF factors (CREB1,
YY1, SRF, USF-1) in the ECC-1 cell line, and could possibly
interact with other genes like ZFP90 and TANGO6 via chromatin
loops.
To expand our existing findings, fine-mapped
genotype imputation analysis within CDH1's target region was
conducted, where 14 newly imputed SNPs were identified to be
significantly associated with endometrial cancer susceptibility.
Among them, 2 imputed SNPs in CDH1 (rs6499197 and
rs10431924) were involved in several best fitting models in our
GMDR analyses, suggesting their roles in the interaction with other
susceptible htSNPs. Functional annotations also revealed the
potential biological functions of these SNPs in DNase I
hypersensitivity, histone modifications, TF binding (especially
with FOXM1, which is involved in inducing EMT and metastasis
(38), and 3D interactions. Two other
imputed SNPs (rs1801026 and rs17690554) were previously found to be
respectively associated with the susceptibility of gastric cardiac
adenocarcinoma, non-small-cell lung cancer, cervical cancer, breast
cancer prognosis, or gastric cancer (39–41), while
none of these studies were focused on EC. The rest of the imputed
SNPs aforementioned have not been studied in any publications yet,
therefore warranting future studies.
Our study has three main strengths. To begin with,
this is the first comprehensive gene-wide association study of
CDH1 with EC risk in Chinese Han population. By using
haplotype-tagging SNPs plus fine-mapped genotype imputation, we
nearly covered all common SNPs of CDH1. Secondly, after
identifying independent susceptible SNPs, we further analyzed
SNP-SNP and SNP-environment interactions to identify the joint
effect of SNPs and environmental risk factors on EC development.
Thirdly, functional annotations using various databases revealed
the potential biological functions of the causal SNPs, giving
possible directions for future research. This study inevitably has
limitations. Due to the small sample size of certain subgroups, we
had to merge several groups or leave out some rare subgroups to
increase efficiency, though we did use various statistical methods
to minimalize false positives. Also, to improve statistical power,
we extracted existing eQTL results in multiform races from GTEx
Portal instead of using the Han Chinese in Beijing (CHB)
population, mainly due to small sample size of the available
unrelated CHB population from the HapMap project.
In summary, this study suggests that the genetic
polymorphisms of CDH1 were associated with endometrial
cancer susceptibility. Our data found susceptible loci that were
independently associated with EC risk, as well as conjoint effects
among themselves and with environmental factors. Furthermore,
several SNPs might carry potential functions regulating CDH1
expression, and additional studies are needed to verify and
identify the truly causal SNPs. If further supportive studies are
validated, these findings may serve to improve personalized
evaluation and early prediction of EC susceptibility in the general
population.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant nos. 81672790 and 81621063).
Availability of data and materials
The datasets used and analyzed for the present study
are available from the corresponding author upon reasonable
request.
Authors' contributions
YHG, XXT, DGL, XHL and WGF conceived and designed
the experiments. YHG, YMJ, ZFW, LYZ and LC performed the
experiments. ZFW and YHG analyzed and interpreted the data, and
wrote the manuscript. XXT, WGF, ZFW and YHG revised the paper. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Peking University IRB
(reference no. IRB00001052-11029). Written informed consents were
obtained from all women in the control group. Genomic DNAs of the
patients were extracted from archived formalin-fixed
paraffin-embedded normal fallopian tube tissues. Since the contact
information of patients treated before 2011 was not obtainable,
Peking University IRB approved our application to waive informed
consent for the archived samples collected before April 2011. This
study only used these samples. All the data/samples were used
anonymously.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Amant F, Moerman P, Neven P, Timmerman D,
Van Limbergen E and Vergote I: Endometrial cancer. Lancet.
366:491–505. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schouten LJ, Goldbohm RA and van den
Brandt PA: Anthropometry, physical activity, and endometrial cancer
risk: Results from the Netherlands cohort study. J Natl Cancer
Inst. 96:1635–1638. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Giardiello FM, Allen JI, Axilbund JE,
Boland CR, Burke CA, Burt RW, Church JM, Dominitz JA, Johnson DA,
Kaltenbach T, et al: Guidelines on genetic evaluation and
management of Lynch syndrome: A consensus statement by the US
Multi-society task force on colorectal cancer. Am J Gastroenterol.
109:1159–1179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tomlinson IP, Webb E, Carvajal-Carmona L,
Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A,
Sullivan K, et al: A genome-wide association study identifies
colorectal cancer susceptibility loci on chromosomes 10p14 and
8q23.3. Nat Genet. 40:623–630. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong LM, Potter JD, White E, Ulrich CM,
Cardon LR and Peters U: Genetic susceptibility to cancer: The role
of polymorphisms in candidate genes. JAMA. 299:2423–2436. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mirantes C, Espinosa I, Ferrer I, Dolcet
X, Prat J and Matias-Guiu X: Epithelial-to-mesenchymal transition
and stem cells in endometrial cancer. Hum Pathol. 44:1973–1981.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kent CN and Reed Guttilla IK: Regulation
of epithelial-mesenchymal transition in endometrial cancer:
Connecting PI3K, estrogen signaling, and microRNAs. Clin Transl
Oncol. 18:1056–1061. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou XM, Zhang H and Han X: Role of
epithelial to mesenchymal transition proteins in gynecological
cancers: Pathological and therapeutic perspectives. Tumour Biol.
35:9523–9530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mohamet L, Hawkins K and Ward CM: Loss of
function of e-cadherin in embryonic stem cells and the relevance to
models of tumorigenesis. J Oncol. 2011:3526162011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schlosshauer PW, Ellenson LH and Soslow
RA: Beta-catenin and E-cadherin expression patterns in high-grade
endometrial carcinoma are associated with histological subtype. Mod
Pathol. 15:1032–1037. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beeghly-Fadiel A, Lu W, Gao YT, Long J,
Deming SL, Cai Q, Zheng Y, Shu XO and Zheng W: E-cadherin
polymorphisms and breast cancer susceptibility: A report from the
Shanghai breast cancer study. Breast Cancer Res Treat. 121:445–452.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Juan W, Shan K, Na W, Rong-Miao Z and Yan
L: The associations of genetic variants in E-cadherin gene with
clinical outcome of epithelial ovarian cancer. Int J Gynecol
Cancer. 26:1601–1607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Govatati S, Tangudu NK, Deenadayal M,
Chakravarty B, Shivaji S and Bhanoori M: Association of E-cadherin
single nucleotide polymorphisms with the increased risk of
endometriosis in Indian women. Mol Hum Reprod. 18:280–287. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ke X and Cardon LR: Efficient selective
screening of haplotype tag SNPs. Bioinformatics. 19:287–288. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jia YM, Xie YT, Wang YJ, Han JY, Tian XX
and Fang WG: Association of genetic polymorphisms in CDH1 and
CTNNB1 with breast cancer susceptibility and patients' prognosis
among Chinese han women. PLoS One. 10:e01358652015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JY, Park AK, Lee KM, Park SK, Han S,
Han W, Noh DY, Yoo KY, Kim H, Chanock SJ, et al: Candidate gene
approach evaluates association between innate immunity genes and
breast cancer risk in Korean women. Carcinogenesis. 30:1528–1531.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alanazi MS, Parine NR, Shaik JP,
Alabdulkarim HA, Ajaj SA and Khan Z: Association of single
nucleotide polymorphisms in Wnt signaling pathway genes with breast
cancer in Saudi patients. PLoS One. 8:e595552013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang H, Xie YT, Han JY, Ruan Y, Song AP,
Zheng LY, Zhang WZ, Sajdik C, Li Y, Tian XX and Fang WG: Genetic
polymorphisms in centrobin and Nek2 are associated with breast
cancer susceptibility in a Chinese Han population. Breast Cancer
Res Treat. 136:241–251. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ruan Y, Song AP, Wang H, Xie YT, Han JY,
Sajdik C, Tian XX and Fang WG: Genetic polymorphisms in AURKA and
BRCA1 are associated with breast cancer susceptibility in a Chinese
Han population. J Pathol. 225:535–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Chen YL, Xie YT, Zheng LY, Han JY,
Wang H, Tian XX and Fang WG: Association study of germline variants
in CCNB1 and CDK1 with breast cancer susceptibility, progression,
and survival among Chinese Han women. PLoS One. 8:e844892013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Porcu E, Sanna S, Fuchsberger C and
Fritsche LG: Genotype imputation in genome-wide association
studies. Curr Protoc Hum Genet Chapter. 1:Unit 1.252013.
|
|
23
|
Marchini J and Howie B: Genotype
imputation for genome-wide association studies. Nat Rev Genet.
11:499–511. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Willer CJ, Ding J, Scheet P and
Abecasis GR: MaCH: Using sequence and genotype data to estimate
haplotypes and unobserved genotypes. Genet Epidemiol. 34:816–834.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Purcell S, Neale B, Todd-Brown K, Thomas
L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ
and Sham PC: PLINK: A tool set for whole-genome association and
population-based linkage analyses. Am J Hum Genet. 81:559–575.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lou XY, Chen GB, Yan L, Ma JZ, Zhu J,
Elston RC and Li MD: A generalized combinatorial approach for
detecting gene-by-gene and gene-by-environment interactions with
application to nicotine dependence. Am J Hum Genet. 80:1125–1137.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu HM, Xu LF, Hou TT, Luo LF, Chen GB, Sun
XW and Lou XY: GMDR: Versatile software for detecting gene-gene and
gene-environ- ment interactions underlying complex traits. Curr
Genomics. 17:396–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
GTEx consortium: The genotype-tissue
expression (GTEx) project. Nat Genet. 45:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ward LD and Kellis M: HaploReg: A resource
for exploring chromatin states, conservation, and regulatory motif
alterations within sets of genetically linked variants. Nucleic
Acids Res. 40(Database Issue): D930–D934. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo L, Du Y, Chang S, Zhang K and Wang J:
rSNPBase: A database for curated regulatory SNPs. Nucleic Acids
Res. 42(Database Issue): D1033–D1039. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rosenbloom KR, Armstrong J, Barber GP,
Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo
L, Haeussler M, et al: The UCSC genome browser database: 2015
update. Nucleic Acids Res. 43(Database Issue): D670–D681. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Korch C, Spillman MA, Jackson TA, Jacobsen
BM, Murphy SK, Lessey BA, Jordan VC and Bradford AP: DNA profiling
analysis of endometrial and ovarian cell lines reveals
misidentification, redundancy and contamination. Gynecol Oncol.
127:241–248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moore JH: Computational analysis of
gene-gene interactions using multifactor dimensionality reduction.
Expert Rev Mol Diagn. 4:795–803. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Muise AM, Walters TD, Glowacka WK,
Griffiths AM, Ngan BY, Lan H, Xu W, Silverberg MS and Rotin D:
Polymorphisms in E-cadherin (CDH1) result in a mis-localised
cytoplasmic protein that is associated with Crohn's disease. Gut.
58:1121–1127. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martinelli M, Palmieri A, Rodia MT, Cura
F, Scapoli L, Ugolini G, Montroni I, De Sanctis P and Solmi R: CDH1
polymorphisms and low expression of e-cadherin and β-catenin in
colorectal cancer patients. J Biol Regul Homeost Agents. 29 3 Suppl
1:S89–S96. 2015.
|
|
36
|
Yoshida K, Yoshihara K, Adachi S, Haino K,
Nishino K, Yamaguchi M, Nishikawa N, Kashima K, Yahata T, Masuzaki
H, et al: Possible involvement of the E-cadherin gene in genetic
susceptibility to endometriosis. Hum Reprod. 27:1685–1689. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Krebs AM, Mitschke J, Losada Lasierra M,
Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D,
Reichardt W, Bronsert P, et al: The EMT-activator Zeb1 is a key
factor for cell plasticity and promotes metastasis in pancreatic
cancer. Nat Cell Biol. 19:518–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kong FF, Qu ZQ, Yuan HH, Wang JY, Zhao M,
Guo YH, Shi J, Gong XD, Zhu YL, Liu F, et al: Overexpression of
FOXM1 is associated with EMT and is a predictor of poor prognosis
in non-small cell lung cancer. Oncol Rep. 31:2660–2668. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Y, Tang Y, Zhou R, Sun D, Duan Y, Wang
N, Chen Z and Shen N: Genetic polymorphism in the 3′-untranslated
region of the E-cadherin gene is associated with risk of different
cancers. Mol Carcinog. 50:857–862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Memni H, Macherki Y, Klayech Z,
Ben-Haj-Ayed A, Farhat K, Remadi Y, Gabbouj S, Mahfoudh W, Bouzid
N, Bouaouina N, et al: E-cadherin genetic variants predict survival
outcome in breast cancer patients. J Transl Med. 14:3202016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhan Z, Wu J, Zhang JF, Yang YP, Tong S,
Zhang CB, Li J, Yang XW and Dong W: CDH1 gene polymorphisms, plasma
CDH1 levels and risk of gastric cancer in a Chinese population. Mol
Biol Rep. 39:8107–8113. 2012. View Article : Google Scholar : PubMed/NCBI
|