Introduction
Basal cell carcinoma (BCC) is the most frequent
malignant tumor of the eyelid, and it accounts for ~80% of all
malignant eyelid tumors (1,2). Well-known risk factors of BCC include
high levels of sunlight exposure and ultraviolet radiation and
increasing age (2,3). The progression of BCC is relatively
slow, and distant metastasis is rare (4–6). Various
treatments have been reported for BCC of the eyelid, including
surgical techniques, radiotherapy, cryotherapy, photodynamic
therapy, laser ablation, chemotherapy, and immunotherapy (7). However, BCC is topically invasive tumor
with destructive growth to surrounding tissue and has a recurrence
rate of 1–10% even if appropriate treatment is performed (8). There is limited understanding of how
alteration of gene expression in BCC of the eyelid is related to
its pathogenesis; therefore, it is important to study the mechanism
underlying carcinogenesis at the level of gene profiling.
Several studies have reported gene mutations
associated with the carcinogenesis of BCC, such as mutations in the
Patched-1 (PTCH1) gene of the sonic hedgehog (SHH) pathway
(9) and mutations in the tumor
suppressor p53 (10). PTCH-1
suppresses Smoothened signaling and plays an important role in the
transcription of cell cycle regulators. p53 is a tumor suppressor
gene and prevents gene replication in the case of DNA damage by
cell cycle arrest and apoptosis. However, limited information is
available at the genetic and molecular levels for BCC of the
eyelid, and only a few gene profiling studies have been performed
on this disease (11). Therefore, the
aim of the current study was to better understand the molecular
mechanisms underlying BCC of the eyelid. For this purpose, we
analyzed gene expression patterns by using a combination of global
microarray analysis and bioinformatics tools.
Patients and methods
Patients and tissue samples
We enrolled two patients, patient 1 (78-year-old
woman) and patient 2 (83-year-old woman) with BCC of the eyelid.
The patients underwent surgical excision of the tumor at Toyama
University Hospital. A part of the tissue samples was frozen and
kept at −80°C after sampling for subsequent RNA extraction. The
remaining part of the tissue samples was fixed in 4%
paraformaldehyde, and paraffin-embedded tissues were stained with
hematoxylin-eosin. The study was approved by the institutional
review board of the University of Toyama (Toyama, Japan; no.
27–51), and the procedures conformed to the tenets of the World
Medical Association's Declaration of Helsinki. Written informed
consent was obtained from the patients after they had been provided
sufficient information about the procedures.
Cell culture
Normal human epidermal keratinocytes (NHEKs) were
obtained from Kurabo Ind., Ltd. (Osaka, Japan; cat. no. KK-4109).
They were cultured with HuMedia-KB2 medium (Kurabo Ind., Ltd.) at
37°C in humidified air containing 5% CO2.
RNA isolation
Total RNA was extracted from cancer tissues and
NHEKs using a NucleoSpin® RNA isolation kit
(Macherey-Nagel GmbH & Co., Düren, Germany) along with
On-column DNase I treatment, as per manufacturer instructions. RNA
quality was analyzed using a Bioanalyzer 2100 (Agilent
Technologies, Inc., Santa Clara, CA, USA).
Microarray gene expression
analysis
Microarray gene expression analysis was performed
using a GeneChip® system with a Human Genome U133-plus
2.0 array, which was spotted with 54,675 probe sets (Affymetrix,
Inc., Santa Clara, CA, USA) according to the manufacturer's
instructions.
The hybridization intensity data obtained were
converted into a presence or an absence call for each gene, and
changes in gene expression levels between experiments were detected
by comparison analysis. The data were further analyzed using the
GeneSpring® GX software (Agilent Technologies, Inc.) to
extract the significant genes. For gene ontology analysis,
including that pertaining to biological processes, cellular
components, molecular functions, and gene networks, the data
obtained were analyzed using Ingenuity® Pathway Analysis
tools (Ingenuity Systems, Inc., Mountain View, CA, USA) (12,13).
Results
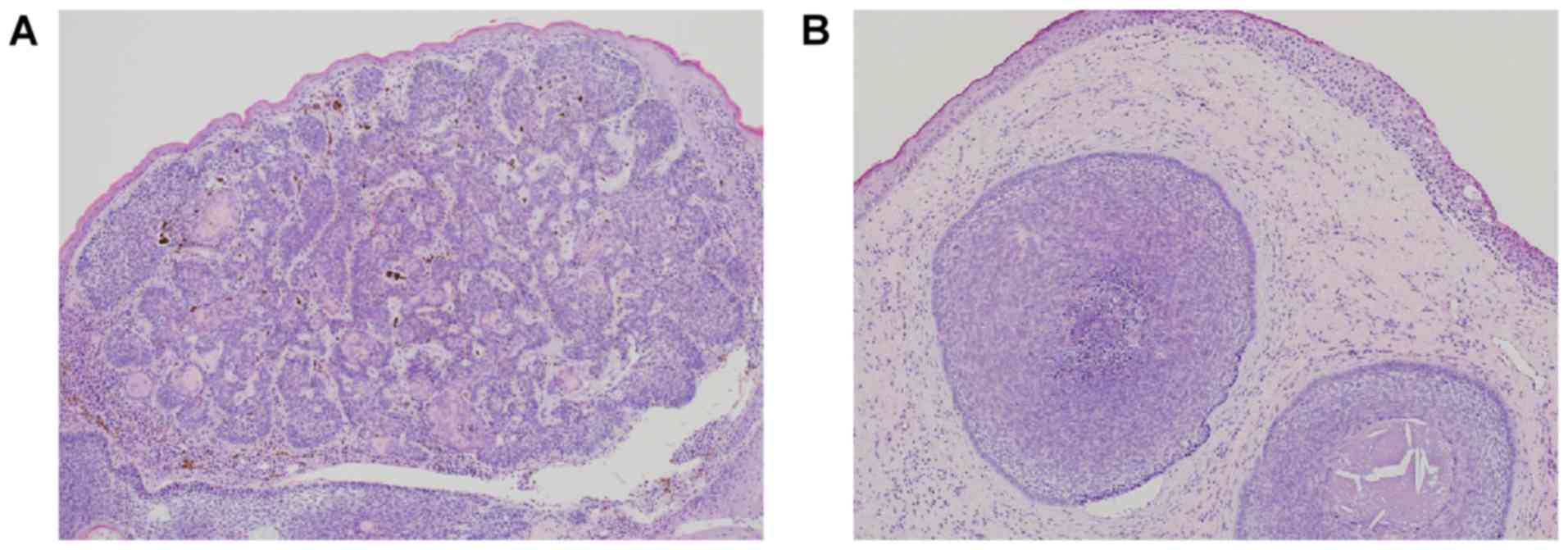
Histopathological analysis
We performed histopathological examination of
surgically removed eyelid tissues from two patients with a lower
eyelid mass. We found that the tumor nest composed with small
basaloid cell was proliferating with peripheral palisading and
mucinous stroma; the histopathological findings were suggestive of
BCC of the eyelid (Fig. 1).
Global gene expression analysis
Global gene expression analysis was performed to
identify the genes involved in BCC of the eyelid. Complete lists of
probe sets for all samples are available on the Gene Expression
Omnibus, a public database (accession no. GSE103439). The
GeneSpring software was used to analyze gene expression in the two
patient samples and the NHEKs and revealed that many genes showed
>5.0-fold difference in expression between the two sample types.
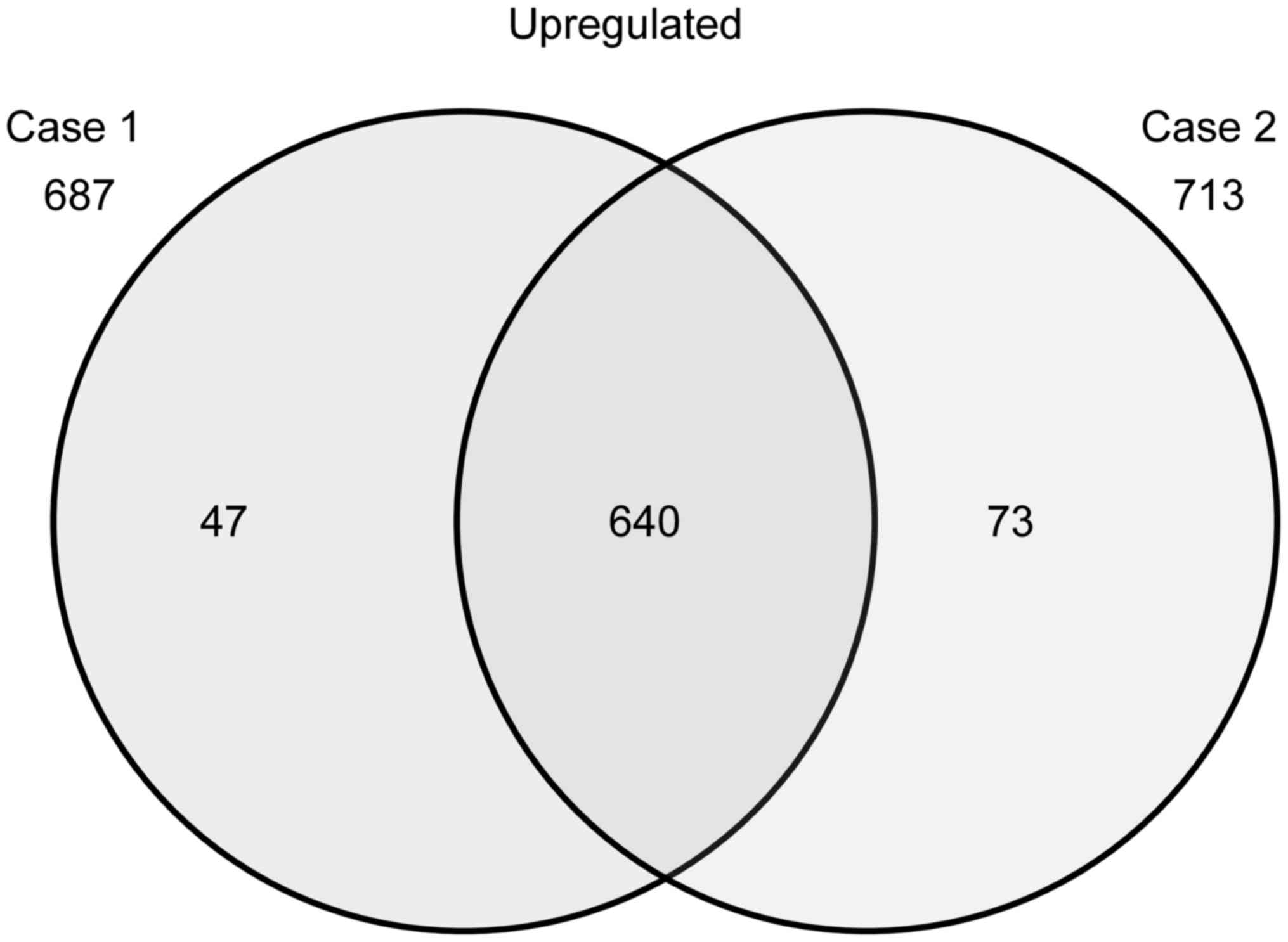
The Venn diagram in Fig. 2 summarizes
the numbers of specifically and commonly expressed genes in each
group. The samples from patients 1 and 2 showed upregulation of the
expression of 687 and 713 genes, respectively, as compared to that
noted in the control cells (NHEKs); 640 genes were upregulated in
both patient samples (Fig. 2). From
these 640 genes, the upregulated genes that were reported to be
associated with BCC are listed as follows: Nkyrin 2 (ANK2),
anoctamin 5 (ANO5), androgen receptor (AR), ATPase Na+/K+
transporting subunit α 2 (ATP1A2), BCL2 apoptosis regulator (BCL2),
calcium voltage-gated channel auxiliary subunit alpha2delta1
(CACNA2D1), complement factor H (CFH), chromogranin B (CHGB),
cardiomyopathy associated 5 (CMYA5), collagen type VI α 3 chain
(COL6A3), dihydropyrimidine dehydrogenase (DPYD), fibroblast growth
factor receptor 1 (FGFR1), filaggrin (FLG), forkhead box C1
(FOXC1), frizzled class receptor 7 (FZD7), heat shock protein
family B member 2 (HSPB2), heat shock protein family B member 3
(HSPB3), heat shock protein family B member 7 (HSPB7), interferon
induced transmembrane protein 1 (IFITM1), keratin 1 (KRT1), keratin
4 (KRT4), keratin 7 (KRT7), keratin 13 (KRT13), keratin 19 (KRT19),
keratin 23 (KRT23), keratin 25 (KRT25), keratin 79 (KRT79), LIM
homeobox 2 (LHX2), LIM and calponin homology domains 1 (LIMCH1),
myosin heavy chain (MYH1), nebulin (NEB), nebulin related anchoring
protein (NRAP), phosphodiesterase 4D interacting protein (PDE4DIP),
PTCH1, SRY-box 9 (SOX9), tropomyosin 2 (TPM2), titin (TTN),
tyrosinase related protein 1 (TYRP1) and xin actin binding repeat
containing 2 (XIRP2).
Identification of biological functions
and gene networks
To identify the biological functions of and the
relevant gene networks in the case of genes that were involved in
BCC of the eyelid and showed differential expression, functional
category and gene network analyses were conducted using the
Ingenuity® Pathways Knowledge Base. In the case of the
640 genes that were upregulated in both patient samples,
Ingenuity® pathway analysis showed that the gene network
included many BCC-associated genes, such as the following (Fig. 3): AR; BCL2; PTCH1; CHGB; heat shock
protein 27 (Hsp27); TPM2; NEB; XIRP2; TTN; CACNA2D1; CMYA5; MYH1;
and SOX9.
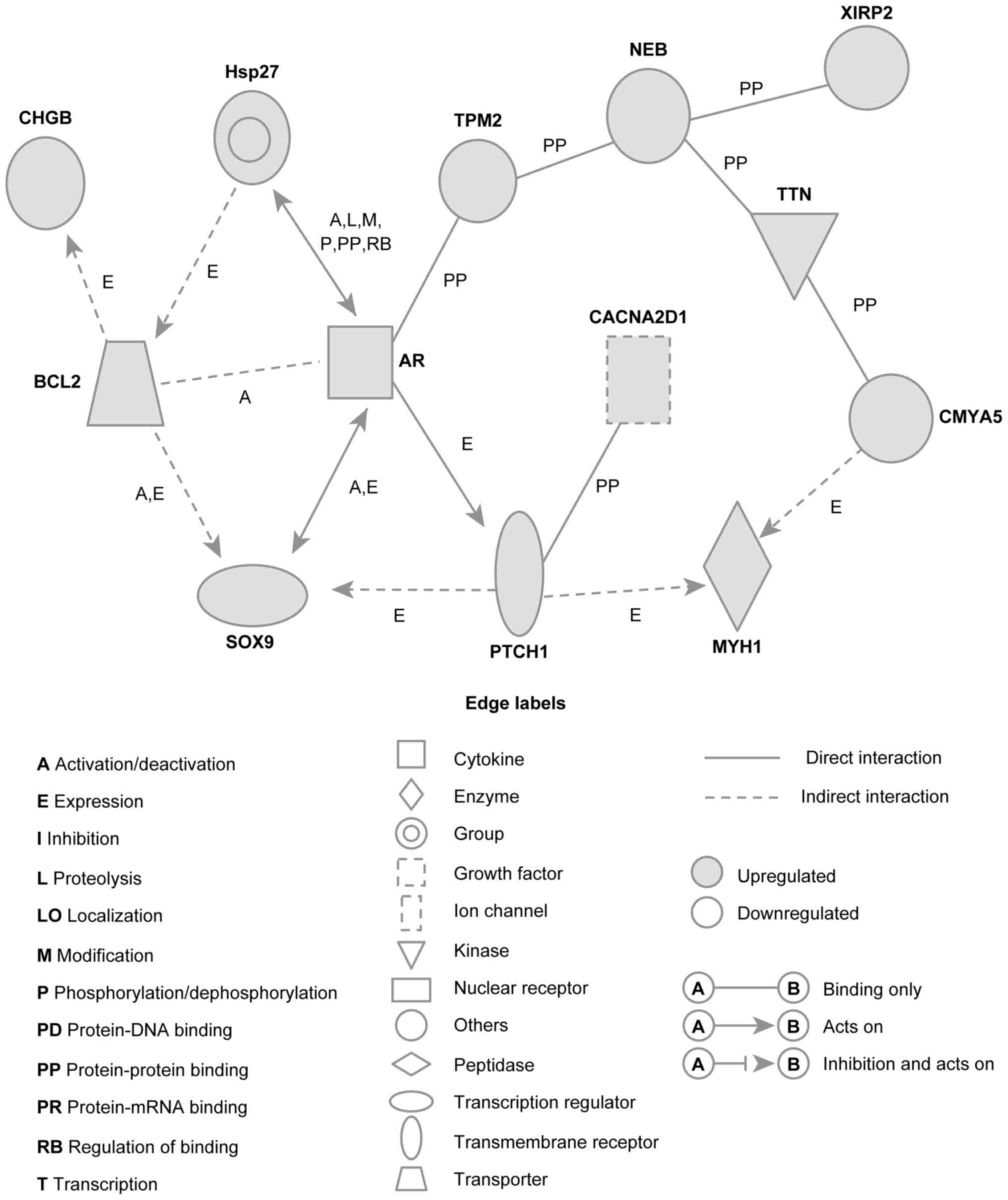 | Figure 3.Networks involving upregulated genes
in BCC of the eyelid. Upregulated genes were analyzed using
Ingenuity pathway tools. The network is represented graphically
with nodes (genes) and edges (biological associations between the
nodes). Nodes and edges are displayed using various shapes and
labels reflecting the functional class of each gene and the nature
of the relationships involved, respectively. BCC, basal cell
carcinoma; CHGB, chromogranin B; Hsp27, heat shock protein 27;
TPM2, tropomyosin 2; NEB, nebulin; XIRP2, xin actin binding repeat
containing 2; BCL2, BCL2 apoptosis regulator; AR, androgen
receptor; TTN, titin; CACNA2D1, calcium voltage-gated channel
auxiliary subunit alpha2delta1; SOX9, SRY-box 9; PTCH1, Patched-1;
MYH1, myosin heavy chain 1; CMYA5, cardiomyopathy associated 5. |
We also identified unique gene networks related to
cancer cell growth, tumorigenesis, and cell survival (Fig. 4). The cancer cell growth gene network
included several genes, such as the following (Fig. 4A): AR, erb-b2 receptor tyrosine kinase
3 (ERBB3), C-X-C motif chemokine ligand 12 (CXCL12), BCL2,
insulin-like growth-factor-binding protein 5 (IGFBP5), matrix
metallopeptidase 2 (MMP2), platelet-derived growth factor receptor
α (PDGFRA), plasminogen activator, tissue type (PLAT), and prostate
stem cell antigen (PSCA). This network was associated with the
biological functions involved in cancer cell growth. The
tumorigenesis gene network included several genes, such as the
following: BCL2, KIT proto-oncogene receptor tyrosine kinase (KIT),
CXCL12, serpin family F member 1 (SERPLNF1), insulin-like growth
factor 2 (IGF2), IGFBP5, PLAT, and neurotrophic receptor tyrosine
kinase 2 (NTRK2). This network was associated with biological
functions involved in tumorigenesis (Fig.
4B). The cell survival gene network included several genes,
such as the following (Fig. 4C): AR;
PSCA; PTCH1; IGF2; fibroblast growth factor receptor 1 (FGFR1);
ERBB3; IGFBP5; versican (VCAN); BCL2; and galanin and GMAP
prepropeptide (GAL). This network was associated with biological
functions involved in cell survival.
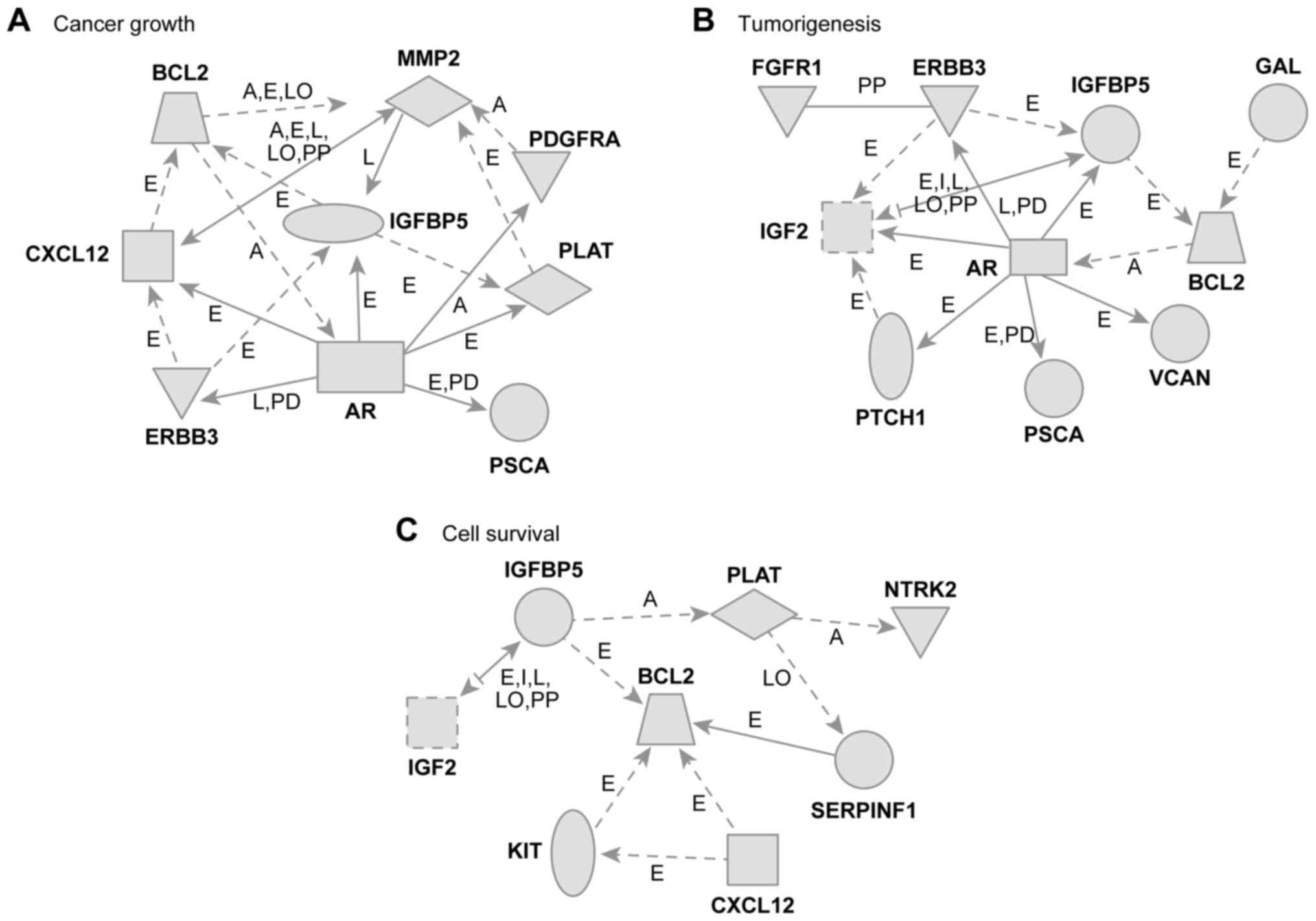 | Figure 4.Gene networks. (A) Cancer growth, (B)
tumorigenesis, and (C) cell survival in relation to the upregulated
genes in patients with BCC of the eyelid. These networks were
analyzed using the Ingenuity pathway analysis tools. The networks
are represented graphically with nodes (genes) and edges
(biological associations between the nodes). BCC, basal cell
carcinoma; BCL2, BCL2 apoptosis regulator; MMP2, matrix
metallopeptidase 2; PDGFRA, platelet-derived growth factor receptor
α; CXCL12, C-X-C motif chemokine ligand 12; IGFBP5, insulin like
growth factor binding protein 5; PLAT, plasminogen activator,
tissue type; AR, androgen receptor; ERBB3, erb-b2 receptor tyrosine
kinase 3; PSCA, prostate stem cell antigen; GAL, galanin and GMAP
prepropeptide; IGF2, insulin like growth factor 2; VCAN, versican;
PTCH1, Patched-1; NTRK2, neurotrophic receptor tyrosine kinase 2;
KIT, KIT proto-oncogene receptor tyrosine kinase. |
Discussion
BCC of the eyelid is the most frequent malignant
tumor of the eyelid; however, to date, only insufficient
information is available regarding the molecular mechanisms
underlying this disease. In the current study, we examined gene
expression patterns associated with BCC of the eyelid by performing
global microarray analysis, and gene network analysis of
differentially expressed genes was performed using computational
gene expression analysis tools.
We successfully identified a unique gene network on
the basis of our data for upregulated gene expression in patients
with BCC of the eyelid. This network included PTCH1 (14,15), which
has been reported as an important player in BCC pathogenesis. The
PTCH1 gene is a negative regulator of the SHH pathway; the
dysfunction of this pathway has been recognized as essential events
for BCC development (15,16). Furthermore, it is reported that PTCH-1
mutations contribute to the development of cutaneous eyelid BCC
(17). In this network, PTCH1 was
found to interact with several genes, such as MYH1, CACNA2D1, AR,
SOX9, BCL2, CHGB, Hsp27, TPM2, NEB, NIRP2, TTN, and CMYA5. These
genes have been reported to be related to BCC (18–20); it is
possible that they play a role at the molecular level in the
pathogenesis of BCC of the eyelid. Among them, SOX9 (21,22) and
BCL2 (23,24) have been reported as biomarkers of BCC
and may play an important role through interactions with PTCH1 in
the pathogenesis of this disease.
Our functional category analysis showed that the
upregulated genes in patients with BCC of the eyelid were related
to biological functions such as cancer cell growth, tumorigenesis,
and cell survival (Fig. 4). The
cancer cell growth gene network consisted of nine genes, that is,
AR (25), BCL2 (26), IGFBP5 (27), CXCL12 (28), ERBB3 (29), MMP2 (30), PDGFRA (31), PLAT (32) and PSCA (33), which have been found to be associated
with the cell growth of various cancers, such as renal cell
carcinoma, neuroblastoma and pancreatic cancer. The tumorigenesis
gene network consisted of 10 genes, that is, PTCH1 (9,34), BCL2
(35), FGFR1 (36), AR (37),
ERBB3 (38), IGF2 (39), IGFBP5 (40), PSCA (41), VCAN (42), and GAL (43), which have been reported to be
associated with the tumorigenesis of various cancers, such as
prostate cancer, neck squamous cell carcinoma and breast cancer.
The cell survival gene network consisted of eight genes, that is,
BCL2 (44), LGF2 (45), IGFBP5 (46), PLAT (47), NTRK2 (48), SERPINF1 (49), KIT (50), and CXCL12 (50), which have been found to be associated
with cell survival in various cancers, such as lung cancer and
prostate cancer. In BCC of the eyelid, the genes upregulated in
these networks may play an important role in BCC cell survival
through the network.
To summarize, from the 640 genes that were
upregulated in both patient samples, Ingenuity® pathway
analysis showed that the gene network of BCC of the eyelid included
many BCC-associated genes, such as BCL2, PTCH1, and SOX9. We
identified unique gene networks related with cancer cell growth,
tumorigenesis, and cell survival. Thus, our results may help
clarify the pathogenesis of BCC of the eyelid at the molecular
level. However, the interaction between gene expression and this
disease needs to be studied further. These results of integrating
microarray analyses provide further insights into the molecular
mechanisms involved in BCC of the eyelid and may lead to a
therapeutic approach for this disease.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by JSPS
KAKENHI (grant no. JP16K20309).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TY, YT and TH conceived the study, designed the
experiments, wrote the manuscript and performed the experiments. SM
and JI also performed experiments. TY and AH provided materials for
microarray analysis and were involved in data analysis.
Ethics approval and consent to
participate
The study was approved by the institutional review
board of the University of Toyama (no. 27–51). All patients
provided written informed consent.
Patient consent for publication
Written informed consent was obtained from the
patients after they were provided with sufficient information about
the procedures and publication.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ANK2
|
ankyrin 2
|
|
ANO5
|
anoctamin 5
|
|
AR
|
androgen receptor
|
|
ATP1A2
|
ATPase Na+/K+ transporting subunit α
2
|
|
BCC
|
basal cell carcinoma
|
|
BCL2
|
BCL2 apoptosis regulator
|
|
CACNA2D1
|
calcium voltage-gated channel
auxiliary subunit alpha2delta1
|
|
CFH
|
complement factor H
|
|
CHGB
|
chromogranin B
|
|
CMYA5
|
cardiomyopathy associated 5
|
|
COL6A3
|
collagen type VI α 3 chain
|
|
CXCL12
|
C-X-C motif chemokine ligand 12
|
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3
|
|
FGFR1
|
fibroblast growth factor receptor
1
|
|
FLG
|
filaggrin
|
|
FOXC1
|
forkhead box C1
|
|
FZD7
|
frizzled class receptor 7
|
|
GAL
|
galanin and GMAP prepropeptide
|
|
Hsp27
|
heat shock protein 27
|
|
HSPB2
|
heat shock protein family B member
2
|
|
HSPB3
|
heat shock protein family B member
3
|
|
HSPB7
|
heat shock protein family B member
7
|
|
IFITM1
|
interferon induced transmembrane
protein 1
|
|
IGF2
|
insulin like growth factor 2
|
|
IGFBP5
|
insulin like growth factor binding
protein 5
|
|
KIT
|
KIT proto-oncogene receptor tyrosine
kinase
|
|
KRT1
|
keratin 1
|
|
KRT4
|
keratin 4
|
|
KRT7
|
keratin 7
|
|
KRT13
|
keratin 13
|
|
KRT19
|
keratin 19
|
|
KRT23
|
keratin 23
|
|
KRT25
|
keratin 25
|
|
KRT79
|
keratin 79
|
|
LHX2
|
LIM homeobox 2
|
|
LIMCH1
|
LIM and calponin homology domains
1
|
|
MMP2
|
matrix metallopeptidase 2
|
|
MYH1
|
myosin heavy chain 1
|
|
NEB
|
nebulin
|
|
NHEK
|
normal human epidermal
keratinocytes
|
|
NRAP
|
nebulin related anchoring protein
|
|
NTRK2
|
neurotrophic receptor tyrosine kinase
2
|
|
PDE4DIP
|
phosphodiesterase 4D interacting
protein
|
|
PDGFRA
|
platelet-derived growth factor
receptor α
|
|
PLAT
|
plasminogen activator, tissue type
|
|
PSCA
|
prostate stem cell antigen
|
|
PTCH1
|
Patched-1
|
|
SERPINF1
|
serpin family F member 1
|
|
SHH
|
sonic hedgehog
|
|
SOX9
|
SRY-box 9
|
|
TPM2
|
tropomyosin 2
|
|
TTN
|
titin
|
|
TYRP1
|
tyrosinase related protein 1
|
|
VCAN
|
versican
|
|
XIRP2
|
xin actin binding repeat containing
2
|
References
|
1
|
Cook BE Jr and Bartley GB: Epidemiologic
characteristics and clinical course of patients with malignant
eyelid tumors in an incidence cohort in Olmsted County, Minnesota.
Ophthalmology. 106:746–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allali J, D'Hermies F and Renard G: Basal
cell carcinomas of the eyelids. Ophthalmologica. 219:57–71. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Neale RE, Davis M, Pandeya N, Whiteman DC
and Green AC: Basal cell carcinoma on the trunk is associated with
excessive sun exposure. J Am Acad Dermatol. 56:380–386. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lindgren G, Lindblom B and Larkö O: Mohs'
micrographic surgery for basal cell carcinomas on the eyelids and
medial canthal area. II. Reconstruction and follow-up. Acta
Ophthalmol Scand. 78:430–436. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malhotra R, Huilgol SC, Huynh NT and Selva
D: The Australian Mohs database, part II: Periocular basal cell
carcinoma outcome at 5-year follow-up. Ophthalmology. 111:631–636.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morris DS, Elzaridi E, Clarke L, Dickinson
AJ and Lawrence CM: Periocular basal cell carcinoma: 5-year outcome
following Slow Mohs surgery with formalin-fixed paraffin-embedded
sections and delayed closure. Br J Ophthalmol. 93:474–476. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Margo CE and Waltz K: Basal cell carcinoma
of the eyelid and periocular skin. Surv Ophthalmol. 38:169–192.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Diepgen TL and Mahler V: The epidemiology
of skin cancer. Br J Dermatol. 146:1–6. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Zwaan SE and Haass NK: Genetics of
basal cell carcinoma. Australas J Dermatol. 51:81–92; quiz 93–94.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Muller PA, Vousden KH and Norman JC: p53
and its mutants in tumor cell migration and invasion. J Cell Biol.
192:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heller ER, Gor A, Wang D, Hu Q, Lucchese
A, Kanduc D, Katdare M, Liu S and Sinha AA: Molecular signatures of
basal cell carcinoma susceptibility and pathogenesis: A genomic
approach. Int J Oncol. 42:583–596. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tabuchi Y, Yunoki T, Hoshi N, Suzuki N and
Kondo T: Genes and gene networks involved in sodium
fluoride-elicited cell death accompanying endoplasmic reticulum
stress in oral epithelial cells. Int J Mol Sci. 15:8959–8978. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yunoki T, Tabuchi Y, Hayashi A and Kondo
T: Network analysis of genes involved in the enhancement of
hyperthermia sensitivity by the knockdown of BAG3 in human oral
squamous cell carcinoma cells. Int J Mol Med. 38:236–242. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reifenberger J, Wolter M, Knobbe CB,
Köhler B, Schönicke A, Scharwächter C, Kumar K, Blaschke B, Ruzicka
T and Reifenberger G: Somatic mutations in the PTCH, SMOH, SUFUH
and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol.
152:43–51. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Palmer CJ, Galan-Caridad JM, Weisberg SP,
Lei L, Esquilin JM, Croft GF, Wainwright B, Canoll P, Owens DM and
Reizis B: Zfx facilitates tumorigenesis caused by activation of the
Hedgehog pathway. Cancer Res. 74:5914–5924. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lupu M, Caruntu C, Ghita MA, Voiculescu V,
Voiculescu S, Rosca AE, Caruntu A, Moraru L, Popa IM, Calenic B, et
al: Gene expression and proteome analysis as sources of biomarkers
in basal cell carcinoma. Dis Markers. 2016:98312372016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Celebi AR, Kiratli H and Soylemezoglu F:
Evaluation of the ‘Hedgehog’ signaling pathways in squamous and
basal cell carcinomas of the eyelids and conjunctiva. Oncol Lett.
12:467–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Astarci HM, Gurbuz GA, Sengul D,
Hucumenoglu S, Kocer U and Ustun H: Significance of androgen
receptor and CD10 expression in cutaneous basal cell carcinoma and
trichoepithelioma. Oncol Lett. 10:3466–3470. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sharpe HJ, Pau G, Dijkgraaf GJ,
Basset-Seguin N, Modrusan Z, Januario T, Tsui V, Durham AB, Dlugosz
AA, Haverty PM, et al: Genomic analysis of smoothened inhibitor
resistance in basal cell carcinoma. Cancer Cell. 27:327–341. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trautinger F, Kindas-Mügge I, Dekrout B,
Knobler RM and Metze D: Expression of the 27-kDa heat shock protein
in human epidermis and in epidermal neoplasms: An
immunohistological study. Br J Dermatol. 133:194–202. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vidal VP, Ortonne N and Schedl A: SOX9
expression is a general marker of basal cell carcinoma and
adnexal-related neoplasms. J Cutan Pathol. 35:373–379. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Youssef KK, Lapouge G, Bouvrée K, Rorive
S, Brohée S, Appelstein O, Larsimont JC, Sukumaran V, Van de Sande
B, Pucci D, et al: Adult interfollicular tumour-initiating cells
are reprogrammed into an embryonic hair follicle progenitor-like
fate during basal cell carcinoma initiation. Nat Cell Biol.
14:1282–1294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zbacnik AP, Rawal A, Lee B, Werling R,
Knapp D and Mesa H: Cutaneous basal cell carcinosarcoma: Case
report and literature review. J Cutan Pathol. 42:903–910. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Regl G, Kasper M, Schnidar H, Eichberger
T, Neill GW, Philpott MP, Esterbauer H, Hauser-Kronberger C,
Frischauf AM and Aberger F: Activation of the BCL2 promoter in
response to Hedgehog/GLI signal transduction is predominantly
mediated by GLI2. Cancer Res. 64:7724–7731. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He D, Li L, Zhu G, Liang L, Guan Z, Chang
L, Chen Y, Yeh S and Chang C: ASC-J9 suppresses renal cell
carcinoma progression by targeting an androgen receptor-dependent
HIF2α/VEGF signaling pathway. Cancer Res. 74:4420–4430. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kannan S, Sutphin RM, Hall MG, Golfman LS,
Fang W, Nolo RM, Akers LJ, Hammitt RA, McMurray JS, Kornblau SM, et
al: Notch activation inhibits AML growth and survival: A potential
therapeutic approach. J Exp Med. 210:321–337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tanno B, Cesi V, Vitali R, Sesti F,
Giuffrida ML, Mancini C, Calabretta B and Raschellà G: Silencing of
endogenous IGFBP-5 by micro RNA interference affects proliferation,
apoptosis and differentiation of neuroblastoma cells. Cell Death
Differ. 12:213–223. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sutton A, Friand V, Brulé-Donneger S,
Chaigneau T, Ziol M, Sainte-Catherine O, Poiré A, Saffar L, Kraemer
M, Vassy J, et al: Stromal cell-derived factor-1/chemokine (C-X-C
motif) ligand 12 stimulates human hepatoma cell growth, migration,
and invasion. Mol Cancer Res. 5:21–33. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gaborit N, Abdul-Hai A, Mancini M, Lindzen
M, Lavi S, Leitner O, Mounier L, Chentouf M, Dunoyer S, Ghosh M, et
al: Examination of HER3 targeting in cancer using monoclonal
antibodies. Proc Natl Acad Sci USA. 112:839–844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kesanakurti D, Chetty C, Dinh DH, Gujrati
M and Rao JS: Role of MMP-2 in the regulation of IL-6/Stat3
survival signaling via interaction with α5β1 integrin in glioma.
Oncogene. 32:327–340. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pettazzoni P, Viale A, Shah P, Carugo A,
Ying H, Wang H, Genovese G, Seth S, Minelli R, Green T, et al:
Genetic events that limit the efficacy of MEK and RTK inhibitor
therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer
Res. 75:1091–1101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Menouny M, Binoux M and Babajko S: Role of
insulin-like growth factor binding protein-2 and its limited
proteolysis in neuroblastoma cell proliferation: Modulation by
transforming growth factor-beta and retinoic acid. Endocrinology.
138:683–690. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Saeki N, Gu J, Yoshida T and Wu X:
Prostate stem cell antigen: A Jekyll and Hyde molecule? Clin Cancer
Res. 16:3533–3538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Northcott PA, Jones DT, Kool M, Robinson
GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P,
Taylor MD and Pfister SM: Medulloblastomics: The end of the
beginning. Nat Rev Cancer. 12:818–834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sánchez-García I and Grütz G: Tumorigenic
activity of the BCR-ABL oncogenes is mediated by BCL2. Proc Natl
Acad Sci USA. 92:5287–5291. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marshall ME, Hinz TK, Kono SA, Singleton
KR, Bichon B, Ware KE, Marek L, Frederick BA, Raben D and Heasley
LE: Fibroblast growth factor receptors are components of autocrine
signaling networks in head and neck squamous cell carcinoma cells.
Clin Cancer Res. 17:5016–5025. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schroeder A, Herrmann A, Cherryholmes G,
Kowolik C, Buettner R, Pal S, Yu H, Müller-Newen G and Jove R: Loss
of androgen receptor expression promotes a stem-like cell phenotype
in prostate cancer through STAT3 signaling. Cancer Res.
74:1227–1237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vaught DB, Stanford JC, Young C, Hicks DJ,
Wheeler F, Rinehart C, Sánchez V, Koland J, Muller WJ, Arteaga CL
and Cook RS: HER3 is required for HER2-induced preneoplastic
changes to the breast epithelium and tumor formation. Cancer Res.
72:2672–2682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Veronese A, Lupini L, Consiglio J, Visone
R, Ferracin M, Fornari F, Zanesi N, Alder H, D'Elia G, Gramantieri
L, et al: Oncogenic role of miR-483-3p at the IGF2/483 locus.
Cancer Res. 70:3140–3149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Butt AJ, Dickson KA, McDougall F and
Baxter RC: Insulin-like growth factor-binding protein-5 inhibits
the growth of human breast cancer cells in vitro and in vivo. J
Biol Chem. 278:29676–29685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chapman EJ, Kelly G and Knowles MA: Genes
involved in differentiation, stem cell renewal, and tumorigenesis
are modulated in telomerase-immortalized human urothelial cells.
Mol Cancer Res. 6:1154–1168. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kunisada M, Yogianti F, Sakumi K, Ono R,
Nakabeppu Y and Nishigori C: Increased expression of versican in
the inflammatory response to UVB- and reactive oxygen
species-induced skin tumorigenesis. Am J Pathol. 179:3056–3065.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tofighi R, Joseph B, Xia S, Xu ZQ,
Hamberger B, Hökfelt T and Ceccatelli S: Galanin decreases
proliferation of PC12 cells and induces apoptosis via its subtype 2
receptor (GalR2). Proc Natl Acad Sci USA. 105:2717–2722. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin Y, Fukuchi J, Hiipakka RA, Kokontis JM
and Xiang J: Up-regulation of Bcl-2 is required for the progression
of prostate cancer cells from an androgen-dependent to an
androgen-independent growth stage. Cell Res. 17:531–536. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Linnerth NM, Baldwin M, Campbell C, Brown
M, McGowan H and Moorehead RA: IGF-II induces CREB phosphorylation
and cell survival in human lung cancer cells. Oncogene.
24:7310–7319. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kashyap AS, Hollier BG, Manton KJ,
Satyamoorthy K, Leavesley DI and Upton Z: Insulin-like growth
factor-I:vitronectin complex-induced changes in gene expression
effect breast cell survival and migration. Endocrinology.
152:1388–1401. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin L, Jin Y and Hu K: Tissue-type
plasminogen activator (tPA) promotes M1 macrophage survival through
p90 ribosomal S6 kinase (RSK) and p38 mitogen-activated protein
kinase (MAPK) pathway. J Biol Chem. 290:7910–7917. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Osborne JK, Larsen JE, Shields MD,
Gonzales JX, Shames DS, Sato M, Kulkarni A, Wistuba II, Girard L,
Minna JD and Cobb MH: NeuroD1 regulates survival and migration of
neuroendocrine lung carcinomas via signaling molecules TrkB and
NCAM. Proc Natl Acad Sci USA. 110:6524–6529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sheikpranbabu S, Haribalaganesh R and
Gurunathan S: Pigment epithelium-derived factor inhibits advanced
glycation end-products-induced cytotoxicity in retinal pericytes.
Diabetes Metab. 37:505–511. 2011.PubMed/NCBI
|
|
50
|
Sun J, Pedersen M and Rönnstrand L: The
D816V mutation of c-Kit circumvents a requirement for Src family
kinases in c-Kit signal transduction. J Biol Chem. 284:11039–11047.
2009. View Article : Google Scholar : PubMed/NCBI
|