Introduction
Colorectal cancer is one of the most common types of
cancer and one of the leading causes of cancer-associated mortality
globally, according to statistical data in 2013 (1). Identifying cancer stem-like cells (CSCs)
may be essential for improving targeted cancer therapy (2,3). However,
detecting purified CSCs in colorectal cancer remains challenging
with existing methods, since few specific markers are known for
CSCs in colorectal cancer (4,5).
CSCs may trigger tumorigenesis, self-renewal,
differentiation and resistance to therapy (6). Previous studies have demonstrated that
CSCs may activate one or more highly conserved signaling pathways
present in normal stem cells that are associated with development
and tissue homeostasis, including the Notch (7), Hedgehog (8) and Wnt (9)
signaling pathways. These pathways are associated with
tumorigenesis, metastasis and the cell cycle, which are required
for cancer proliferation (10).
Crosstalk between signaling pathways also increases the complexity
of cellular external stimuli response networks (11). Upregulating secreted frizzled related
protein (SFRP)1 expression in the Hedgehog signaling pathway
inhibited the Wnt signaling pathway (12), whereas activating the Hedgehog
signaling pathway resulted in increased jagged 2 expression and the
upregulation of the Notch signaling pathway (11). A network-level view of signaling
pathways in cancer stem cells is required to identify shared
features among malignant cells and provide means for clinical
therapy to develop.
Previously, multiple studies have developed
multigene classifiers for determining the prognosis of patients
with colon cancer, including the 12-gene based Oncotype DX
(13,14), 18-gene based ColoPrint (15) and the 13-gene based classifier
ColoGuideEx (16). Compared with
conventional pathological criteria alone, genomic classifiers,
including Oncotype DX, Coloprint and ColoGuideEx, provide more
accurate information on the risk of recurrence and may assist in
selecting patients who can benefit more from chemotherapy. However,
these gene-based classification systems may not assist in
developing a patient selection tool for specific inhibitor-targeted
therapy. Therefore, establishing a signaling pathway- or multiple
signaling pathways-based gene expression signature to facilitate
treatment decisions is crucial.
The present study assessed the cancer stem cell
signaling pathways and the gene expression profiles of 1,198
patients with colorectal cancer from The Cancer Genome Atlas (TCGA)
and the Gene Expression Omnibus (GEO) database. By analyzing the
association between gene expression profiling and the clinical
outcome of patients with colorectal cancer, the present study
identified an eight-gene signature associated with the
Hedgehog-Notch-Wnt signaling pathways, which was associated with
the prognosis of the patients. The results of the present study may
assist in developing therapeutic strategies for treating colorectal
cancer.
Materials and methods
Colorectal cancer data
Colorectal cancer gene expression datasets and the
corresponding clinical data were downloaded from TCGA and the GEO
database. Integrated gene expression and clinical data of TCGA were
downloaded from the University of California, Santa Cruz Cancer
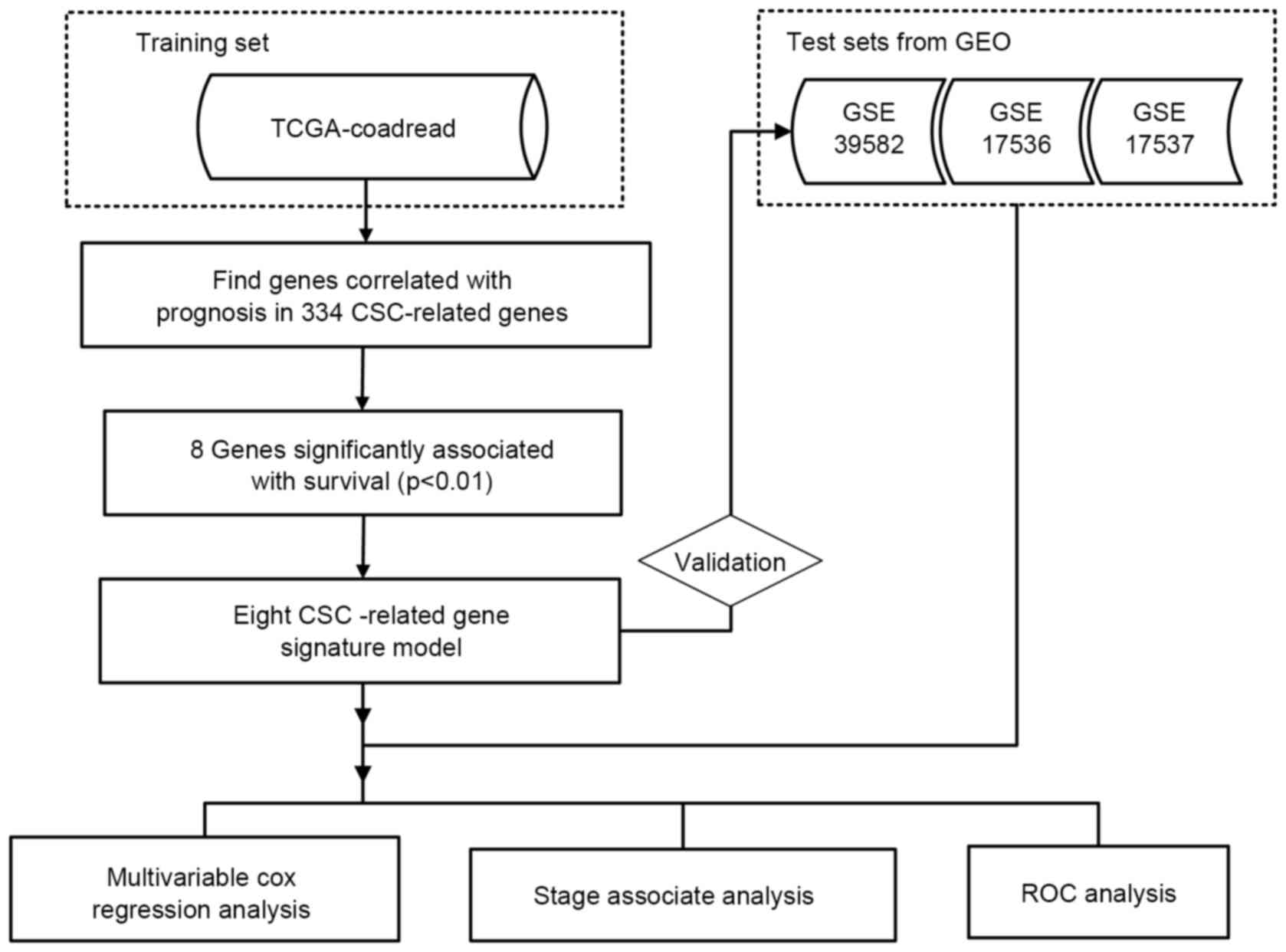
Genomics Browser (Santa Cruz, CA, USA; http://genome-cancer.ucsc.edu) (17). In the present study, 383 colorectal
tumors from patients with detailed gene expression information were
chosen from TCGA according to parameters defined in a previous
study (18); three datasets GSE39582
(n=582) (19), GSE17536 (n=177)
(20) and GSE17537 (n=55) (21) from the GEO database (http://www.ncbi.nlm.nih.gov/geo) were selected as
testing datasets and contained overall survival and
recurrence/disease-free survival information. A total of 1,198
patients were analyzed in the present study. Clinical information
was extracted from the original publications (19–21). The
workflow of the present study is presented in Fig. 1.
Candidate gene selection
The present study selected genes that are associated
with the signaling or transcriptional regulators of CSCs in the
Hedgehog, Notch and Wnt signaling pathways. To identify more genes
associated with these signaling pathways, SABiosciences (http://www.sabiosciences.com/PCRArrayPlate.php) and
polymerase chain reaction (PCR) array gene lists were used to form
a gene list and group functional genes (22). The gene tables contained
Hedgehog/Notch/Wnt ligands, receptors and regulators, and
downstream signaling molecules and target proteins associated with
these signaling pathways.
Statistical analysis
The association between gene expression and patient
survival rate was assessed with univariate Cox's regression
analysis and a permutation test using Biometric Research Branch
(BRB)-Array Tools edition 4.5.0 (https://brb.nci.nih.gov/BRB-ArrayTools/) (23). Genes were considered statistically
significant if their permutation P-value was ≤0.01. Selected genes
were fitted using a multivariable Cox regression model in the
training set as described. A risk score formula was constructed by
including statistically significant genes, weighted by their
estimated Cox's regression coefficients (24). Patients with assigned risk score were
classified into high-risk or low-risk groups by using the median as
the threshold. Kaplan-Meier estimator survival analysis using the R
package survival (v2.41-3; http://cran.r-project.org/web/packages/survival/)
was performed to estimate the survival distributions between
stratified survival groups in each set (25,26). The
two-sided log-rank test was used to assess the survival differences
between high-risk and low-risk groups. A two-way ANOVA was used to
analyze the association between the eight-gene signature and the
American Joint Committee on Cancer stage.
The receiver operating characteristic (ROC) curve
was constructed using the R package pROC to evaluate the
sensitivity and specificity of the survival prediction for the CSC
signature risk score, age and ColoGuideEx, a reported prognostic
predictor. According to ColoGuideEx, a 13-gene prognostic predictor
developed by Agesen et al (16), patients were stratified according to
the number of genes exceeding the 80% high-risk genes and below the
20% level of low-risk genes. Area under the curve values were
calculated from the ROC curves.
Results
Identifying prognostic CSC-associated
genes from the TCGA COADREAD dataset
A total of 334 CSC-associated genes in the Hedgehog,
Notch and Wnt signaling pathways and their targets were identified
from the colorectal tumors in the TCGA cohorts and the GEO
datasets. The 383 TCGA patients cohort was defined as the training
set, and was used to select the prognostic CSC-associated genes.
Using BRB-Array Tools, univariate Cox's proportional hazards
regression analysis was performed for the CSC-associated gene
expression data, and eight CSC-associated genes were identified as
significantly associated with overall survival (P≤0.01). Of these
genes, a hazard ratio >1, which was associated with low-density
lipoprotein-related protein 2 (LRP2), hairy/enhancer-of-split
associated with YRPW motif-like protein (HEYL), cubilin (CUBN),
SFRP2, growth arrest and DNA-damage-inducible 45β (GADD45B),
insulin-like growth factor-binding protein 3 (IGFBP3) and lymphoid
enhancer-binding factor 1 (LEF1), indicated that high expression of
that gene was associated with poor survival; a hazard ratio <1,
which was associated with cyclin E1 (CCNE1), indicated that
increased expression of that gene was associated with good survival
(Table I).
 | Table I.Eight genes associated with overall
survival in the training-set patients. |
Table I.
Eight genes associated with overall
survival in the training-set patients.
| Gene symbol | Full name | Parametric
P-value | Hazard ratio | Coefficient |
|---|
| LRP2 | Low-density
lipoprotein-related protein 2 | 0.0005083 | 1.355 | 0.2530 |
| HEYL |
Hairy/enhancer-of-split associated with
YRPW motif-like protein | 0.0011791 | 1.483 | 0.0617 |
| CUBN | Cubilin | 0.0020518 | 1.461 | 0.0477 |
| SFRP2 | Secreted
frizzled-related protein 2 | 0.0025242 | 1.13 | 0.0360 |
| GADD45B | Growth arrest and
DNA-damage-inducible 45β | 0.0033662 | 1.466 | 0.0582 |
| IGFBP3 | Insulin-like growth
factor-binding protein 3 | 0.0041609 | 1.352 | 0.0841 |
| LEF1 | Lymphoid
enhancer-binding factor 1 | 0.0044962 | 1.314 | 0.1389 |
| CCNE1 | Cyclin E1 | 0.005372 | 0.627 | −0.1902 |
Association of the eight-gene
signature and patient survival in the training set
A risk-score formula was established according to
the expression of these significant CSC-associated genes and their
respective coefficients: Risk score=0.253 × LRP2 + 0.062 × HEYL +
0.048 × CUBN + 0.036 × SFRP2 + 0.058 × GADD45B + 0.084 × IGFBP3 +
0.139 × LEF1-0.190 × CCNE1. The present study calculated the
eight-gene signature risk score for each patient in the training
set. Patients with an assigned risk score were divided into
high-risk or low-risk groups using the median risk score in the
training set as the threshold. The distribution of patient risk
scores, survival status and significant gene expression level were
analyzed for the TCGA training set (Fig.
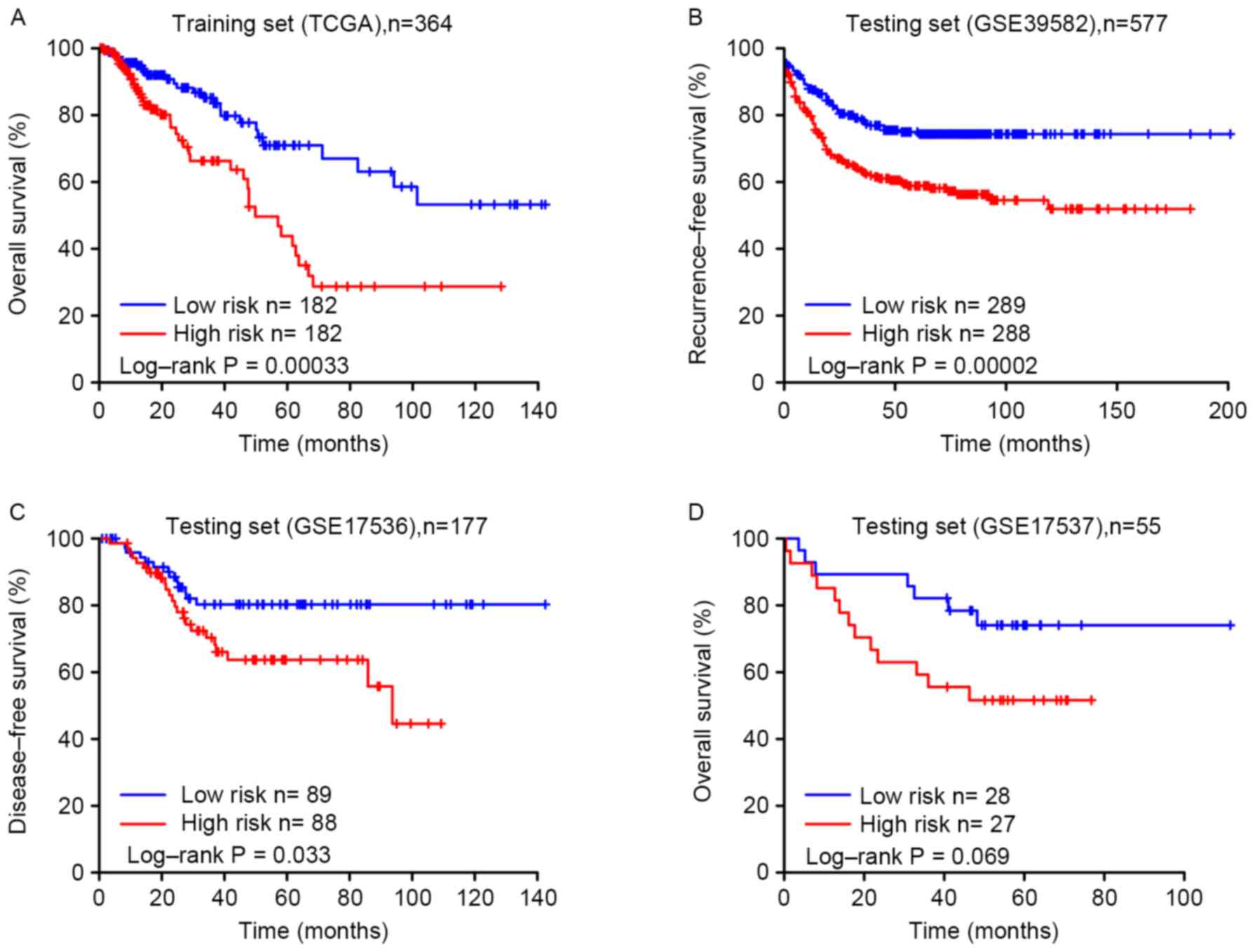
2). Patients in the high-risk group were associated with
significantly decreased overall survival compared with patients in
the low-risk group (P=0.00033; Fig.
3A). The association of the eight-gene signature risk score
with clinical outcome was significant when it was analyzed as a
continuous variable in the univariate Cox's regression model:
Overall survival (hazard ratio, 2.72; P<0.0001) and
recurrence-free survival (hazard ratio, 2.29; P<0.0001).
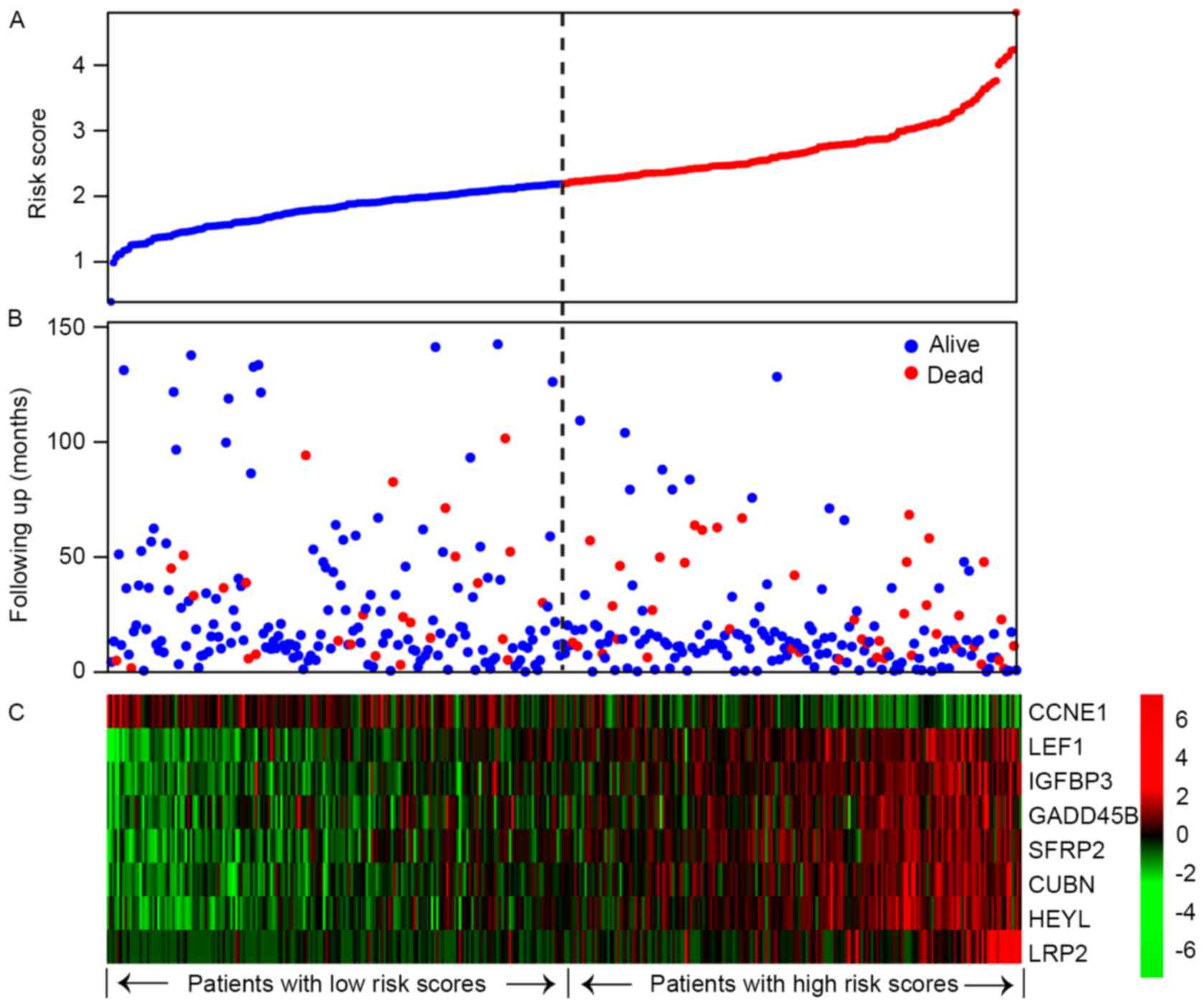 | Figure 2.Eight gene-based risk score analysis
of patient data from The Cancer Genome Atlas. The distributions of
eight gene-based risk score, patient survival status and gene
expression signature were analyzed for the training set patients
(n=364). (A) Eight gene-based risk score distribution. (B) Patient
overall survival status and time. (C) Heat map of the eight gene
expression profiles. Rows represent genes; columns represent
patients. The black dotted line represents the risk score median
threshold, which divided patients into low-risk and high-risk
groups. CCNE1, cyclin E1; LEF1, lymphoid enhancer-binding factor 1;
IGFBP3, insulin-like growth factor-binding protein 3; GADD45B,
growth arrest and DNA-damage-inducible 45β; SFRP2, secreted
frizzled-related protein 2; CUBN, cubilin; HEYL,
hairy/enhancer-of-split associated with YRPW motif-like protein;
LRP2, low-density lipoprotein-related protein 2. |
Validating the eight-gene signature
for survival prediction in the testing sets
To confirm the results of the present study, the
risk score for the testing sets, including GSE39582, GSE17536 and
GSE17537, were calculated. Similar to the TCGA training set,
patients in the high-risk group were associated with decreased
survival time compared with patients in the low-risk group
(Fig. 3B-D). In addition, patient
survival throughout the follow-up in the low-risk group was
improved compared with that in the high-risk group. The univariate
Cox's regression model revealed a similar association between risk
score and overall survival, with the high-risk group associated
with decreased overall survival compared with the low-risk
group.
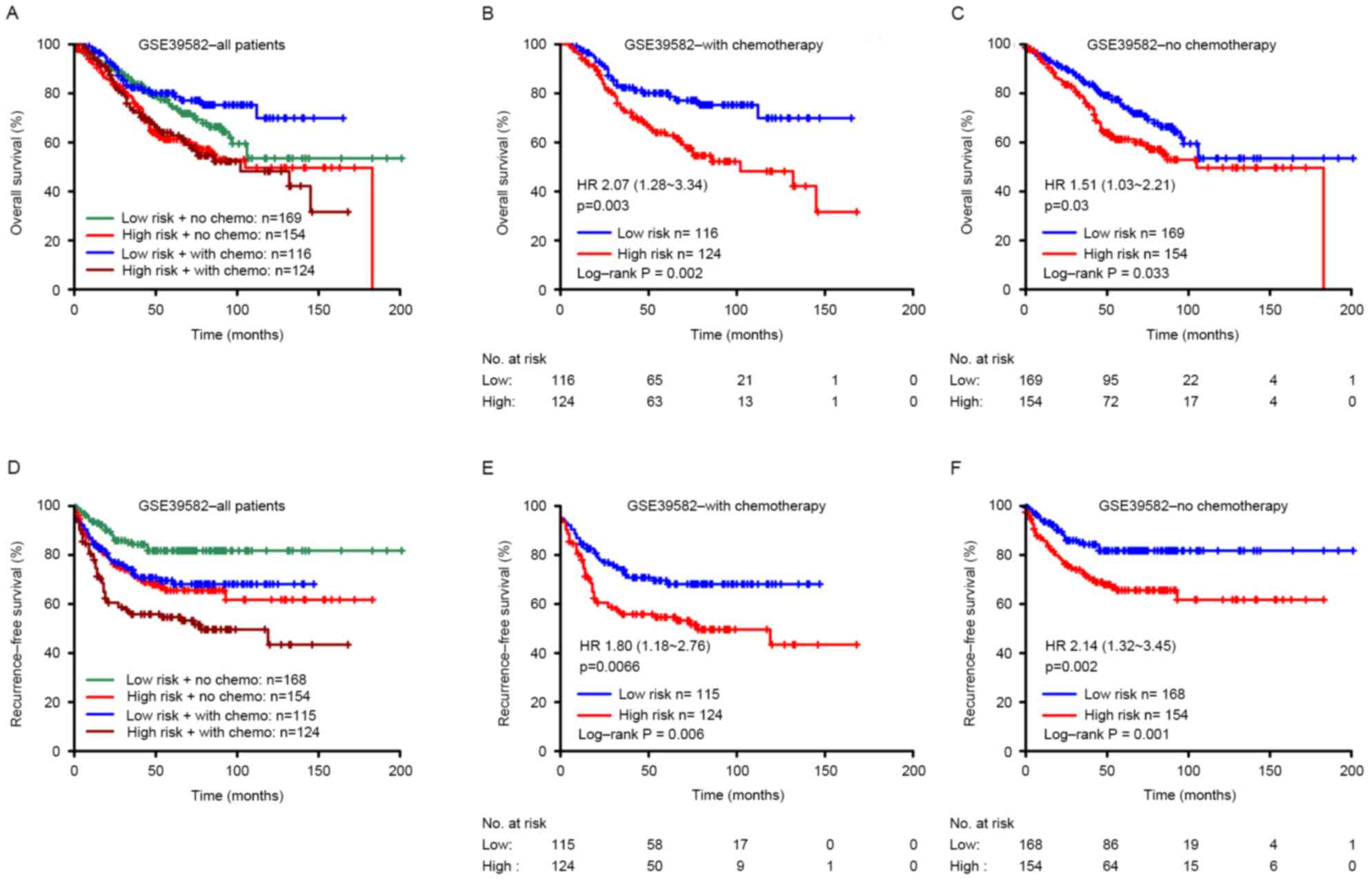
The prognostic value of the signature for the
patients with or without post-operative chemotherapy was also
assessed, according to the treatment records provided in the
original raw data in GEO. Adjuvant chemotherapy information was
available for the GSE39582 series; 240 patients received adjuvant
chemotherapy, and 323 did not. The present study revealed that the
high-risk score was significantly associated with unfavorable
overall and recurrence-free survival in patients with or without
post-operative chemotherapy (Fig.
4).
Eight-gene signature represents an
independent predictor in colorectal cancer
The present study performed Cox's univariate and
multivariate analysis to ascertain whether the eight-gene signature
represented an independent predictor of overall survival in
patients with colorectal cancer (Table
II). The effect of risk score, age, sex and stage on patient
survival time was analyzed further using a multivariate Cox's
proportional hazard model for each cohort. The results indicated
that risk score could represent an independent predictor of overall
survival when adjusted for age, sex or stage in three cohorts. In
GSE17537 (n=55), univariate Cox's analysis demonstrated that the
eight-gene model was statistically significant for prognosis,
although multivariate Cox's analysis revealed it was not.
 | Table II.Univariate and multivariate Cox
regression analyses of overall survival in each data set. |
Table II.
Univariate and multivariate Cox
regression analyses of overall survival in each data set.
|
| Univariate
model | Multivariate
model |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value |
|---|
| Training set (TCGA)
(N=364) |
|
|
|
|
|
|
| 8 gene
risk score (high risk vs. low risk) | 2.38 | 1.46–3.87 | 0.00048 | 2.19 | 1.27–3.76 | 0.0045 |
| Age
(≥65 vs. <65) | 1.85 | 1.11–3.09 | 0.0183 | 3.04 | 1.68–5.49 | 0.0002 |
| Gender
(male vs. female) | 1.52 | 0.94–2.47 | 0.0903 | 1.28 | 0.77–2.14 | 0.3465 |
| Stage
(I/II/III/IV) | 1.93 | 1.44–2.58 | 1.00E-05 | 1.91 | 1.40–2.61 | 5.01E-05 |
| Testing set
(GSE39582) (N=562) |
|
|
|
|
|
|
| 8 gene
risk score (high risk vs. low risk) | 1.59 | 1.19–2.12 | 0.0016 | 1.49 | 1.11–1.99 | 0.0072 |
| Age
(≥65 vs. <65) | 1.47 | 1.08–2.00 | 0.0146 | 1.71 | 1.25–2.33 | 0.0007 |
| Gender
(male vs. female) | 1.31 | 0.98–1.75 | 0.0684 | 1.39 | 1.04–1.86 | 0.0251 |
| Stage
(I/II/III/IV) | 1.92 | 1.57–2.34 | 1.93E-10 | 2.00 | 1.63–2.45 | 2.95E-11 |
| Testing set
(GSE17536) (N=177) |
|
|
|
|
|
|
| 8 gene
risk score (high risk vs. low risk) | 1.58 | 0.99–2.52 | 0.0555 | 1.60 | 0.99–2.59 | 0.0566 |
| Age
(≥65 vs. <65) | 0.96 | 0.60–1.52 | 0.848 | 1.33 | 0.81–2.17 | 0.2483 |
| Gender
(male vs. female) | 1.10 | 0.69–1.76 | 0.674 | 0.98 | 0.60–1.60 | 0.9405 |
| Stage
(I/II/III/IV) | 2.85 | 2.11–3.86 | 9.20E-12 | 3.00 | 2.19–4.12 | 1.02E-11 |
| Testing set
(GSE17537) (N=55) |
|
|
|
|
|
|
| 8 gene
risk score (high risk vs. low risk) | 2.30 | 0.91–5.77 | 0.0766 | 1.53 | 0.57–4.11 | 0.3992 |
| Age
(≥65 vs. <65) | 1.43 | 0.59–3.44 | 0.425 | 1.86 | 0.73–4.78 | 0.1958 |
| Gender
(male vs. female) | 0.68 | 0.28–1.66 | 0.395 | 1.16 | 0.44–3.02 | 0.7642 |
| Stage
(I/II/III/IV) | 2.97 | 1.56–5.65 | 0.0009 | 2.98 | 1.54–5.78 | 0.0012 |
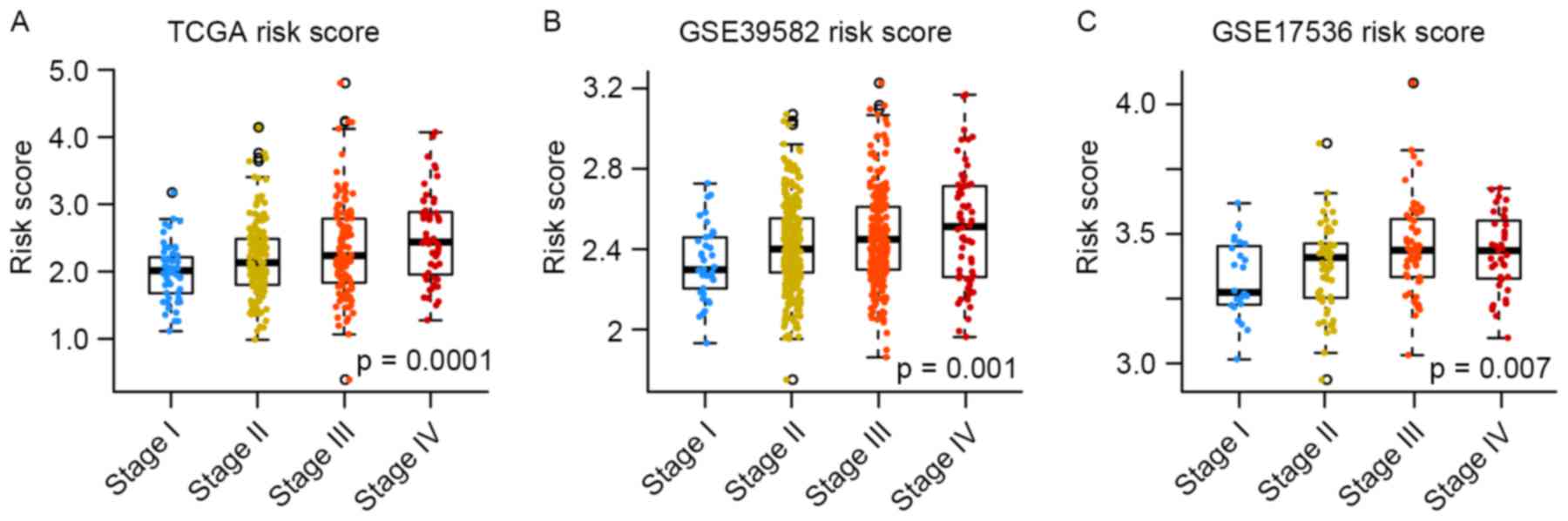
The present study also analyzed the association
between the eight-gene signature and the American Joint Committee
on Cancer stage (27). Mean risk
score increased with tumor malignance in the training and testing
sets (Fig. 5). Therefore, the risk
score model of the present study may facilitate stratifying
patients with colorectal cancer.
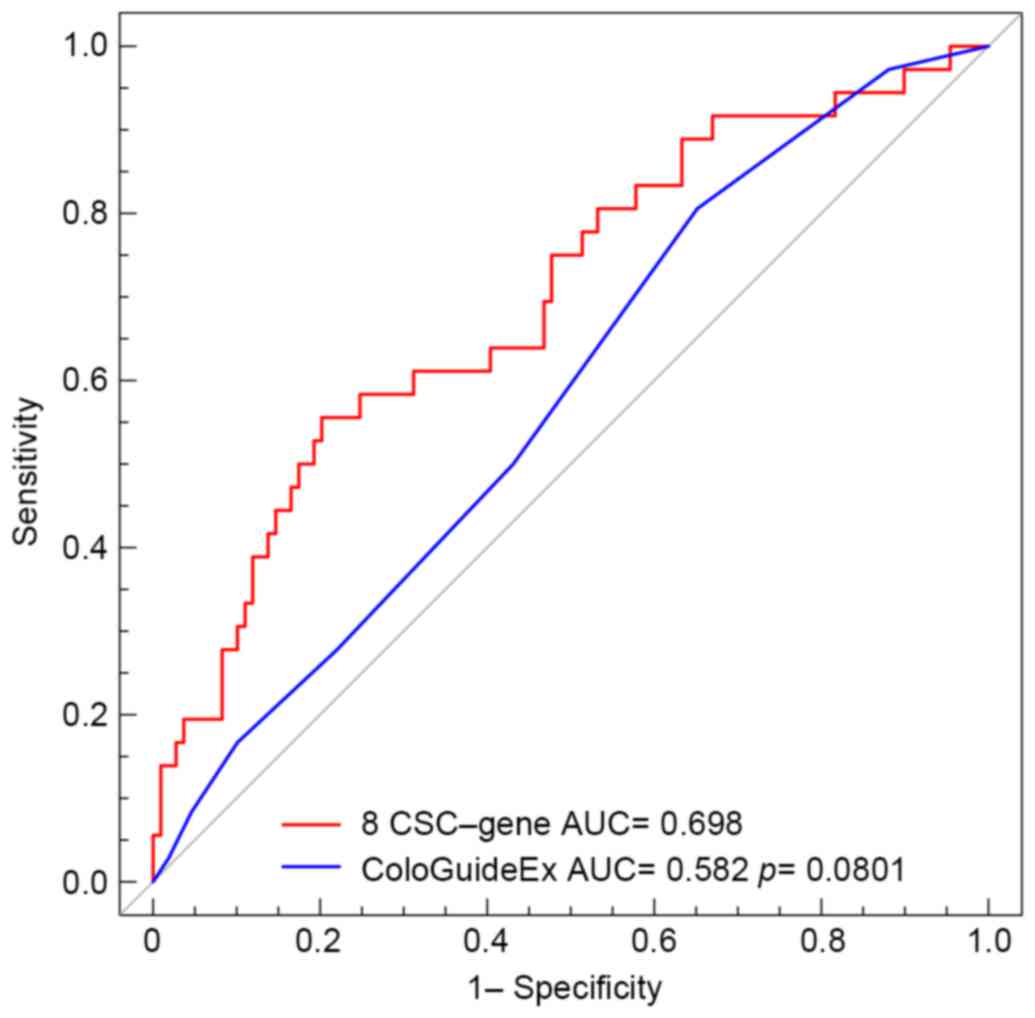
Evaluating the risk score using ROC
analysis
ROC analysis was performed to compare the
sensitivity and specificity of disease-free survival prediction of
the eight-gene signature model and the ColoGuideEx risk model for
the GSE17536 dataset. The area under ROC (AUROC) was determined and
compared between these two prognostic factors (Fig. 6). The ROC curves indicated that the
AUROCs of the eight-gene signature and the ColoGuideEx were 0.698
and 0. 582, respectively (P=0.0801). These results indicated that
the eight-gene signature model may possess increased predictive
power compared with the ColoGuideEx model.
Discussion
The present study analyzed cancer stem cell
signaling gene expression profiles from TCGA and the GEO database.
By assessing the association between gene expression profiling and
the clinical outcome of patients with colorectal cancer, the
present study identified an eight-gene Hedgehog-Notch-Wnt signaling
signature that was associated with the overall survival of the
patients. The present study further validated the prognostic power
of the eight-gene signature in independent cohorts and demonstrated
that the eight-gene signature represents an independent predictor
in colorectal cancer. This eight-gene signature requires further
study in larger cohorts.
Characteristics of the eight
CSC-associated genes
According to the predictive model of the present
study, CCNE1 expression level was positive associated with good
survival, whereas the expression of seven other genes were
upregulated in the high-risk group compared with that in the
low-risk group. CCNE1 has been reported as a tumor suppressor gene
in multiple types of cancer (28).
However, in the present study, high expression of CCNE1 was
associated with increased survival in patients with colorectal
cancer in TCGA and the GEO datasets (data not shown). The
non-robustness could be due to the same gene functioning
differently in different types or stages of cancer (29).
LRP2 is a component and an auxiliary Hedgehog
signaling receptor (30). Andersen
et al (31) reported that LRP2
regulated melanoma cell proliferation and survival rates, and that
knocking down LRP2 induced apoptosis. LRP2 may serve a similar
function in colorectal cancer. HEYL is regarded as a Notch
effector, and upregulated HEYL expression has been reported in
ovarian, breast and colon cancer (32). Han et al (33) demonstrated that HEYL was associated
with Smad protein expression, transforming growth factor β
signaling, and initiating and progressing breast cancer. SFRP2 may
augment Wnt16B signaling to promote a malignant phenotype and
therapeutic resistance in the damaged tissue microenvironment, and
increased SFRP2 expression has been reported to be associated with
a poorer clinical outcome in colorectal cancer (34). LEF1, IGFBP3 and GADD45B, which were
also reported to participate in stem cell signaling, have been
reported to be upregulated in multiple types of cancer tissue, and
possess growth-promoting functions in colon cancer cells (35–37).
Cubilin was reported as a Wnt signaling target and modulator of the
fibroblast growth factor (FGF) 8-FGF receptor signaling pathway,
although this has not been demonstrated to be associated with a
type of tumor (38).
The present study also performed a network analysis
based on the eight signature genes in TCGA colorectal cancer
cohorts downloaded from cBioPortal (see http://www.cbioportal.org/ for further pre-processing
information). Default parameters were used to construct an
interaction network of eight signature genes (data not shown).
Certain well-studied genes in colorectal cancer, including APC,
B-Raf proto-oncogene, tumor protein p53, MYC proto-oncogene, RB
transcriptional corepressor 1 and SMAD family member 4 exhibited
closed interaction with the eight signature genes. Further study of
the function of these genes is required to understand colorectal
cancer tumorigenesis and development.
Eight-gene signature possesses
prognostic power in patients with colorectal cancer undergoing
adjuvant chemotherapy
Since the majority of the previous prognostic gene
signatures were dominated by proliferation genes, the present study
focused on cancer stem cell signaling pathways and provided novel
insights into how cancer stem cell signaling contributes to
colorectal cancer development. The eight-gene signature predicted
unfavorable overall survival in those undergoing post-operative
chemotherapy in the GSE39582 cohorts. Patients with an increased
CSC-associated gene risk score were associated with adjuvant
chemotherapy resistance. The present study proposed that this
eight-gene signature reflects the activation state of tumor stem
cell signaling, which may serve a crucial function in drug
resistance and treatment outcome. Patients who have tumors with a
high-risk CSC-associated gene signature could benefit from combined
stem cell-targeted adjuvant therapy using this model, although
further studies are required to verify this.
ROC and sensitivity
Further survival ROC analysis demonstrated that the
eight-gene signature was comparable with the ColoGuideEx model
(P=0.0801) for prognosis in GSE17536 sets, which were used in
ColoGuideEx as the largest external validation datasets. Although
the ColoGuideEx model is not typically used in deciding whether
adjuvant chemotherapy is appropriate for a given patient with
colorectal cancer (16), the test has
been validated and rendered financially feasible for clinical
practice using reverse transcription-PCR (39). The eight-gene signature of the present
study could suffice in predicting the outcome of colorectal cancer,
and predicts survival with increased accuracy compared with that
predicted using ColoGuideEx.
Limitations of the present study
The limitations of the present study should be
acknowledged. First, gene expression profile data were obtained
from different platforms: TCGA for RNA-seq and GEO for microarray.
Normalization methods differed between TCGA and GEO datasets.
Secondly, only 334 cancer stem cell-associated genes were included
in the present study, and the prognostic genes identified may not
represent all cancer stem cell-associated gene candidates
associated with overall survival in patients with colorectal
cancer. Thirdly, more patient information and larger prospective
patient cohorts are required to confirm the prognostic power of the
eight-gene signature.
The present study developed a CSC-associated
eight-gene signature that predicted colorectal cancer prognosis in
four independent datasets. Further analysis revealed that the
eight-gene signature may represent an independent prognostic factor
with respect to age and stage. The eight-gene signature may assist
molecular classification and outcome prediction in colorectal
cancer. Further study is required to improve this eight-gene
signature model in colorectal cancer.
Acknowledgements
The present study was supported by the National High
Technology Research and Development Program of China (863 Program;
grant no. 2015AA020104).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. Ca Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schneider M, Huber J, Hadaschik B, Siegers
GM, Fiebig HH and Schüler J: Characterization of colon cancer
cells: A functional approach characterizing CD133 as a potential
stem cell marker. BMC Cancer. 12:962012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Asfaha S, Hayakawa Y, Muley A, Stokes S,
Graham TA, Ericksen RE, Westphalen CB, von Burstin J, Mastracci TL,
Worthley DL, et al: Krt19(+)/Lgr5(−) cells are radioresistant
cancer-initiating stem cells in the colon and intestine. Cell Stem
Cell. 16:627–638. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pang R, Law WL, Chu AC, Poon JT, Lam CS,
Chow AK, Ng L, Cheung LW, Lan XR, Lan HY, et al: A subpopulation of
CD26+ cancer stem cells with metastatic capacity in human
colorectal cancer. Cell Stem Cell. 6:6032010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pattabiraman DR and Weinberg RA: Tackling
the cancer stem cells-what challenges do they pose? Nat Rev Drug
Discov. 13:497–512. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Varnat F, Siegl-Cachedenier I, Malerba M,
Gervaz P and Ruiz i Altaba A: Loss of WNT-TCF addiction and
enhancement of HH-GLI1 signalling define the metastatic transition
of human colon carcinomas. EMBO Mol Med. 2:440–457. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad
A, Banerjee S, Azmi AS, Miele L and Sarkar FH: Notch-1 induces
epithelial-mesenchymal transition consistent with cancer stem cell
phenotype in pancreatic cancer cells. Cancer Lett. 307:26–36. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao C, Chen A, Jamieson CH, Fereshteh M,
Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, et al:
Hedgehog signalling is essential for maintenance of cancer stem
cells in myeloid leukaemia. Nature. 458:776–779. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vermeulen L, De Sousa E, Melo F, van der
Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M,
Merz C, Rodermond H, et al: Wnt activity defines colon cancer stem
cells and is regulated by the microenvironment. Nat Cell Biol.
12:468–476. 2015. View
Article : Google Scholar
|
|
10
|
Takebe N, Miele L, Harris PJ, Jeong W,
Bando H, Kahn M, Yang SX and Ivy SP: Targeting Notch, Hedgehog, and
Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin
Oncol. 12:445–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takebe N, Harris PJ, Warren RQ and Ivy SP:
Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog
pathways. Nat Rev Clin Oncol. 8:97–106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He J, Sheng T, Stelter AA, Li C, Zhang X,
Sinha M, Luxon BA and Xie J: Suppressing Wnt signaling by the
hedgehog pathway through sFRP-1. J Biol Chem. 281:35598–35602.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clark-Langone KM, Sangli C, Krishnakumar J
and Watson D: Translating tumor biology into personalized treatment
planning: Analytical performance characteristics of the Oncotype DX
colon cancer assay. BMC Cancer. 10:6912010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gray RG, Quirke P, Handley K, Lopatin M,
Magill L, Baehner FL, Beaumont C, Clark-Langone KM, Yoshizawa CN,
Lee M, et al: Validation study of a quantitative multigene reverse
transcriptase-polymerase chain reaction assay for assessment of
recurrence risk in patients with stage II colon cancer. J Clin
Oncol. 29:4611–4619. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salazar R, Roepman P, Capella G, Moreno V,
Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga
J, et al: Gene expression signature to improve prognosis prediction
of stage II and III colorectal cancer. J Clin Oncol. 29:17–24.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agesen TH, Sveen A, Merok MA, Lind GE,
Nesbakken A, Skotheim RI and Lothe RA: ColoGuideEx: A robust gene
classifier specific for stage II colorectal cancer prognosis. Gut.
61:1560–1567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cline MS, Craft B, Swatloski T, Goldman M,
Ma S, Haussler D and Zhu J: Exploring TCGA pan-cancer data at the
UCSC cancer genomics browser. Sci Rep. 3:26522013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu X, Wan F, Zhang H, Shi G and Ye D:
ITGA2B and ITGA8 are predictive of prognosis in clear cell renal
cell carcinoma patients. Tumor Biol. 37:253–262. 2016. View Article : Google Scholar
|
|
19
|
Marisa L, Reyniès AD, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Freeman TJ, Smith JJ, Chen X, Washington
MK, Roland JT, Means AL, Eschrich SA, Yeatman TJ, Deane NG and
Beauchamp RD: Smad4-mediated signaling inhibits intestinal
neoplasia by inhibiting expression of β-catenin. Gastroenterology.
142:562–571.e2. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo T, Dunphy PS, Lina TT and McBride JW:
Ehrlichia chaffeensis exploits canonical and noncanonical host wnt
signaling pathways to stimulate phagocytosis and promote
intracellular survival. Infect Immun. 84:686–700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Simon R, Lam A, Li MC, Ngan M, Menenzes S
and Zhao Y: Analysis of gene expression data using BRB-ArrayTools.
Cancer Inform. 3:11–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kawaguchi A, Iwadate Y, Komohara Y, Sano
M, Kajiwara K, Yajima N, Tsuchiya N, Homma J, Aoki H, Kobayashi T,
et al: Gene expression signature-based prognostic risk score in
patients with primary central nervous system lymphoma. Clin Cancer
Res. 18:5672–5681. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK,
Ho AS, Lui WM, Fung CF, Wong TS and Leung GK: A long non-coding RNA
signature in glioblastoma multiforme predicts survival. Neurobiol
Dis. 58:123–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin M, Li P, Zhang Q, Yang Z and Fu S: A
four-long non-coding RNA signature in predicting breast cancer
survival. J Exp Clin Cancer Res. 33:842014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lan YT, Yang SH, Chang SC, Liang WY, Li
AF, Wang HS, Jiang JK, Chen WS, Lin TC and Lin JK: Analysis of the
seventh edition of American joint committee on colon cancer
staging. Int J Colorectal Dis. 27:657–663. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Keyomarsi K, Tucker SL, Buchholz TA,
Callister M, Ding Y, Hortobagyi GN, Bedrosian I, Knickerbocker C,
Toyofuku W, Lowe M, et al: Cyclin E and survival in patients with
breast cancer. N Engl J Med. 347:1566–1575. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pils D, Bachmayr-Heyda A, Auer K, Svoboda
M, Auner V, Hager G, Obermayr E, Reiner A, Reinthaller A, Speiser
P, et al: Cyclin E1 (CCNE1) as independent positive prognostic
factor in advanced stage serous ovarian cancer patients-a study of
the OVCAD consortium. Eur J Cancer. 50:99–110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Christ A, Christa A, Kur E, Lioubinski O,
Bachmann S, Willnow TE and Hammes A: LRP2 is an auxiliary SHH
receptor required to condition the forebrain ventral midline for
inductive signals. Dev Cell. 22:268–278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Andersen RK, Hammer K, Hager H,
Christensen JN, Ludvigsen M, Honoré B, Thomsen MB and Madsen M:
Melanoma tumors frequently acquire LRP2/megalin expression, which
modulates melanoma cell proliferation and survival rates. Pigment
Cell Melanoma Res. 28:267–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu C, Bonome T, Li Y, Kamat AA, Han LY,
Schmandt R, Coleman RL, Gershenson DM, Jaffe RB, Birrer MJ and Sood
AK: Gene alterations identified by expression profiling in
tumor-associated endothelial cells from invasive ovarian carcinoma.
Cancer Res. 67:1757–1768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han L, Diehl A, Nguyen NK, Korangath P,
Teo W, Cho S, Kominsky S, Huso DL, Feigenbaum L, Rein A, et al: The
Notch pathway inhibits TGFβ signaling in breast cancer through
HEYL-mediated crosstalk. Cancer Res. 74:6509–6518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun Y, Zhu D, Chen F, Qian M, Wei H, Chen
W and Xu J: SFRP2 augments WNT16B signaling to promote therapeutic
resistance in the damaged tumor microenvironment. Oncogene.
35:4321–4334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
O'Connell MJ, Lavery I, Yothers G, Paik S,
Clark-Langone KM, Lopatin M, Watson D, Baehner FL, Shak S, Baker J,
et al: Relationship between tumor gene expression and recurrence in
four independent studies of patients with stage II/III colon cancer
treated with surgery alone or surgery plus adjuvant fluorouracil
plus leucovorin. J Clin Oncol. 28:3937–3944. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Georges RB, Adwan H, Hamdi H, Hielscher T,
Linnemann U and Berger MR: The insulin-like growth factor binding
proteins 3 and 7 are associated with colorectal cancer and liver
metastasis. Cancer Biol Ther. 12:69–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hsieh TH, Hsu CY, Tsai CF4, Chiu CC, Liang
SS, Wang TN, Kuo PL, Long CY and Tsai EM: A novel cell-penetrating
peptide suppresses breast tumorigenesis by inhibiting
β-catenin/LEF-1 signaling. Sci Rep. 6:191562016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cases O, Perea-Gomez A, Aguiar DP, Nykjaer
A, Amsellem S, Chandellier J, Umbhauer M, Cereghini S, Madsen M,
Collignon J, et al: Cubilin, a high affinity receptor for
fibroblast growth factor 8, is required for cell survival in the
developing vertebrate head. J Biol Chem. 288:16655–16670. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Park YY, Lee SS, Lim JY, Kim SC, Kim SB,
Sohn BH, Chu IS, Oh SC, Park ES, Jeong W, et al: Comparison of
prognostic genomic predictors in colorectal cancer. PLoS One.
8:e607782013. View Article : Google Scholar : PubMed/NCBI
|