Introduction
Peritoneal metastasis is a key process during the
development of chemo-resistance and postoperative relapse in
ovarian cancer and leads to a poor prognosis and a high mortality
rate (1,2). Based on observations in clinical
practice, ~80% of patients with ovarian cancer were diagnosed with
peritoneal metastasis with a considerable proportion of the cases
developed during early or medium stages of ovarian cancer
progression (3). Changes in gene
expression, intracellular signal transduction and cell morphology
during ovarian cancer cell metastasis are closely associated with
the gain of stemness, maintenance and resistance to chemotherapy-
or radiation-induced apoptosis, suggesting that these tumor
biological characteristics are governed by number of conserved
mechanisms (4–6).
Somatic copy number variation and change in the
level of transcription are well-recognized key steps in the
progression of ovarian cancer and other types of cancer (7–9). MicroRNAs
(miRNAs/miRs) together with other non-coding RNAs comprise a
sophisticated gene expression regulatory network. The disorder or
alteration of the gene expression regulatory network often leads to
the pathogenesis of different diseases, particularly those largely
affected by changes in gene expression, including inflammatory
disease and cancer (10–12). The alterations in the expression of
different miRNAs have been linked to ovarian cancer progression in
different aspects, such as proliferation, invasion and metastasis
(13–15). However, current investigations are
mainly in vitro experiments that are focused on individual
miRNAs using cancer cell lines, yielding results unable to
accurately reflect the effects in patients and hard to be
translated into therapeutic strategy (13–15).
Another limitation of the majority of current
studies focusing on miRNAs in ovarian cancer is that microRNAs not
only deposit in cells but also in extracellular vesicles, including
microvesicles and exosomes (16).
Exosomes as important extracellular microenvironment components can
be secreted from and received by different cells, transporting
contents, including effector proteins and RNAs between cells, and
thus mediating intercellular signal transduction (16). Focusing only on miRNAs in cancer cells
may therefore miss important information. It was raised by several
studies that exosomes may have an important role in regulating
ovarian cancer metastasis and development (17–19).
The aim and scope of the present study was to
identify miRNAs in exosome that may function as promoters of
oncogenesis or peritoneal metastasis of ovarian cancer. By
comparing the miRNA expression patterns in exosomes that were
isolated from 10 samples of ascites from patients with epithelial
ovarian cancer (EOC) and 10 non-cancerous (NC) peritoneal lavage
samples. It was revealed that two exosomal miRNAs, miR-149-3p and
miR-222-5p, were upregulated in ovarian cancer ascites-derived
exosomes compared with the non-cancerous samples. It was further
confirmed that the increased expression levels of these two miRNAs
significantly correlated with the survival of patients with ovarian
cancer, suggesting the prognostic value of the two miRNAs for
malignant ovarian cancer.
Materials and methods
Participants and isolation of
peritoneal exosomes
The present study was approved by the Medical Ethics
Committee of the Second Affiliated Hospital of Zhengzhou University
(Henan, China). A total of 10 patients with peritoneal metastatic
epithelial ovarian cancer (EOC group) and 10 subjects without
epithelial ovarian cancer with acute pelvic peritonitis (NC group)
were enrolled in the present study. The clinical data of the
participants are described in Table
I. All participants were recruited at The Second Affiliated
Hospital of Zhengzhou University (Zhengzhou, China), and written
informed consent was obtained from each participant prior to
enrolment. Peritoneal exosomes were isolated from a sample of
ascites from each patient with EOC or a peritoneal lavage sample
from subjects in the NC group using ExoQuick™ exosome
precipitation solution (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) following the manufacturer's instructions.
Briefly, each sample of ascites or peritoneal lavage specimen was
pre-cleared by centrifugation at 3,000 × g for 15 min under 4°C to
remove cells or cell debris followed by mixing with precipitation
solution on a 4:1 ratio (v/v). The mixture was then refrigerated
under 4°C for 12 h, after which the exosomes in the mixture were
pelleted by centrifugation at 1,500 × g for 30 min under 4°C. The
exosome pellet extracted from each sample was then re-suspended in
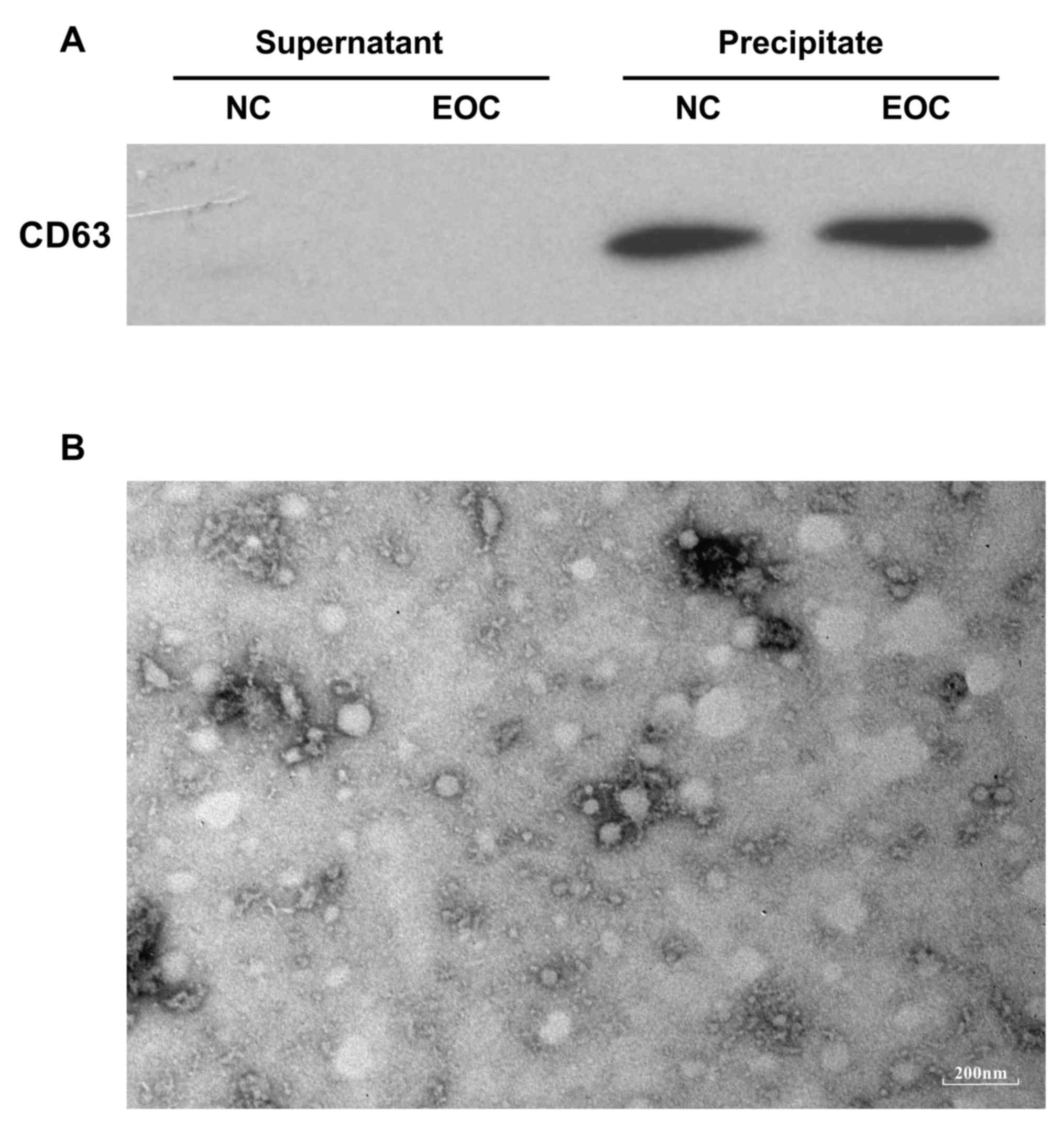
200 µl PBS, validated by a transmission electron microscopy and
stored under 4°C for ≤1 h before further analysis. A total of 30 µl
supernatant from exosome isolation and 20 µl of each exosome sample
suspended in PBS were subjected to western blotting using a mouse
monoclonal antibody against human CD63 (dilution of 1:1,000)
(catalog no. ab213090; Abcam, Cambridge, UK) to evaluate the
efficiency of exosome precipitation. In western blotting, the
ProteoPrep® Total Extraction Sample kit (Sigma-Aldrich,
Merck KGaA Darmstadt, Germany) was to separate proteins from
exosomes according to the manufacturer's protocol. Samples were
normalized for protein concentration by a BCA assay. The volumes
(20 µg) of each sample were loaded on a acrylamide-bisacrylamide
(10 or 12.5%) gel. Following electrophoresis, the protein was
transfected with PVDF membrane. Skimmed milk powder (5%) blocked
PVDF for 3 h at room temperature. The secondary antibody (Goat
Anti-mouse IgG H&L, catalog no. ab6785, Abcam, Cambridge, UK)
has a dilution of 1:5,000, incubated in TBS-T buffer (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) for 1
h at room temperature. Proteins were visualized using an enhanced
chemiluminescence solution (Life Sciences; Thermo Fisher
Scientific, Inc.).
 | Table I.Thermocycling conditions for
quantitative polymerase reaction. |
Table I.
Thermocycling conditions for
quantitative polymerase reaction.
| Cycles | Steps | Temperature
(°C) | Time | Detection |
|---|
| 1 | Initial
denaturation | 95 | 10 min | No |
| 40 | Denaturation | 95 | 10 sec | No |
|
| Annealing | 75 | 20 sec | No |
|
| Extension | 72 | 20 sec | Yes |
High-throughput sequencing of
peritoneal exosomal miRNAs
A total of 2 sequencing libraries of peritoneal
exosomal miRNAs from EOC- or NC-group specimens were constructed
using a sequencing library construction kit provided by Gene Denovo
(Guangzhou, China) following manufacturer's instructions. Briefly,
10 samples of PBS-suspended exosomes (80 µl) from participants in
each group were mixed, and total RNA was extracted from the mixture
using TRIzol. Small RNAs with 18–30 nucleotides were separated by
SDS-PAGE (15%). After the ligation of 5′ and 3′ adapters to both
ends of small RNAs by nested-polymerase chain reaction (PCR), the
ligation products were reversely transcribed and amplified by PCR.
The products (size, 140–160 bp) were further separated by agarose
gel (3.5%) to generate the cDNA library. A total of 2 cDNA
libraries generated from EOC or NC samples were then subjected to
high throughput sequencing by Gene Denovo using the Illumina
HiSeqTM 2500 system (Illumina, Inc., San Diego, CA, USA).
Pre-processing of raw data and miRNA
identification
To obtain small RNA clean reads, raw reads data were
first filtered using the steps as follows: i) Removal of
low-quality reads containing >1 low-quality (quality score ≤20)
base or containing unknown nucleotides (N); ii) removal of reads
without 3′ or 5′ adapters; iii) Removal of reads containing 3′ and
5′ adapters but no small RNA fragments; iv) removal of reads
containing ploy-A sequence within small RNA fragments and v)
removal of reads that are shorter than 18 nucleotides.
All clean reads were then aligned to human small RNA
sequence data that were obtained from GeneBank (release 209.0,
http://www.ncbi.nlm.nih.gov/genbank/)
and Rfam databases (version 11.0, http://rfam.xfam.org/) to filter any ribosomal RNA,
small cytoplasmic RNA, small nucleolar RNA, small nuclear RNA or
transfer RNA. The clean reads were also aligned to a reference
human genome [Homo sapiens (assembly GRCh38.p12)] to remove reads
mapped to exons or introns that might be fragments from mRNA
degradation or reads mapped to repeat sequences. Filtered small RNA
clean reads were then referred to as clean tags, which were mapped
in miRBase database (release 21, http://www.mirbase.org/index.shtml) to annotate any
known human miRNA.
Analysis of miRNA expression
The expression level of every annotated miRNA in EOC
or NC sample group was first normalized to transcripts per million
(TPM) using the following formula:
TPM=counts of miRNATotal counts of clean
tags×106
and A log2 (FC) value of each annotated
miRNA in NC or EOC sample group was calculated using the following
formula:
log2(FC)=log2(TPMEOCTPMNC)
where TPMEOC and TPMNC were
TPM of a miRNA in EOC and NC samples, respectively, and all TPM=0
were adjusted to TPM=0.01 to avoid infinity error. miRNAs with
log2 (FC) >1 OR <-1 between EOC and NC samples
were considered upregulated or downregulated, respectively. The
annotated miRNAs were clustered according to their expression
patterns to generate the heat map of miRNA expression in NC and EOC
samples.
Gene set enrichment analysis of the
target genes of miRNAs
Target gene candidates of annotated miRNAs in each
sample group were first predicted using RNAhybrid (version 2.1.2,
http://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/),
miRanda (version. 3.3a, http://miranda.org.uk/) and TargetScan (version. 7.0,
http://genes.mit.edu/targetscan.test/ucsc.html)
databases. The genes predicted using all three databases were
considered potential target genes. The preliminary miRNA pathway
enrichment analysis was performed using the DIANA-miRPath (version
3, http://snf-515788.vm.okeanos.grnet.gr/) online
software. DIANA-miRPath performs GO and KEGG enrichment analysis of
genes in TarBase (version 7, http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index)
that have been validated as targets of given miRNAs. For Gene
Ontology (GO) enrichment analysis, predicted target genes were
mapped to GO terms in the Gene Ontology database (http://www.geneontology.org/). Significantly enriched
GO terms were determined by an adjusted P<0.05 that was
calculated by hyper-geometric distribution and false discovery rate
correction. For Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis, predicted miRNAs target genes were
mapped to KEGG annotation in the KEGG database (http://www.genome.jp/kegg/pathway.html),
and significantly enriched KEGG annotations were determined using
the same method as that for Gene Ontology enrichment analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) of miR-149-3p and miR-222-5p in
exosome samples
The expression levels of miR-149-3p or miR-222-5p in
exosomes from 10 patients with epithelial ovarian cancer (EOC
group) and 10 subjects without epithelial ovarian cancer (NC group)
were evaluated by RT-qPCR using a custom-made microRNA qPCR kit
(GeneCopoeia, Inc., Rockville, MD, USA) following the
manufacturer's instructions. miRNAs were first reverse transcribed
using a reverse transcription reaction (miRNA First-Strand cDNA
Synthesis kit, GeneCopoeia, Inc., Rockville, MD, USA) mix at 37°C
for 60 min followed by inactivation at 85°C for 5 min. The primer
sequence for has-miR-149-3p detection was
5′-GGCUCCGUGUCUUCACUCCCAAA-3′ and for has-miR-222-5p was
5′-AGUAGCCAGUGUAGAUCCUAAA-3′. qPCR reaction was performed following
a standard three-step method as described in Table I.
The qPCR reaction was performed using a SimpliAmp
thermocycler (Thermo Fisher Scientific, Inc.) and was monitored by
a SYBR Green fluorescent dye. Semi-quantitative analysis was first
performed to normalize the expression level of detected microRNAs
to snRNA U6 (GeneCopoeia, Inc.) in each sample using the
2−ΔΔCq method. The fold change (FC) of microRNA
expression level in each EOC-group sample was compared to the mean
expression level in the NC group samples (20).
Kaplan-Meier curve analysis and
statistical analysis
The associations between the expression of specific
miRNAs and the survival of patients with ovarian cancer were
investigated by Kaplan-Meier curve (with a log-rank test) that was
generated using the UCSC Xena browser (http://xena.ucsc.edu/). The sequencing data from 595
publically available samples from patients with ovarian cancer and
clinical record from The Cancer Genome Atlas ovarian cancer
database (https://cancergenome.nih.gov/) were analyzed. Graphpad
Prism software (version 7.0; GraphPad Software, Inc., La Jolla, CA,
USA) was employed for statistical analysis unless otherwise
indicated. Student's t-test was used for significance test, and
P<0.05 was considered statistically significant.
Results
Differentially expressed peritoneal
exosomal miRNAs between participants without cancer and patients
with epithelial ovarian cancer
To characterize the differences in peritoneal
exosomal miRNA expression patterns between NC participants and
patients with EOC, exosomes were isolated from 10 samples of
ascites from patients with ovarian cancer and 10 peritoneal lavage
samples from participants without cancer. The clinical information
of these patients is shown in Table
II. Following total exosome precipitation as verified by
western blotting and transmission electron microscopy (Fig. 1), small RNAs from two groups of
exosome samples were extracted, and differentially expressed miRNA
between NC and EOC sample were detected by next-generation
sequencing. Subsequently, bioinformatic analysis was performed. As
described in the materials and method section, annotated miRNAs
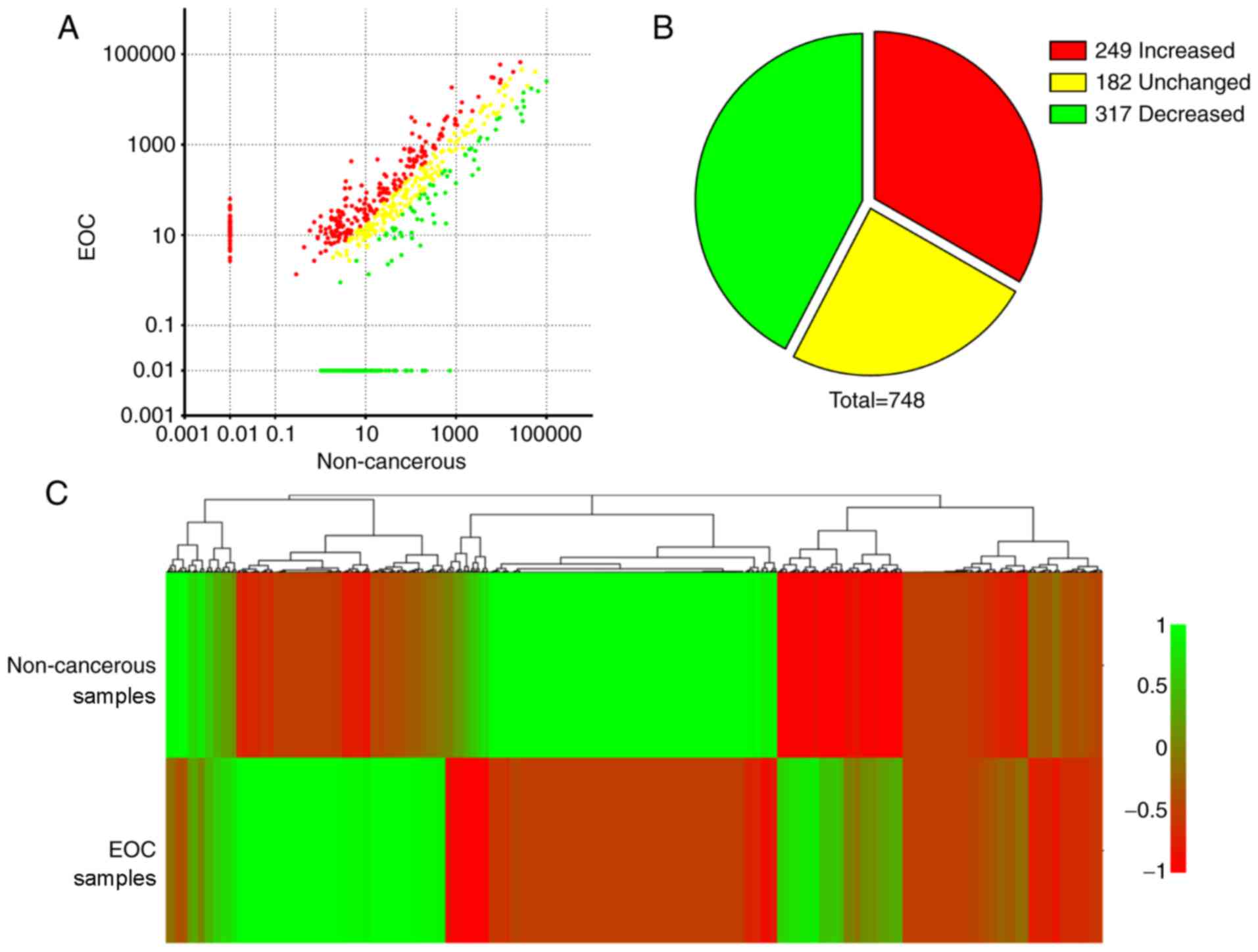
with TPMEOC two-fold higher compared with
TPMNC with log2 (FC)>1 were considered
upregulated in EOC samples compared to NC samples. Similarly,
miRNAs with log2 (FC)<-1 were determined to be
downregulated. Among the 748 annotated miRNAs, 249 miRNAs were
upregulated and 317 were downregulated in EOC samples compared with
NC samples (Fig. 2A and B). These
differentially expressed miRNAs were clustered according to changes
in expression level in a heat map (Fig.
2C) to further illustrate that there are different patterns of
miRNA expression between NC and EOC samples. These data indicated
that miRNAs in peritoneal exosomes were differentially expressed
between NC participants and patients with EOC.
 | Table II.Clinical data of patients with
epithelial ovarian cancer (n=10) and subjects without epithelial
ovarian cancer (n=10). |
Table II.
Clinical data of patients with
epithelial ovarian cancer (n=10) and subjects without epithelial
ovarian cancer (n=10).
| Variables | EOC | NC |
|---|
| Nο. of
participants | 10 | 10 |
| Age |
|
|
|
<48 | 3 | 6 |
|
≥48 | 7 | 4 |
| Pathology |
| N/A |
|
Benign | 0 | N/A |
|
Borderline | 3 | N/A |
|
Malignant | 7 | N/A |
| Histological
grade |
| N/A |
|
Well-differentiated | 1 | N/A |
|
Moderately differentiated | 7 | N/A |
| Poorly
differentiated | 2 | N/A |
| FIGO stages | | N/A |
|
I–II | 3 | N/A |
|
III–VI | 7 | N/A |
| Volume of
ascites | | N/A |
|
Nonesmall | 2 | N/A |
|
Moderate | 3 | N/A |
|
Large | 5 | N/A |
GO enrichment and KEGG pathway
enrichment analysis of the target genes of differentially expressed
miRNAs
miRNAs regulate the expression level of their
specific target genes mostly by decreasing the efficiency of
protein translation, therefore affecting different biological
processes in cells. Considering that peritoneal exosomes are
secretory extracellular vesicles from cells within the abdominal
cavity and that exosomal contents may affect the biological
processes in recipient cells when fused with exosome, changes in
the expression of peritoneal exosomal miRNAs may therefore reflect
or cause physiopathological changes in peritoneal cells and the
associated microenvironment.
To investigate the potential effect of changes in
the expression pattern of peritoneal exosomal miRNAs on EOC
oncogenesis or abdominal cavity metastasis, GO enrichment and KEGG
pathway enrichment analysis were performed on the predicted target
genes of upregulated and downregulated miRNAs. A total of 44,955
genes were identified as target genes of all differentially
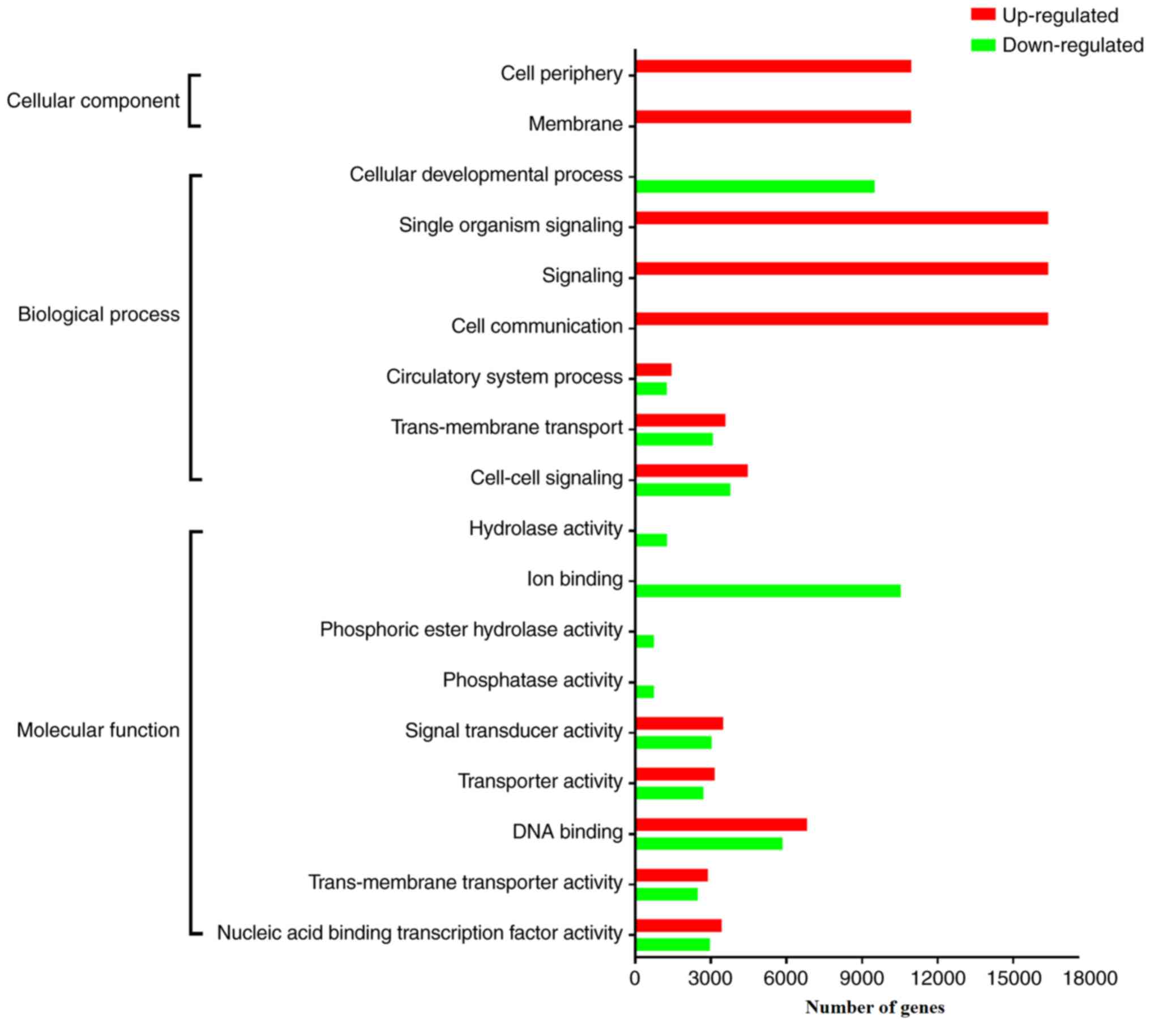
expressed miRNAs. The significantly enriched GO terms of the genes
targeted by upregulated and downregulated miRNAs are summarized in
Tables III and IV, respectively, and further compared in
Fig. 3. Notably, the genes targeted
by upregulated miRNAs encrypt proteins that are mostly located on
the cell membrane or in the extracellular microenvironment (EM)
that is involved in signaling and cell communication, implying that
the upregulation of these miRNAs might inhibit cell-to-cell or
cell-to-EM communication. To get a better understanding of the
effect of the change in the expression levels of peritoneal
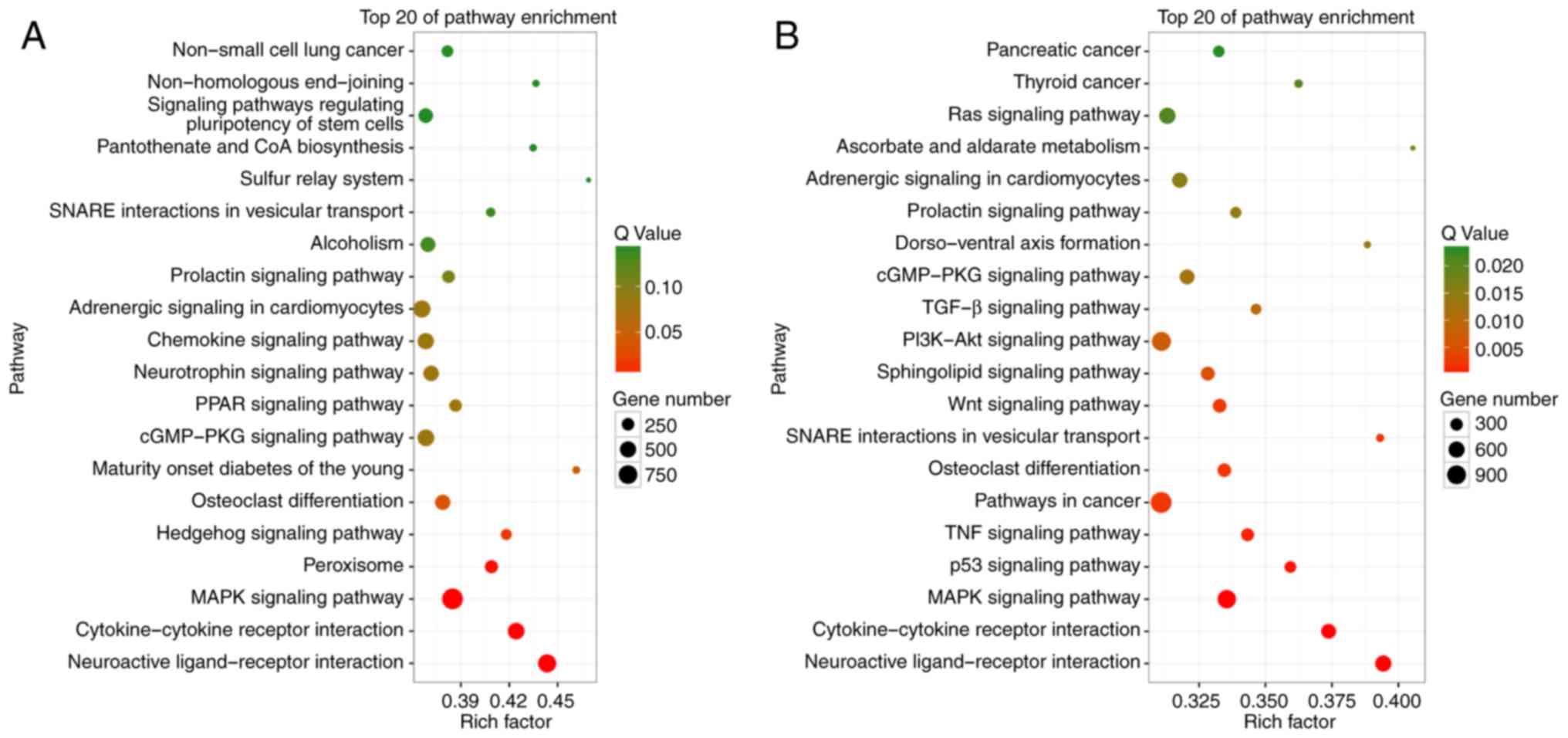
exosomal miRNAs between NC and EOC samples, KEGG pathway enrichment
analysis of the target genes of upregulated and downregulated
miRNAs was performed. The top 20 significantly enriched KEGG
pathways are highlighted and summarized in Fig. 4. The genes that are targeted by miRNAs
that were downregulated in the EOC group compared with the NC group
were most significantly involved in pathways with cancer-promoting
potential, such as phosphoinositide 3-kinase-Akt, Wnt or
mitogen-activated protein kinase signaling pathways. These results
suggested that changes in the expression of peritoneal exosomal
miRNAs might reflect and have potential effects on the
physiopathological features of ovarian cancer.
 | Table III.GO enrichment of the target genes of
upregulated miRNAs (P<0.05). |
Table III.
GO enrichment of the target genes of
upregulated miRNAs (P<0.05).
| GO ID | Description | Gene ratio: 44955
(total number of differential genes) (%) | P-value |
|---|
| Cellular
component |
|
GO:0005886 | Plasma
membrane | 10947 (27.42) | 0.018589 |
|
GO:0016020 | Membrane | 10947 (27.42) | 0.018589 |
|
GO:0071944 | Cell periphery | 10955 (27.44) | 0.018807 |
| Molecular
function |
|
GO:0001071 | Nucleic acid
binding transcription factor activity | 3440 (7.65) | P<0.1 |
|
GO:0022857 | Trans-membrane
transporter activity | 2887 (6.42) | 0.000001 |
|
GO:0003677 | DNA binding | 6824 (15.18) | 0.000001 |
|
GO:0005215 | Transporter
activity | 3149 (7) | 0.00003 |
|
GO:0004871 | Signal transducer
activity | 3485 (7.75) | 0.034531 |
| Biological
process |
|
GO:0007267 | Cell-cell
signaling | 4472 (10.12) | P<0.1 |
|
GO:0055085 | Trans-membrane
transport | 3575 (8.09) | 0.000132 |
|
GO:0003013 | Circulatory system
process | 1446 (3.27) | 0.000211 |
|
GO:0007154 | Cell
communication | 16385 (37.06) | 0.018231 |
|
GO:0023052 | Signaling | 16385 (37.06) | 0.018231 |
|
GO:0044700 | Single organism
signaling | 16385 (37.06) | 0.018231 |
 | Table IV.Top 10-upregulated miRNA with the
highest fold change and TPM >50 in EOC exosomes. |
Table IV.
Top 10-upregulated miRNA with the
highest fold change and TPM >50 in EOC exosomes.
| MicroRNA | Total reads in
NC | Total reads in
EOC | NC count | EOC count |
TPMNC |
TPMEOC |
log2(FC) |
|---|
| hsa-miR-149-3p | 6893108 | 2217219 | 0 | 140 | 0.0100 | 63.1422 | 12.62438881 |
| hsa-miR-150-3p | 6893108 | 2217219 | 33 | 958 | 4.7874 | 432.0728 | 6.49588825 |
| hsa-miR-1246 | 6893108 | 2217219 | 25 | 345 | 3.6268 | 155.6003 | 5.42300384 |
|
hsa-miR-1228-5p | 6893108 | 2217219 | 24 | 240 | 3.4817 | 108.2437 | 4.95834735 |
| hsa-miR-194-5p | 6893108 | 2217219 | 845 | 7238 | 122.5862 | 3264.4497 | 4.73497133 |
| hsa-miR-215-5p | 6893108 | 2217219 | 125 | 1046 | 18.1341 | 471.7621 | 4.70128247 |
| hsa-miR-671-3p | 6893108 | 2217219 | 31 | 244 | 4.4972 | 110.0478 | 4.61295945 |
| hsa-miR-192-5p | 6893108 | 2217219 | 5508 | 41272 | 799.0590 | 18614.3092 | 4.54196624 |
| hsa-miR-1197 | 6893108 | 2217219 | 24 | 138 | 3.4817 | 62.2401 | 4.15998057 |
| hsa-miR-222-5p | 6893108 | 2217219 | 26 | 147 | 3.7719 | 66.2993 | 4.13563031 |
Upregulation of miR-149-3p and
miR-222-5p is associated with the progression of ovarian
cancer
To further investigate the effect of changes in the
expression of peritoneal exosomal miRNAs on the pathophysiology of
ovarian cancer, the top 10 most significantly upregulated miRNAs
with TPMEOC >50 were sorted (Table IV). A total of three miRNAs with an
increased expression that was associated with a decreased survival
in patients with ovarian cancer were identified. As indicated in
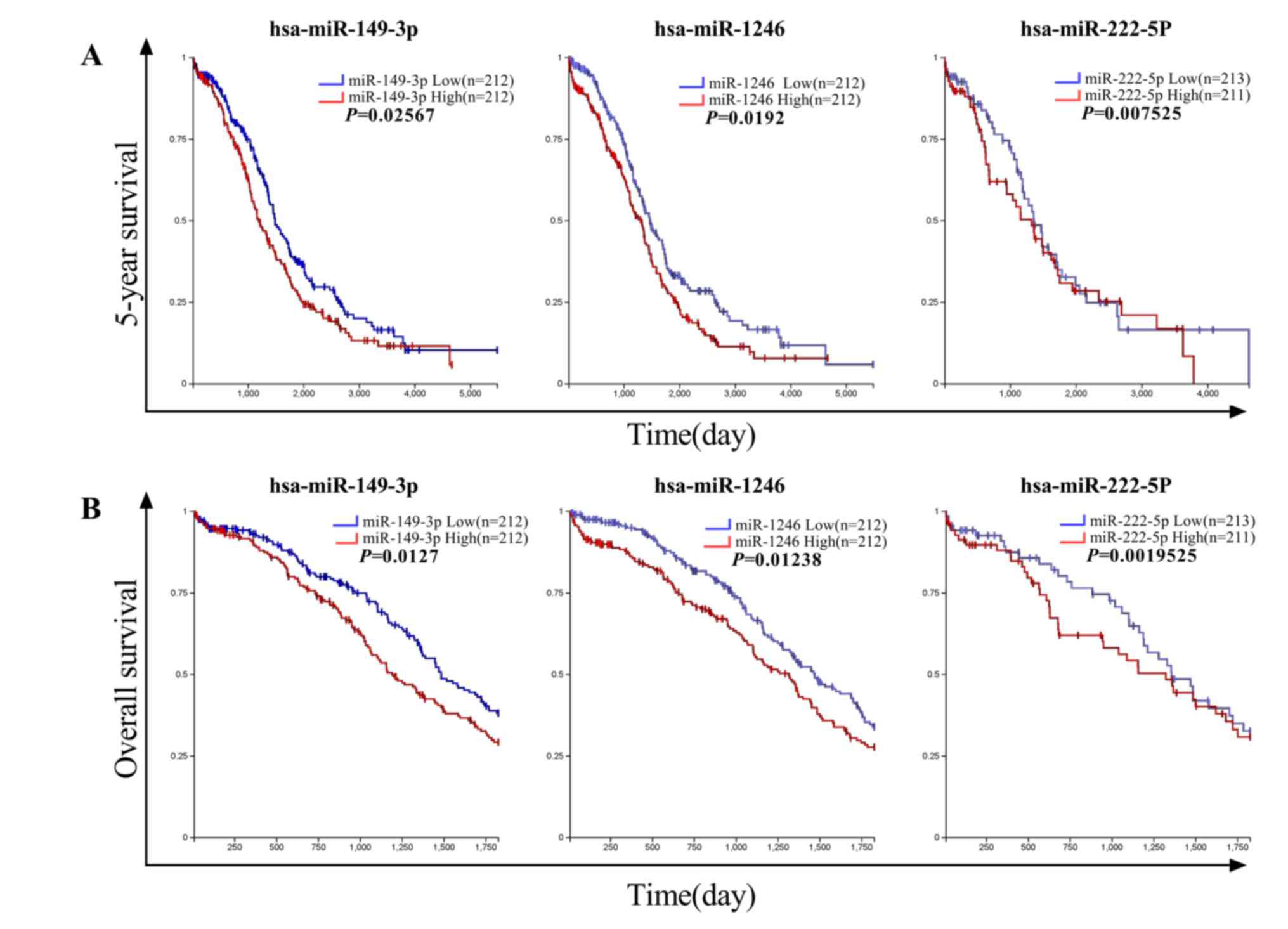
Fig. 5, a high expression of
miR-149-3p and miR-222-5p was significantly correlated with a
decrease in the 5-year and overall survival of patients with EOC as
revealed by Kaplan-Meier curve analysis. A high expression of
miR-1246 was also associated with a poor survival in patients with
EOC, but there was no statistical significance (P>0.05). To the
best of our knowledge, the effects of miR-149-3p and miR-222-5p on
the pathophysiology of EOC have not been adequately characterized
by other studies. Considering that these two miRNAs were
significantly upregulated in EOC samples compared with the NC
samples, it was inferred from these results that miR-149-3p and
miR-222-5p may participate in the oncogenesis or progression of
EOC.
The association between EOC and the upregulation of
miR-149-3p or miR-222-5p in peritoneal exosomes was further
verified by evaluating the expression level of these two miRNAs in
the peritoneal exosomes that were obtained from the 10 patients
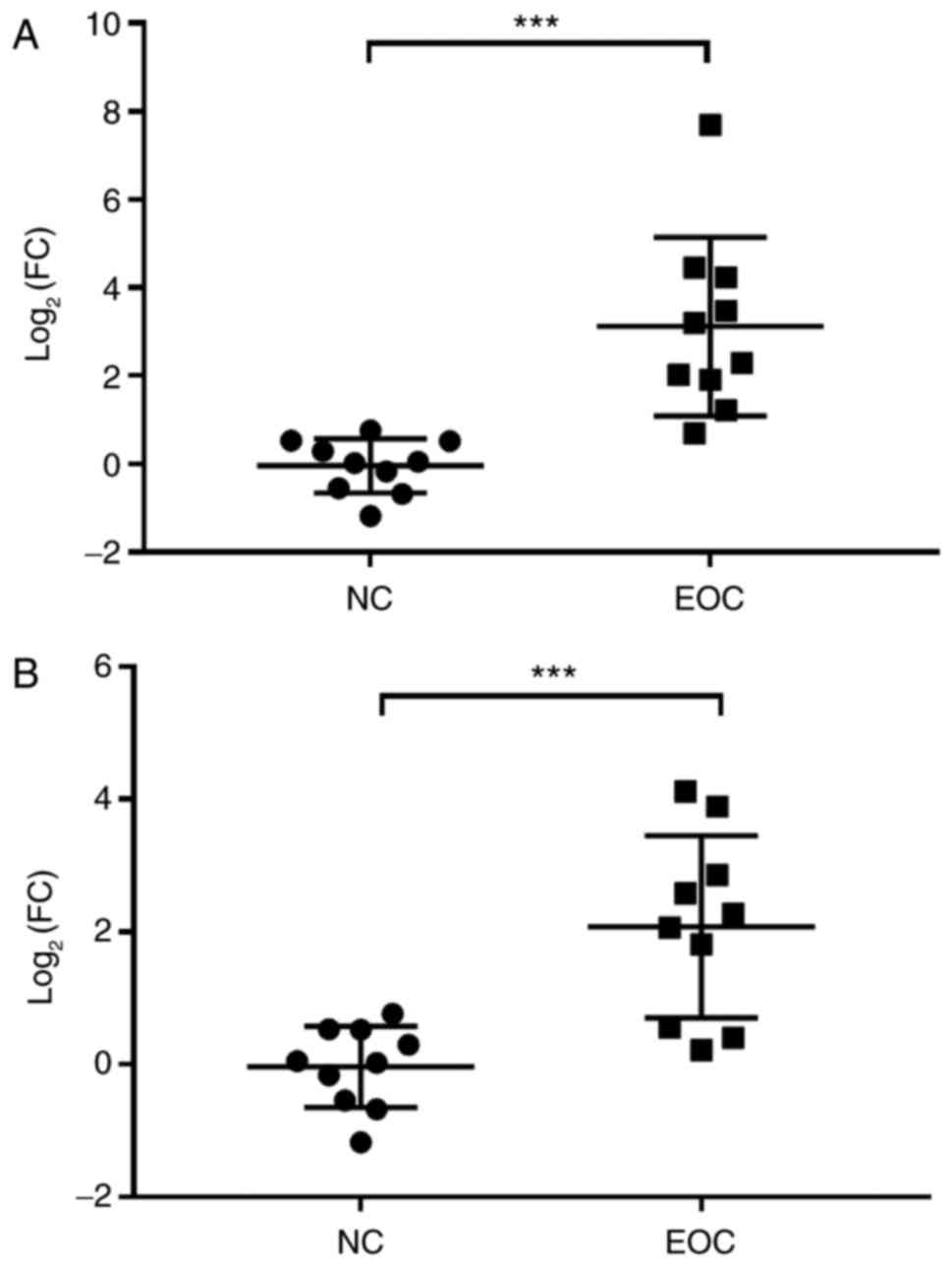
with EOC and 10 NC participants. As indicated in Fig. 6, RT-qPCR results indicated that the
expression level of miR-149-3p and miR-222-5p was significantly
increased in EOC samples compared with the NC samples. These data
suggested that miR-149-3p and miR-222-5p were associated with EOC
and the miRNAs might be involved in the progression of EOC with the
potential to serve as a biomarker for estimating the prognosis of
patients with EOC.
Discussion
The aim and scope of the present research was to
compare the peritoneal exosomal miRNA profile between patients with
ovarian cancer and participants without ovarian cancer in order to
identify differentially expressed miRNAs that are potentially
associated with peritoneal metastasis of ovarian cancer with
considerable prognostic value. A recent research by Azais et
al (21) reported that peritoneal
metastases were diagnosed in 70–75% of all ovarian cancer cases.
Abdominal cavity is a preferential metastatic site for ovarian
cancer, and peritoneal exosomes or other extracellular vesicles
have been characterized as important mediators of metastasis or
development of ovarian cancer by transporting substances including
proteins, mRNAs and miRNAs in an intercellular fashion and
affecting the biological features of recipient cells (18,22,23).
Of note, two major types of exosomes are supposed to
be in the tumor microenvironment and of great relevance to tumor
development, including tumor cell-derived exosomes and exosomes
derived from non-tumoral cells (24).
It is not possible to distinguish these two types of exosomes by
using body fluids, including blood or ascites as specimens
(25). The contents loaded in these
exosomes may therefore be affected by different factors, including
cellular origin or physiopathological condition, yielding different
analysis results. The sequencing result in the present study
indicated an upregulation of miR-149-3p and miR-222-5p in
peritoneal exosomes that were acquired from patients with ovarian
cancer compared with that from participants without cancer. The
upregulation of miR-149-3p and miR-222-5p was significantly
associated with ovarian cancer and poor patient survival, thereby
strongly indicating their prognostic and therapeutic values. There
appears to be no previous reports that link these two miRNAs to
ovarian cancer progression. Vaksman et al (26) reported in 2011 that miR-222 was
upregulated in the pleural effusion of patient with ovarian cancer;
therefore miR-222 may aid the survival of ovarian cancer cells by
targeting P21 (RAC1) activated kinase 1 and phosphatase and tensin
homolog. However, the present study failed to determine whether the
miR-222 described in the report by Vaksman et al (26) was miR-222-5p or miR-222-3p as no
sequence related to this miRNA was provided in their report.
Interestingly, a number of studies have proposed that miR-149-3p
may act as a tumor suppressor in gastric, pancreatic and renal cell
cancer (27–29). Considering the significant association
between miR-149-3p expression and poor prognosis in patients with
ovarian cancer revealed in the present study, these contradicting
results may be a reflection of the heterogeneity in cancer types
with different tissue origins.
To examine the potential effect of the upregulation
of miR-149-3p and miR-483-5p in the development of ovarian cancer,
using the DIANA-miRPath online software performs GO and KEGG
enrichment analysis of genes in TarBase that have been validated as
targets of given miRNAs. The result suggested that the target genes
of miR-149-3p and miR-222-5p were significantly enriched in 19
pathways, including the antigen processing and presentation
pathway, where it was identified that the two miRNAs target major
histocompatibility complex class I (MHC I) gene (data not shown).
Considering the GO enrichment analysis of predicted target genes of
all upregulated miRNAs and the survival analysis in the present
study, it was hypothesized that the upregulation of miR-149-3p and
miR-222-5p might downregulate MHC I expression, impairing antigen
presentation and therefore reducing the susceptibility of ovarian
cancer cells to T cell cytotoxicity. This potential mechanism will
be further investigated in our future research.
Collectively, the present study identified
significant differences in peritoneal exosomal miRNA components
between patients with ovarian cancer with peritoneal metastasis and
participants without cancer, and it suggested the prognostic value
of two peritoneal exosomal miRNAs, namely miR-149-3p and
miR-222-5p. However, the relatively low number of samples might
undermine these results. To further validate these results, a
large-scale screening in patients with ovarian cancer is required
in the future.
Acknowledgements
Not applicable.
Funding
The present study is supported by the ‘Henan
Provincial Higher Education Key Research Project Guidance Program’,
project number: 19B320022.
Availability of data and materials
All data used in this study are included in this
published article.
Authors' contributions
YKL wrote the manuscript. YKL and CHL completed the
experiments including extraction of extraperitoneal exosomes,
exosomes identification and western blot. YML and WLW performed the
experiments including extraction of exosomal miRNAs, construction
of sequencing libraries and pre-processing of raw data. BH and XQL
participated in analysis of miRNA expression, gene set enrichment
analysis of the target genes of miRNAs. JQC participated in RT-qPCR
assay, Kaplan-Meier curve analysis and statistical analysis. YKL
and JQC revised the manuscript. All authors read and approved the
final the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of the Second Affiliated Hospital of Zhengzhou University
(Henan, China). Written informed consent was provided from each
patient.
Patient consent for publication
Patients provided consent for the publication of
this data and any associated images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thibault B, Castells M, Delord JP and
Couderc B: Ovarian cancer microenvironment: Implications for cancer
dissemination and chemoresistance acquisition. Cancer Metastasis
Rev. 33:17–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nieman KM, Kenny HA, Penicka CV, Ladanyi
A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB,
Hotamisligil GS, et al: Adipocytes promote ovarian cancer
metastasis and provide energy for rapid tumor growth. Nat Med.
17:1498–1503. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bapat SA, Mali AM, Koppikar CB and Kurrey
NK: Stem and progenitor-like cells contribute to the aggressive
behavior of human epithelial ovarian cancer. Cancer Res.
65:3025–3029. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alvero AB, Chen R, Fu HH, Montagna M,
Schwartz PE, Rutherford T, Silasi DA, Steffensen KD, Waldstrom M,
Visintin I and Mor G: Molecular phenotyping of human ovarian cancer
stem cells unravels the mechanisms for repair and chemoresistance.
Cell Cycle. 8:158–166. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kajiyama H, Shibata K, Terauchi M,
Yamashita M, Ino K, Nawa A and Kikkawa F: Chemoresistance to
paclitaxel induces epithelial-mesenchymal transition and enhances
metastatic potential for epithelial ovarian carcinoma cells. Int J
Oncol. 31:277–283. 2007.PubMed/NCBI
|
|
7
|
Zack TI, Schumacher SE, Carter SL,
Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J,
Mermel CH, et al: Pan-cancer patterns of somatic copy number
alteration. Nat Genet. 45:1134–1140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patch AM, Christie EL, Etemadmoghadam D,
Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey
PJ, et al: Whole-genome characterization of chemoresistant ovarian
cancer. Nature. 521:489–494. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Malek JA, Mery E, Mahmoud YA, Al-Azwani
EK, Roger L, Huang R, Jouve E, Lis R, Thiery JP, Querleu D and
Rafii A: Copy number variation analysis of matched ovarian primary
tumors and peritoneal metastasis. PLoS One. 6:e285612011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bracken CP, Scott HS and Goodall GJ: A
network-biology perspective of microRNA function and dysfunction in
cancer. Nat Rev Genet. 17:719–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O'Connell RM, Rao DS and Baltimore D:
microRNA regulation of inflammatory responses. Annu Rev Immunol.
30:295–312. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deb B, Uddin A and Chakraborty S: miRNAs
and ovarian cancer: An overview. J Cell Physiol. 233:3846–3854.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mihanfar A, Fattahi A and Nejabati HR:
MicroRNA-mediated drug resistance in ovarian cancer. J Cell
Physiol. Jun 19–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Palma Flores C, García-Vázquez R, Gallardo
Rincón D, Ruiz-García E, Astudillo de la Vega H, Marchat LA,
Salinas Vera YM and López-Camarillo C: MicroRNAs driving invasion
and metastasis in ovarian cancer: Opportunities for translational
medicine (Review). Int J Oncol. 50:1461–1476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X and Wang X: The emerging roles and
therapeutic potential of exosomes in epithelial ovarian cancer. Mol
Cancer. 16:922017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakamura K, Sawada K, Kinose Y, Yoshimura
A, Toda A, Nakatsuka E, Hashimoto K, Mabuchi S, Morishige KI,
Kurachi H, et al: Exosomes promote ovarian cancer cell invasion
through transfer of CD44 to peritoneal mesothelial cells. Mol
Cancer Res. 15:78–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang MK and Wong AS: Exosomes: Emerging
biomarkers and targets for ovarian cancer. Cancer Lett. 367:26–33.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Armstrong RN, Colyer HA and Mills KI:
Screening for miRNA expression changes using quantitative PCR
(Q-PCR). Methods Mol Biol. 863:293–302. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Azaïs H, Estevez JP, Foucher P, Kerbage Y,
Mordon S and Collinet P: Dealing with microscopic peritoneal
metastases of epithelial ovarian cancer. A surgical challenge. Surg
Oncol. 26:46–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa
M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F
and Ochiya T: Malignant extracellular vesicles carrying MMP1 mRNA
facilitate peritoneal dissemination in ovarian cancer. Nature
Commun. 8:144702017. View Article : Google Scholar
|
|
23
|
Keller S, König AK, Marmé F, Runz S,
Wolterink S, Koensgen D, Mustea A, Sehouli J and Altevogt P:
Systemic presence and tumor-growth promoting effect of ovarian
carcinoma released exosomes. Cancer Lett. 278:73–81. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Chen JQ, Liu JL and Tian L:
Exosomes in tumor microenvironment: Novel transporters and
biomarkers. J Transl Med. 14:2972016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumar D, Gupta D, Shankar S and Srivastava
RK: Biomolecular characterization of exosomes released from cancer
stem cells: Possible implications for biomarker and treatment of
cancer. Oncotarget. 6:3280–3291. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vaksman O, Stavnes HT, Kaern J, Trope CG,
Davidson B and Reich R: miRNA profiling along tumour progression in
ovarian carcinoma. J Cell Mol Med. 15:1593–1602. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao D, Jia Z, You L, Wu Y, Hou Z, Suo Y,
Zhang H, Wen S, Tsukamoto T, Oshima M, et al: 18β-glycyrrhetinic
acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1
signaling. Oncotarget. 7:71960–71973. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okato A, Arai T, Yamada Y, Sugawara S,
Koshizuka K, Fujimura L, Kurozumi A, Kato M, Kojima S, Naya Y, et
al: Dual strands of pre-miR-149 inhibit cancer cell migration and
invasion through targeting FOXM1 in renal cell carcinoma. Int J Mol
Sci. 18(pii): E19692017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Si L, Xu L, Yin L, Qi Y, Han X, Xu Y, Zhao
Y, Liu K and Peng J: Potent effects of dioscin against pancreatic
cancer via miR-149-3P-mediated inhibition of the Akt1 signalling
pathway. Br J Pharmacol. 174:553–568. 2017. View Article : Google Scholar : PubMed/NCBI
|