Introduction
Nasopharyngeal carcinoma (NPC) is highly prevalent
in Southern China and Southeast Asia with an incidence rate of
15–50 cases/100,000 people (1–3). NPC has
the highest metastasis rate among the various types of head and
neck cancer, with 74.5 and 19.9% of patients presenting with
regional lymph node and distant metastasis respectively, including
liver and lung metastasis at the time of diagnosis (1,4). The
dysregulation of oncogenes or tumour suppressor genes in NPC is
associated with the initiation and progression of cancer, and
resistance to therapy (1,3,5–7). Our group previously identified that a
transcription factor Telomere length regulation protein TEL2, and a
cell membrane protein carbonic anhydrase IX, served a key role in
cell proliferation and metastasis in NPC (1,3). Although
some progress, including microRNAs, long non-coding RNAs and
kinases serving key roles in NPC progression, has been made
recently (8–11), the molecular mechanisms of NPC are
poorly understood.
Human mortality factor 4-like 1 (MORF4L1) is a
member of a subgroup of histone acetyltransferases and belongs to
the mortality factor on chromosome 4 (MORF4) class of proteins
(12). MORF4L1 and MORF4 share a 96%
homology in their amino acid sequences (13). Unlike the majority of histone
acetyltransferases that are known for activating gene transcription
and promoting cell proliferation, MORF4 was initially cloned and
characterized as a senescence gene; this is because the cellular
expression of MORF4 induces cell death and senescence (14–16).
MORF4L1 homodimerization is essential for facilitating the
formation and functionality of a complex that represses cell
proliferation (16). Additionally,
MORF4L1 functions during embryonic development via chromatin
remodeling and transcriptional regulation (17), and MORF4L1 mediates epithelial cell
death in a mouse model of pneumonia (18). In cancer, MORF4L1 acts as a nuclear
ligand for the fungal galectin Agrocybe aegerita lectin and serves
a key role in its antitumour activity (19). MORF4L1 stimulates homology-directed
repair of chromosomal breaks by interacting with BRCA2 DNA repair
associated, partner and localizer of BRCA2, RAD51 recombinase and
replication protein A1 (20,21). However, little is known regarding the
role of MORF4L1 in NPC.
In the present study, the expression of MORF4L1 in
clinical NPC tissues was examined and compared with adjacent,
non-cancerous tissues, and also sought to evaluate the effects of
MORF4L1 expression on NPC cell phenotypes in vitro. The
findings suggest that MORF4L1 inhibits cell proliferation and
migration by increasing the expression levels of p21 and
E-cadherin.
Materials and methods
Cells and reagents
The cell lines NP69, 5-8F, 6-10B and SUNE1 were
obtained from the group of Tiebang Kang (Sun Yat-sen University
Cancer Center, Guangzhou, China). The cell lines 6-10B, 5-8F and
SUNE1 were cultured in Dulbecco's modified Eagle medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and maintained at 37°C in an incubator containing
5% CO2. The NP69 cell line was maintained in
keratinocyte serum-free medium supplemented with 4 µg/ml human
epidermal growth factor and 40 µg/ml bovine pituitary extract
(Gibco; Thermo Fisher Scientific, Inc.; catalogue no. 17005-042)
and maintained at 37°C in an incubator containing 5%
CO2. All cell lines used in this study were
authenticated through short tandem repeat profiling within 6 months
of initiating this project, and the cells were cultured for <2
months.
Plasmids
The full-length cDNA of human MORF4L1 was cloned
into the pSin-puro vector (Focus Bioscience Co., Ltd., Nanchang,
China). Short hairpin (sh) RNA-NC targeting EGFP (with no known
targets in the human genome) was cloned into the pFCL2.0 vector
with the following sequence: 5′-GGGCGAGGAGCTGTTCACCG-3′. The
oligonucleotide sequence for the human MORF4L1 short hairpin RNA
(shRNA)-MORF4L1 was 5′-GTTGCCATAAAGGACAAACAA-3′ (Focus Bioscience
Co., Ltd.).
Antibodies
Mouse monoclonal anti-β-actin (catalogue no. FB075)
antibody was obtained from Nanchang Focus Bioscience Co., Ltd, and
the species specificity is human, mouse, rat and pig.
Anti-E-cadherin (catalogue no. 14472) and Anti-p21 (catalogue no.
2947) antibodies were obtained from Cell Signalling Technology,
Inc. (Danvers, MA, USA), the species specificity of anti-E-cadherin
is human, mouse and rat, and the species specificity of anti-p21 is
human and monkey. Anti-MORF4L1 (catalogue no. HPA042360) antibody
was obtained from Sigma-Aldrich (Merck KGaA; Darmstadt, Germany),
the species specificity of anti-MORF4L1 is human and rat.
Stable cell lines
Subsequently, 3 µg pSin-puro-MORF4L1 (Focus
Bioscience Co., Ltd.), 3 µg pSin-puro-empty vector (Focus
Bioscience Co., Ltd.), 3 µg shRNA-NC (Focus Bioscience Co., Ltd.)
or 3 µg shRNA-MORF4L1 (Focus Bioscience Co., Ltd.) was
co-transfected with 3 µg pMD2.G and 3 µg psPAX2 into 293 cells for
48 h. The recombinant viruses were subsequently collected and added
to NPC cells, which were cultured with 8 µg/ml polybrene for 24 h
at 37°C. The stable lines were selected by treating the cells with
1 µg/ml puromycin for 2 weeks (Focus Bioscience Co., Ltd.).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA extraction and RT-qPCR procedures were performed
as previously described (22,23). Briefly, total RNA obtained from cell
lines was isolated using TRIzol reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. First-strand cDNA
was synthesized using the RevertAid™ First Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.). Subsequently, the
qPCR reaction was performed in a CFX96 Real-Time PCR Detection
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using
SYBR® Green mix (Tiangen Biotech Co., Ltd., Beijing,
China). Thermal cycling of the qPCR reaction was initiated with a
denaturation step at 95°C for 15 min, and consisted of 40 cycles
(denaturation at 95°C 15 sec, annealing at 60°C for 30 sec and
elongation at 72°C for 30 sec). The amplified products were
examined using the 2∆∆Cq method (24), and each sample was calibrated to the
expression level of the housekeeping gene GAPDH. The primers
employed for amplifying MORF4L1, p21, E-cadherin and GAPDH were
validated. The sequences for the primers were as follows: MORF4L1
forward (F), 5′-AGCAATGTTGGCTTATACACCTC-3′ and reverse (R),
5′-AGCTTTCCGATGGTACTCAGG-3′; GAPDH F, 5′-ACAGTCAGCCGCATCTTCTT-3′
and R, 5′-GACAAGCTTCCCGTTCTCAG-3′; E-cadherin F,
5′-AATAGTGCCTAAAGTGCTGC-3′ and R, 5′-AGACCCACCTCAATCATCCT-3′; P21
F, 5′-CGATGGAACTTCGACTTTGTCA-3′ and R,
5′-GCACAAGGGTACAAGACAGTG-3′.
Cell proliferation and cell viability
assays
In vitro cell proliferation and cell
viability assays were performed using a Cell Counting Kit-8 (CCK-8)
assay. To assess cell proliferation, the cell lines in the present
study were seeded in 96-well plates at a density of
1×103 cells/well and incubated for 1, 2, 3, 4 and 5
days. Subsequently, 10 µl of the CCK-8 reagent (catalogue no.
C0038; Beyotime Institute of Biotechnology, Haimen, China) was
added to each well and incubated for 1.5 h. The absorbance value of
each well was measured at 450 nm. For each experimental condition,
six wells were evaluated.
Transwell assays
For the Transwell migration assay, 5×104
cells in the present study suspended in 200 µl of serum-free DMEM
were added to cell culture inserts, which had 8-µm microporous
filters and no extracellular matrix coating (BD Biosciences,
Franklin Lakes, NJ, USA). DMEM supplemented 10% FBS was then added
to the bottom chamber. Following a 24 h incubation, the cells on
the lower surface of the filter were fixed with 10% formalin for 15
min at room temperature, stained with 0.1% crystal violet for 60
min at room temperature, and examined using a light microscope
(magnification, ×100). For the in vitro invasion assay, a
total of 8×104 cells were resuspended in 200 µl of
serum-free DMEM and added to the cell culture inserts, which had
8-µm microporous filters and were coated with Matrigel (BD
Biosciences). DMEM supplemented with 10% FBS was then added to the
bottom chamber and the chamber was incubated for 24 h. The mean
number of migrated or invaded cells in three randomly selected
optical fields (×100 magnification) from triplicate filters was
calculated.
Wound-healing assay
Cell motility was assessed by measuring the movement
of NP69, 5-8F, 6-10B and SUNE1 cells from into a scraped cellular
area created using a 200-µl pipette tip, and the wound closure was
observed at 0, 24 and 48 h. The cells were imaged under a light
microscope (magnification, ×40).
Western blotting
Western blotting procedures were performed as
described previously (1,22). Briefly, cultured cells from all cell
lines were lysed in ice-cold radioimmunoprecipitation assay lysis
buffer (cat. no., P0013C; Beyotime Institute of Biotechnology) at
4°C for 30 min. Following centrifugation (12,000 × g) at 4°C for 20
min, the lysates were obtained and protein concentration was
determined with the BCA method. Equal amounts of protein (30
µg/lane) were separated by 10% SDS-PAGE gels; the conditions were:
Voltage of 100 V for 2.5 h at room temperature. Proteins were then
transferred to polyvinylidene difluoride membranes; the conditions
were: Electrical current of 250 mA at 4°C for 2 h. To block the
non-specific binding sites, the membranes were incubated with 5%
non-fat milk (in Tris-buffered saline with 0.1% Tween-20) at room
temperature for 60 min, and then probed with the following primary
antibodies: β-actin (cat. no., FB075; Focus Bioscience, Nanjing,
China), E-cadherin (cat. no., 14472; Cell Signalling Technology,
Inc.), p21 (cat. no., 2947; Cell Signalling Technology, Inc.),
MORF4L1 (cat. no., HPA042360; Sigma-Aldrich; Merck KGaA) at a
dilution of 1:1,000 overnight at 4°C. Subsequently, the membranes
were incubated with anti-Rabbit IgG Secondary Antibody Peroxidase
Conjugated (cat. no., W401B; Promega Corporation, Madison, WI, USA)
and anti-Mouse IgG Secondary Antibody Peroxidase Conjugated (cat.
no., W402B; Promega Corporation) with the dilution 1:10,000 at room
temperature for 1 h. Specific protein bands were visualized using
the BeyoECL Moon detection system (catalogue no. P0018F; Beyotime
Institute of Biotechnology) and exposed to radiographic film.
Densitometry analysis of the protein bands was performed using
ImageJ 1.42 software (National Institutes of Health, Bethesda, MD,
USA).
Colony formation assay
Cells used from the aforementioned cell lines in the
present study were plated in six-well culture plates at a density
of 2.5×102 cells/well. Each group included three wells.
The cells were incubated for 15 days at 37°C, washed twice with PBS
and were fixed with 10% formalin at room temperature for 15 min and
stained with 0.1% crystal violet at room temperature for 60 min. A
cluster containing ≥50 cells was counted as a colony, and was
performed with a light microscope (magnification, ×100).
RNA interference
Effective small interfering (si)RNA oligonucleotides
targeting p21 and E-cadherin with sequences of
5′-CCCUGUUAGUAACGGCAAA-3′ and 5′-AUACCAGAACCUCGAACUAUA−3′,
respectively, were synthesized by Guangzhou RiboBio Co., Ltd.,
(Guangzhou, China). Approximately 2×105 NPC cells/well
were seeded in a six-well tissue culture dish the day prior to
transfection. Transfection was performed according to the
manufacturer's protocol using Lipofectamine RNAiMAX transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and 50 nM
siRNA. At 48 h following transfection, RT-qPCR assays were
performed.
Study approval
The use of human NPC tissues was reviewed and
approved by the Ethical Committee of Jiangxi Cancer Hospital
(Nanchang, China) and was performed in accordance with the approved
guidelines. Written informed consent was obtained from all
patients. A total of 19 tumor specimens were collected from
patients with NPC (median age, 46 years; age range, 29–69 years;
male:female ratio, 10:9) with resection between January 2013 and
December 2017, and inclusion criteria were as follow: Initial
diagnosis of NPC. Exclusion criteria included: Previous
radiotherapy, chemotherapy or surgery, with the exception of
diagnostic procedures, to the primary tumor or nodes; and previous
malignancy or other concomitant malignant disease. A total of 13
normal nasopharyngeal epithelium specimens were collected from
healthy participants (median age, 41 years; age range, 32–66 years;
male: female ratio, 6:7) with resection between January 2013 and
December 2017. All clinical samples were obtained from the
Department of Radiation Oncology at the Jiangxi Cancer Hospital
(Nanchang, China).
DNA methylation analysis
Methylation data of MORF4L1 were obtained from the
MethHC database (http://methhc.mbc.nctu.edu.tw/php/index.php). All the
online data was used for analyzing the methylation rate of MORF4L1
promoter with MethHC 1.0.3. software (http://methhc.mbc.nctu.edu.tw/php/index.php).
Statistical analysis
All statistical analyses were performed using SPSS
for Windows, version 16.0 (SPSS, Inc., Chicago, IL, USA). All
values from the in vitro assays are expressed as the mean ±
standard deviation or mean ± standard error of the mean of at least
three independent experiments or replicates. Paired Student's
t-test was used to compare two groups while comparisons between
multiple groups were performed by using one-way analysis of
variance followed by a Tukey-Kramer post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
MORF4L1 expression levels are
decreased in various cancer types
MORF4L1 expression levels were significantly lower
in NPC tissues compared with normal nasopharyngeal epithelial
tissues at the mRNA level as quantified by RT-qPCR (Fig. 1A). Likewise, MORF4L1 mRNA levels were
significantly downregulated in NPC cell lines when compared with
the normal nasopharyngeal epithelial cell line (NP69) (Fig. 1B). Whether the pattern of MORF4L1
expression in normal vs. tumor tissues could be extended to other
cancer types was subsequently investigated. MethHC is an integrated
and beneficial database, which includes DNA methylation, gene
expression, microRNA methylation and microRNA expression data, and
the association between DNA methylation and gene expression from
The Cancer Genome Atlas (25). As
illustrated in Fig. 1C-L, with the
data from MethHC, MORF4L1 was significantly downregulated in
various types of cancer, including in breast, colon and lung
cancer, suggesting that MORF4L1 expression is associated with
cancer initiation or progression. To investigate the mechanisms
underlying the reduction of MORF4L1 levels in cancer cells, the
promoter sequence of MORF4L1 was analyzed and a CpG island was
identified in this region. The methylation rate of the promoter was
significantly higher in tumor cells, when compared with normal
cells according to the online database (http://methhc.mbc.nctu.edu.tw/php/index.php)
(Fig. 1M), which indicated that
promoter hypermethylation may promote the downregulation of MORF4L1
in cancer types.
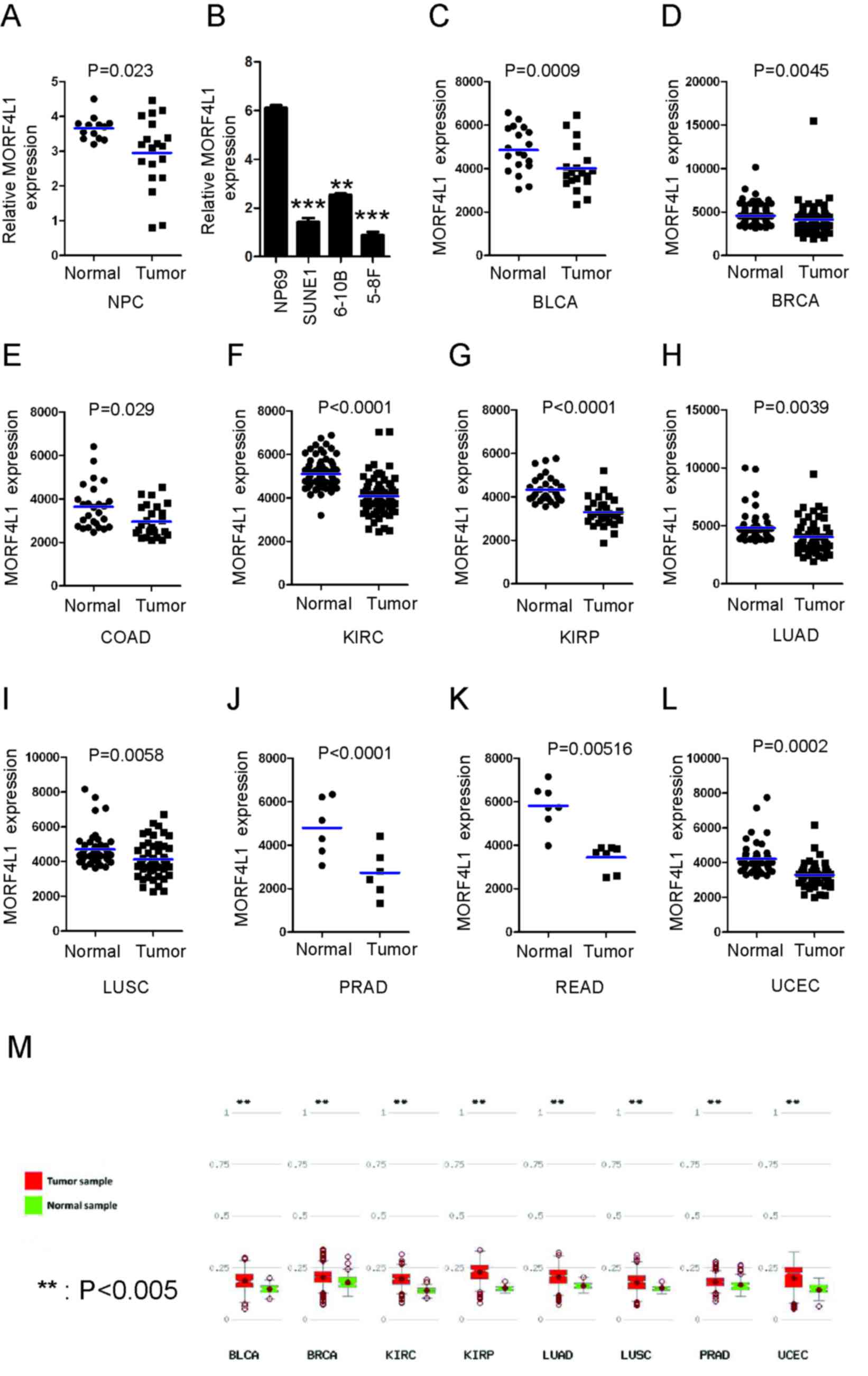 | Figure 1.MORF4L1 expression is decreased in
cancer. (A) RT-qPCR analysis of MORF4L1 expression in clinical NPC
samples of both tumour (n=19) and normal tissues (n=13). Blue bars
indicate mean values. (B) The mRNA levels of MORF4L1 in cell lines
were determined by RT-qPCR. The mean ± SEM of NP69 group is
6.10±0.11, the mean ± SEM of 6-10B group 2.53±0.06; the mean ± SEM
of SUNE1 group 1.42±0.17; the mean ± SEM of 5-8F group 0.88±0.13.
**P<0.01 and ***P<0.001. Meta-analysis of MORF4L1 mRNA levels
in clinical samples from the MethHC database in (C) BLCA, (D) BRCA,
(E) COAD, (F) KIRC, (G) KIRP, (H) LUAD, (I) LUSC, (J) PRAD, (K)
READ, (L) UCEC. (M) The methylation rate of MORF4L1 promoter is
significantly higher in tumor cells, when compared with normal
cells according to the online database (http://methhc.mbc.nctu.edu.tw/php/index.php).
Blue bars indicate mean values. MORF4L1, Mortality factor 4-like 1;
SEM, standard error; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; BLCA, bladder urothelial carcinoma,
n=19; BRCA, breast invasive carcinoma, n=112; COAD, colon
adenocarcinoma, n=26; KIRC, kidney renal clear cell carcinoma,
n=72; KIRP, kidney renal papillary cell carcinoma, n=30; LUAD, lung
adenocarcinoma, n=57; LUSC, lung squamous cell carcinoma, n=50;
PRAD, prostate adenocarcinoma, n=50; READ, rectum adenocarcinoma,
n=6; UCEC, uterine corpus endometrial carcinoma, n=7. |
MORF4L1 inhibits cell proliferation
and colony formation in NPC
Given that MORF4L1 expression is downregulated in
NPC, the present study subsequently evaluated whether MORF4L1 is
involved in regulating the biological behaviour of NPC cells. Based
on the expression levels of MORF4L1 in NPC cells (Fig. 1B), stable cell lines with were
constructed with ectopic expression of MORF4L1 (in 5-8F cells) or
silenced MORF4L1 (in 6-10B cells) (Fig.
2A-C). The effects of MORF4L1 on the biological behaviour of
these cell lines was subsequently investigated. Cell proliferation
and colony formation assays indicated that the ectopic expression
of MORF4L1 significantly inhibited cell proliferation and colony
formation abilities. Conversely, silencing MORF4L1 increased cell
proliferation in 6-10B cells (Fig.
2D-G).
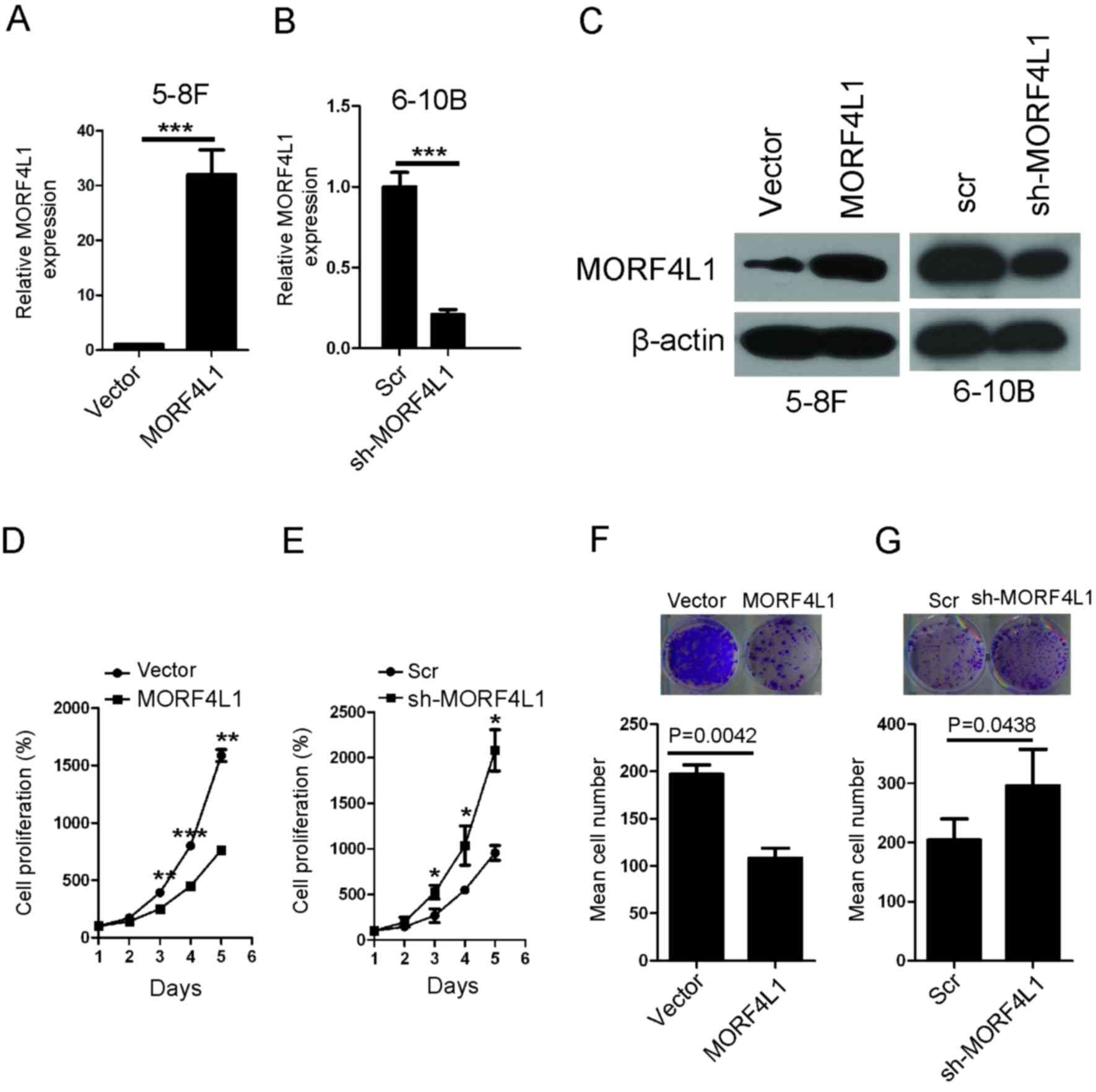 | Figure 2.MORF4L1 inhibits NPC cell
proliferation. The establishment of stable cell lines in (A) 5-8F
and (B) 6-10B cells in which MORF4L1 was (A) overexpressed or (B)
silenced was confirmed by RT-qPCR. GAPDH was used as the internal
control. The bars represent the mean ± standard error values. (C)
The generation of stable cell lines in 5-8F and 6-10B cells in
which MORF4L1 was overexpressed or silenced was confirmed via
western blotting. β-actin was used as the internal control. (D and
E) Cell proliferation of the indicated stable cell lines in
vitro was measured at different time points, using the CCK-8
assay. The bars represent mean ± standard error values, and the
P-values were calculated using the Student's t-test. *P<0.05,
**P<0.01 and ***P<0.001. (F and G) The colony formation
ability of the indicated stable cell lines in vitro was
measured at 14 days, as described in the Methods section. The bars
represent mean ± standard error values, and the P-values were
calculated using the paired Student's t-test. The top two images of
F and G are representative images. MORF4L1, Mortality factor 4-like
1; RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; NPC, nasopharyngeal carcinoma; CCK-8, cell counting
kit-8; Scr, scramble; sh-MORFL1, short hairpin RNA against
MORFL1. |
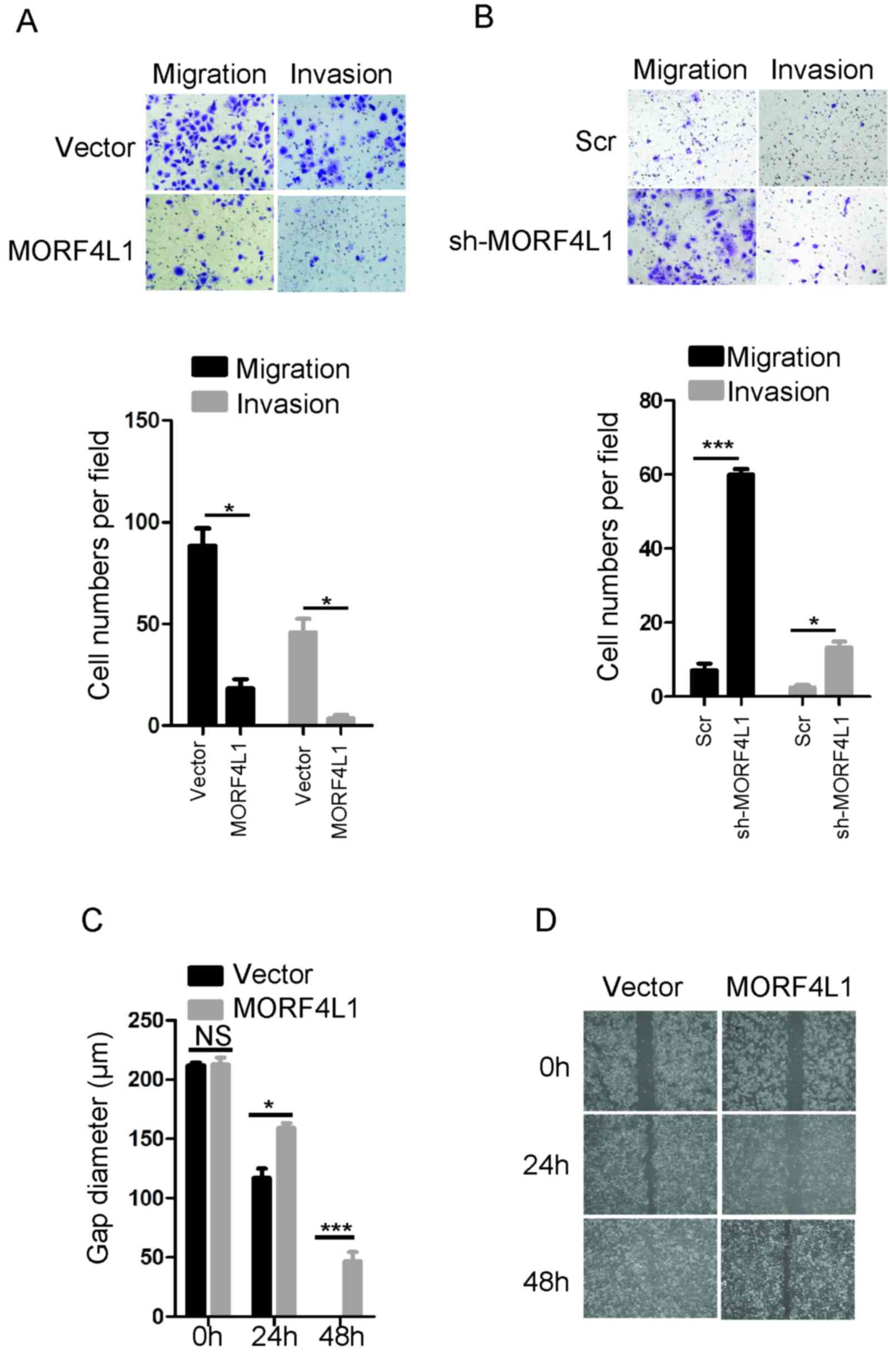
MORF4L1 inhibits NPC cell migration
and invasion
The present study then examined whether MORF4L1
inhibited the migration and invasion of NPC cells. Using Transwell
assays, a significant reduction in the number of migrated and
invaded 5-8F cells following the overexpression of MORF4L1
(*P<0.05) (Fig. 3A), and an
increased number of migrated and invaded 6-10B cells following
MORF4L1 silencing were observed (*P<0.05, ***P<0.001;
Fig. 3B). In addition, the ectopic
expression of MORF4L1 significantly suppressed 5-8F cell wound
healing after 24 h (P<0.05) and 48 h later (P<0.001)
(Fig. 3C and D). Taken together,
these findings indicated that MORF4L1 inhibited NPC cell migration
and invasion.
MORF4L1 increases p21 and E-cadherin
expression in NPC
In order to investigate the mechanisms underlying
MORF4L1 suppressing cell proliferation, migration and invasion, the
present study investigated the expression of p21 and E-cadherin.
P21 and E-cadherin are key regulators in the progression of cancer,
including NPC (26–29). As illustrated in Fig. 4A and B, p21 and E-cadherin protein
levels were significantly higher in 5-8F cells that stably
expressed ectopic MORF4L1. Conversely, silencing MORF4L1 in 6-10B
cells significantly decreased the p21 and E-cadherin protein
levels. To further verify MORF4L1 functions through p21 and
E-cadherin, rescue experiments were performed with p21 siRNA and
E-cadherin siRNA transfection (Fig.
4C). Compared with overexpression of MORF4L1 alone, the
combined use of p21 siRNA and E-cadherin siRNA significantly
increased the cancer cell proliferative (Fig. 4D), colony formation (Fig. 4E), migratory and invasive (Fig. 4F), and wound-healing abilities
(Fig. 4G). These results suggest that
MORF4L1 may regulate cell proliferation, migration and invasion by
increasing the expression levels of p21 and E-cadherin in NPC.
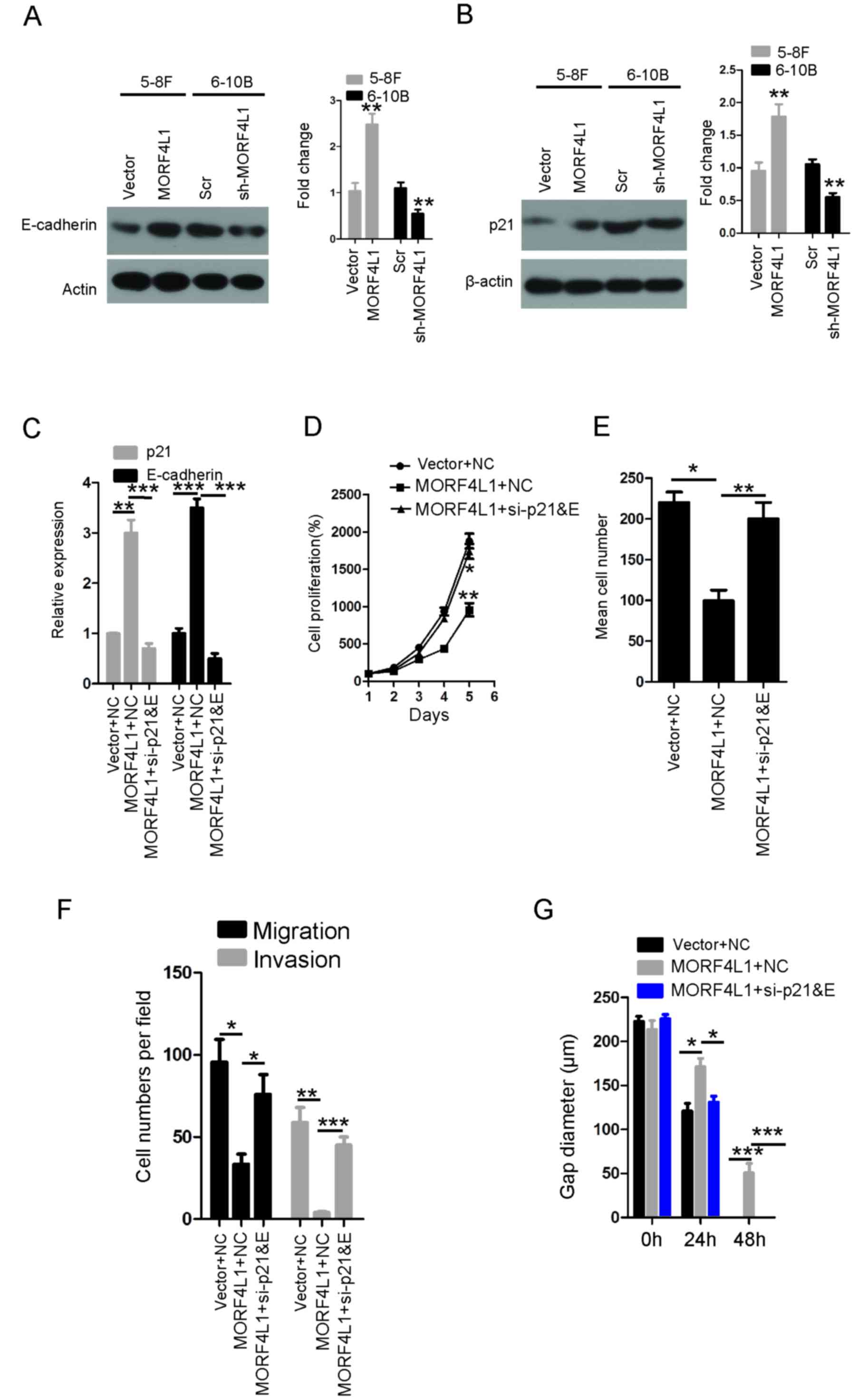 | Figure 4.MORF4L1 increase sp21 and E-cadherin
expression levels in NPC. Immunoblots of whole-cell lysates for (A)
E-cadherin and (B) p21 from 5-8F and 6-10B cell lines. **P<0.01.
(C) The levels of p21 and E-cadherin were detected via RT-qPCR.
**P<0.01 and ***P<0.001. (D) Cell proliferation of the
indicated cell lines in vitro was measured at different time
points using a CCK-8 assay. The bars represent mean ± standard
error values. *P<0.05 and **P<0.01 vs. Vector + NC. (E) The
colony formation ability of the indicated cell lines in
vitro was measured at 14 days, as described in the Methods
section. The bars represent mean ± standard error values,
*P<0.05 and **P<0.01. (F) The migration and invasion
abilities of the indicated stable cell lines were measured using
the Transwell assay. The bars represent mean ± standard error
values. *P<0.05, **P<0.01 and ***P<0.001. (G) The ability
of gap closure rescued following knockdown of p21 and E-cadherin in
cells overexpressing MORF4L1. The bars represent mean ± standard
error values. *P<0.05, **P<0.01 and ***P<0.001. NC,
negative control; si-p21&E, p21 siRNA and E-cadherin siRNA;
siRNA, small interfering RNA; MORF4L1, Mortality factor 4-like 1;
NPC, nasopharyngeal carcinoma; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
Discussion
An improved understanding of the mechanisms
underlying NPC development, progression and therapy resistance is
required in order to design novel effective therapies for this
cancer. The present study, to the best of our knowledge,
demonstrated for the first time that MORF4L1 exhibits a key
function in NPC cell proliferation, migration and invasion by
upregulating the expression levels of p21 and E-cadherin.
Using RT-qPCR analysis and data mining, the present
study demonstrated that the expression of MORF4L1 was significantly
decreased in numerous types of cancer, including breast invasive
carcinoma, colon adenocarcinoma and kidney renal clear cell
carcinoma. In order to explore the mechanisms underlying the
decreased expression of MORF4L1 in cancer cells, the methylation
status of the MORF4L1 promoter was examined, as previous studies
have reported that the hypermethylation of numerous tumour
suppressor gene promoters leads to their decreased expression
levels in cancer (30,31). The results of the present study
demonstrated that the methylation rate of MORF4L1 promoter was
higher in the majority of cancer tissues when compared with normal
tissues, which suggests that promoter hypermethylation is one of
the negative regulators of MORF4L1 in different types of
cancer.
Chromatin structure may be dynamically altered by
covalent modifications of histone tails through acetylation and
methylation, which are involved in numerous important biological
processes, including transcription, DNA replication and DNA damage
repair (32). The results of the
present study demonstrated that MORF4L1 increased the expression
levels of p21 and E-cadherin to inhibit cell proliferation,
migration and invasion. In summary, these findings demonstrate that
MORF4L1 serves a role in tumour suppression by significantly
increasing p21 and E-cadherin expression levels. However, there was
little alteration to E-cadherin protein expression in the western
blot analysis following MORF4L1 overexpression or knockdown. This
may have been due to the transfection technique requiring further
optimization to improve the level of knockdown or overexpression.
In future studies, other methods of gene editing may be used, such
as CRISPR, in order to improve knockdown. In addition, the cell
lines in the present study are suitable for using in studying NPC,
and their results in vitro are consistent with those in
vivo (33–35). Together, these data provide novel
insights into understanding the molecular basis underlying this
malignancy.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Science
Foundation of China (grant no. 81660449 to YS; grant no. 81460430
to SWL; grant no. 81660452 to LZ) and Jiangxi Provincial Natural
Science Foundation of China (grant no. 20161ACB21001, to YS; grant
no. 20161BAB215188, to GH).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YS, RZ, LZ, GH and LS wrote the manuscript and
performed experiments. SL, LX, KC and YP performed analysis and
interpretation of data. All authors read and approved the final
study.
Ethics approval and consent to
participate
The use of human NPC tissues was reviewed and
approved by the Ethical Committee of Jiangxi Cancer Hospital
(Nanchang, China) and was performed in accordance with the approved
guidelines. Written informed consent was obtained from all
patients.
Patient consent for publication
Written informed consent was obtained from all
patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sang Y, Chen MY, Luo D, Zhang RH, Wang L,
Li M, Luo R, Qian CN, Shao JY, Zeng YX and Kang T: TEL2 suppresses
metastasis by down-regulating SERPINE1 in nasopharyngeal carcinoma.
Oncotarget. 6:29240–29253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang L, Sang Y, Tang J, Zhang RH, Luo D,
Chen M, Deng WG and Kang T: Down-regulation of prostate stem cell
antigen (PSCA) by Slug promotes metastasis in nasopharyngeal
carcinoma. J Pathol. 237:411–422. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sang Y, Wang L, Tang JJ, Zhang MF, Zhang
MX, Liu X, Zhang RH, Kang TB and Chen MY: Oncogenic roles of
carbonic anhydrase IX in human nasopharyngeal carcinoma. Int J Clin
Exp Pathol. 7:2942–2949. 2014.PubMed/NCBI
|
|
4
|
Chen QY, Wen YF, Guo L, Liu H, Huang PY,
Mo HY, Li NW, Xiang YQ, Luo DH, Qiu F, et al: Concurrent
chemoradiotherapy vs radiotherapy alone in stage II nasopharyngeal
carcinoma: Phase III randomized trial. J Natl Cancer Inst.
103:1761–1770. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li N, Feng L, Han HQ, Yuan J, Qi XK, Lian
YF, Kuang BH, Zhang YC, Deng CC, Zhang HJ, et al: A novel Smac
mimetic APG-1387 demonstrates potent antitumor activity in
nasopharyngeal carcinoma cells by inducing apoptosis. Cancer Lett.
381:14–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bei JX, Li Y, Jia WH, Feng BJ, Zhou G,
Chen LZ, Feng QS, Low HQ, Zhang H, He F, et al: A genome-wide
association study of nasopharyngeal carcinoma identifies three new
susceptibility loci. Nat Genet. 42:599–603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He
J, Peng JY, Chen QY, Mo HY, Jun-Cui, et al: Exosomal miR-24-3p
impedes T-cell function by targeting FGF11 and serves as a
potential prognostic biomarker for nasopharyngeal carcinoma. J
Pathol. 240:329–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wen X, Tang X, Li Y, Ren X, He Q, Yang X,
Zhang J, Wang Y, Ma J and Liu N: Microarray expression profiling of
long non-coding RNAs involved in nasopharyngeal carcinoma
metastasis. Int J Mol Sci. 17:19562016. View Article : Google Scholar
|
|
10
|
Li W, Li J, Wang Y, Zhang K, Li N, Tian Z,
Ni B, Wang H and Ruan Z: Sphingosine kinase 1 is a potential
therapeutic target for nasopharyngeal carcinoma. Oncotarget.
7:80586–80598. 2016.PubMed/NCBI
|
|
11
|
Zhang L, Su B, Sun W, Li W, Luo M, Liu D,
Mei Q, Long G and Hu G and Hu G: Twist1 promotes radioresistance in
nasopharyngeal carcinoma. Oncotarget. 7:81332–81340.
2016.PubMed/NCBI
|
|
12
|
Zhang P, Du J, Sun B, Dong X, Xu G, Zhou
J, Huang Q, Liu Q, Hao Q and Ding J: Structure of human MRG15
chromo domain and its binding to Lys36-methylated histone H3.
Nucleic Acids Res. 34:6621–6628. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pardo PS, Leung JK, Lucchesi JC and
Pereira-Smith OM: MRG15, a novel chromodomain protein, is present
in two distinct multiprotein complexes involved in transcriptional
activation. J Biol Chem. 277:50860–50866. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bryce SD, Forsyth NR, Fitzsimmons SA,
Clark LJ, Bertram MJ, Cuthbert AP, Newbold RF, Pereira-Smith OM and
Parkinson EK: Genetic and functional analyses exclude mortality
factor 4 (MORF4) as a keratinocyte senescence gene. Cancer Res.
59:2038–2040. 1999.PubMed/NCBI
|
|
15
|
Yochum GS and Ayer DE: Role for the
mortality factors MORF4, MRGX and MRG15 in transcriptional
repression via associations with Pf1, mSin3A, and Transducin-Like
Enhancer of Split. Mol Cell Biol. 22:7868–7876. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Li J, Dunn S, Xiong S, Chen W,
Zhao Y, Chen BB, Mallampalli RK and Zou C: Histone deacetylase 2
(HDAC2) protein-dependent deacetylation of mortality factor 4-like
1 (MORF4L1) protein enhances its homodimerization. J Biol Chem.
289:7092–7098. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tominaga K, Kirtane B, Jackson JG, Ikeno
Y, Ikeda T, Hawks C, Smith JR, Matzuk MM and Pereira-Smith OM:
MRG15 regulates embryonic development and cell proliferation. Mol
Cell Biol. 25:2924–2937. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou C, Li J, Xiong S, Chen Y, Wu Q, Li X,
Weathington NM, Han S, Snavely C, Chen BB and Mallampalli RK:
Mortality factor 4 like 1 protein mediates epithelial cell death in
a mouse model of pneumonia. Sci Transl Med. 7:311ra1712015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang Y, Lin JC, Wang K, Chen YJ, Liu HH,
Luan R, Jiang S, Che T, Zhao Y, Li de F, et al: A nuclear ligand
MRG15 involved in the proapoptotic activity of medicinal fungal
galectin AAL (Agrocybe aegerita lectin). Biochim Biophys Acta.
1800:474–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garcia SN, Kirtane BM, Podlutsky AJ,
Pereira-Smith OM and Tominaga K: Mrg15 null and heterozygous mouse
embryonic fibroblasts exhibit DNA-repair defects post exposure to
gamma ionizing radiation. FEBS Lett. 581:5275–5281. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martrat G, Maxwell CM, Tominaga E,
Porta-de-la-Riva M, Bonifaci N, Gómez-Baldó L, Bogliolo M, Lázaro
C, Blanco I, Brunet J, et al: Exploring the link between MORF4L1
and risk of breast cancer. Breast Cancer Res. 13:R402011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin Y, Zhong J, Li SW, Li JZ, Zhou M, Chen
Y, Sang Y and Liu L: TRIM11, a direct target of miR-24-3p, promotes
cell proliferation and inhibits apoptosis in colon cancer.
Oncotarget. 7:86755–86765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lv XB, Liu L, Cheng C, Yu B, Xiong L, Hu
K, Tang J, Zeng L and Sang Y: SUN2 exerts tumor suppressor
functions by suppressing the Warburg effect in lung cancer. Sci
Rep. 5:179402015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res. 43:(Database
Issue). D856–D861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang RL, Peng LX, Yang JP, Zheng LS, Xie
P, Wang MY, Huang BJ, Zhao HR, Bao YX and Qian CN: IL-8 suppresses
E-cadherin expression in nasopharyngeal carcinoma cells by
enhancing E-cadherin promoter DNA methylation. Int J Oncol.
48:207–214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao L, Lin L, Pan C, Shi M, Liao Y, Bin J
and Liao W: Flotillin-2 promotes nasopharyngeal carcinoma
metastasis and is necessary for the epithelial-mesenchymal
transition induced by transforming growth factor-beta. Oncotarget.
6:9781–9793. 2015.PubMed/NCBI
|
|
28
|
Chen C, Lu Z, Yang J, Hao W, Qin Y, Wang
H, Xie C and Xie R: MiR-17-5p promotes cancer cell proliferation
and tumorigenesis in nasopharyngeal carcinoma by targeting p21.
Cancer Med. 5:3489–3499. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yi C, Wang Q, Wang L, Huang Y, Li L, Liu
L, Zhou X, Xie G, Kang T, Wang H, et al: MiR-663, a microRNA
targeting p21(WAF1/CIP1), promotes the proliferation and
tumorigenesis of nasopharyngeal carcinoma. Oncogene. 31:4421–4433.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sang Y, Cheng C, Tang XF, Zhang MF and Lv
XB: Hypermethylation of TET1 promoter is a new diagnosic marker for
breast cancer metastasis. Asian Pac J Cancer Prev. 16:1197–1200.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang B, Li Y, Qi G, Yuan S, Wang Z, Yu S,
Li B and He S: Clinicopathological significance of CDKN2A promoter
hypermethylation frequency with pancreatic cancer. Sci Rep.
5:135632015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen M, Pereira-Smith OM and Tominaga K:
Loss of the chromatin regulator MRG15 limits neural stem/progenitor
cell proliferation via increased expression of the p21 Cdk
inhibitor. Stem Cell Res. 7:75–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen J, Hao Y, Chen J, Huang L, Ao W, Yang
J, Li L, Heng J, Chen Z, Liang W, et al: Colony stimulating
factor-1 receptor promotes proliferation, migration and invasion in
the human nasopharyngeal carcinoma 6-10B cell line via the
phosphoinositide 3-kinase/Akt pathway. Oncol Lett. 16:1205–1211.
2018.PubMed/NCBI
|
|
34
|
Nie GH, Li Z, Duan HF, Luo L, Hu HY, Yang
WQ, Nie LP, Zhu RF, Chen XF and Zhang W: lncRNA C22orf32-1
contributes to the tumorigenesis of nasopharyngeal carcinoma. Oncol
Lett. 13:4487–4492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan Y, Du Y, Hu XY, Liu MY, Du JK, Liu
XM, Yu HE, Wang TZ, Pu JX, Zhong Q and Zou QF: Longikaurin A, a
natural ent-kaurane, suppresses stemness in nasopharyngeal
carcinoma cells. Oncology Lett. 13:1672–1680. 2017. View Article : Google Scholar
|