Introduction
Gastric cancer is one of the most common diseases
and the third leading cause for cancer-associated mortality in the
world (1). It was reported that in
2010 gastric cancer cases in China lead to a mortality rate of
21.89/100,000 (2). According to the
cancer control program of the World Health Organization, the
incidence and mortality rate of gastric cancer in China are >2
times of the global mean values (3).
Presently, common treatments for gastric cancer include surgery,
chemotherapy and radiation therapy. However, none of these methods
have resulted in a satisfactory decrease of morbidity or mortality
rates since diagnosis is usually made once the disease has reached
an advanced stage (4). Therefore, it
is necessary to find novel treatment approaches, including
biological therapies, to treat advanced gastric cancers.
Understanding the molecular basis of this disease is critical for
developing novel strategies for the prevention and treatment of
gastric cancer.
It is well established that cancer stem cells share
multiple characteristics with embryonic stem cells (ESCs),
including self-renewal and differentiation potential (5). Previous progression in stem tumor cell
research has revealed that tumorigenesis may be associated with
stem cells (6). Nanog homeobox
(NANOG) is a transcription factor that serves a vital function in
maintaining the pluripotency and self-renewal capacity of ESCs
(7,8).
NANOG is also highly expressed in certain somatic tumors, including
breast (9), prostate (10) and cervical cancers (11), thus indicating that NANOG is of vital
importance in tumor transformation and progression. In addition to
NANOG1, an authentic gene that encodes NANOG, there are 10
pseudogenes for NANOG in the human genome (12). NANOGP8, one of these pseudogenes, is
expressed together with NANOG in multiple tumor tissues and cell
lines (13). As such, NANOG/NANOGP8
may be a key component in tumor malignancy. However, the underlying
molecular mechanism of NANOG/NANOGP8 during tumorigenesis remains
unclear. The processes governing genesis and development of tumors
are complicated, and it is therefore critical to identify
downstream targets of NANOG/NANOGP8 in order to discover effective
treatment methods. Deleted in Breast Cancer 1 (DBC1) is a nuclear
protein encoded by a gene initially cloned from chromosome 8p21
that is homozygously deleted in breast cancer (14). Indeed, DBC1 mRNA is lost in several
breast, lung and colon cancer cell lines (14). This has led to the suggestion that
DBC1 is responsible for suppressing tumor development (14,15).
However, other studies have demonstrated that DBC1 is overexpressed
in breast, gastric and other types of cancer, and this over
expression was associated with poor prognosis (16–19). In
addition, the downregulation of DBC1 has been demonstrated to
inhibit gastric cancer cell proliferation and invasiveness
(20). Given these conflicting
findings, the function of DBC1 in tumorigenesis remains unclear,
however it is clear that DBC1 expression serves a key function in
the development and/or progression of many types of human
cancer.
The underlying molecular mechanisms of
NANOG/NANOGP8- or DBC1-mediated effects on gastric cancer are not
well known. In the present study, NANOG and DBC1 expression were
evaluated in gastric cancer cell lines and surgical biopsies. The
effects of NANOGP8 and DBC1 suppression or upregulation on cell
proliferation and apoptosis were investigated using the MKN-45 cell
line. DNA microarrays and dual-luciferase assays were performed to
examine the association between NANOGP8 and DBC1. The present
findings indicated that NANOGP8 promoted gastric cancer progression
by blinding to DBC1 promote region and upregulating DBC1
expression. Thus, NANOGP8 and DBC1 may serve as potential
therapeutic targets for human gastric cancer.
Materials and methods
Cell culture
Human teratocarcinoma cells (N-tera), the human
gastric carcinoma cell lines MKN-45, SGC-7901, HGC-27, NCI-N87,
MGC803 and BGC823, as well as the human normal gastric epithelial
cell line GES-1 were obtained from the Type Culture Collection of
the Chinese Academy of Sciences (Shanghai, China). Cells were
cultured in RPMI-1640 medium (HyClone; GE Healthcare, Logan, UT,
USA) containing 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), under standard conditions at
37°C in a humidified atmosphere containing 5% CO2.
Human tissue samples
Tissue samples were collected from 25 patients (10
male patients and 15 female patients, median, 60 years of age;
range, 45–78 years of age) with gastric cancer who underwent tumor
resection from May 2014 to May 2015 in the Affiliated Hospital of
Xuzhou Medical University (Xuzhou, China). These samples were
frozen in liquid nitrogen. None of the patients had been treated
with chemotherapy or radiotherapy prior to surgery. Written
informed consent was obtained from each patient and The Ethics
Committee of Xuzhou Medical University affiliated with the Hospital
approved the present study, in accordance with the Declaration of
Helsinki.
Animals
A total of 10 male nude mice aged 4–6 weeks (weight,
18–20 g) were used in the present study. The nude mice were
obtained from Shanghai Laboratory Animal Research Center (Shanghai,
China). The mice were kept in a 12 h light/12 h dark cycle and were
provided standard lab chow and tap water ad libitum. The
mice were housed at approximately 22–25°C with a humidity of
~40–70%. The nude mice were sacrificed by cervical dislocation on
the 30th day subsequent to inoculation (weight, 24–26 g) following
an intraperitoneal injection of 2.5% sodium pentobarbital with 0.2
ml/100 g. All experiments were carried out in accordance with the
guidelines established by the Institutional Animal Care and
approved by the Ethics Committee of Xuzhou Medical University
(Xuzhou, China).
Lentivirus package and stable cell
construction
NANOG shRNA, negative control shRNA (21), and DBC1shRNA (22) were generated by Invitrogen (Thermo
Fisher Scientific, Inc.) using sequences outlined in Table I. The concentration of shRNA was
adjusted to 100 nmol/l. shRNAs were then inserted into pLB plasmid
vector (Addgene, Inc., Cambridge, MA, USA). pLB lentiviral
particles containing shRNA were generated by transiently
transfecting 293T cells. Lentivirus production, concentration, and
titration were each performed according to standard procedures
(23).
 | Table I.shRNA design sequences. |
Table I.
shRNA design sequences.
| Gene | Sequence
(5′-3′) |
|---|
| NANOG shRNA-1 |
5′-AACCCTGGAACAGTCCCTTCTATATTCAAGAGATATAGAAGGGACTGTTCCAGGTTTTTTC-3′ |
|
|
5′-TCGAGAAAAAACCTGGAACAGTCCCTTCTATATCTCTTGAATATAGAAGGGACTGTTCCAGGGTT-3′ |
| NANOG shRNA-2 |
5′-AACGGGTTAAGCTGTAACATACTTTTCAAGAGAAAGTATGTTACAGCTTAACCCTTTTTTC-3′ |
|
|
5′-TCGAGAAAAAAGGGTTAAGCTGTAACATACTTTCTCTTGAAAAGTATGTTACAGCTTAACCCGTT-3′ |
| Control shRNA |
5′-AACTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTC-3′ |
|
|
5′-TCGAGAAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAGTT-3′ |
| DBC1 shRNA |
5′-AACCCCATCTGTGACTTCCTAGAATTCAAGAGATTCTAGGAAGTCACAGATGGGTTTTTC-3′ |
|
|
5′-TCGAGAAAAAACCCATCTGTGACTTCCTAGAATCTCTTGAATTCTAGGAAGTCACAGATGGGTT-3′ |
Lipofectamine 2000 was supplied by Invitrogen. For
infection, 2×105 MKN-45 cells were divided into four
groups and cultured in 6-well plates overnight. Cells were
maintained in RPMI-1640 medium, supplemented with 10% FBS (Gibco)
at 37°C in 5% CO2. For transduction, cell culture media
was removed and cells were washed twice with PBS. Then a
5×107 IU lentiviral suspension containing 8 µg/ml
polybrene was added to each well and cells were incubated at 37°C
for 24 h. Subsequently, the media was replaced with RPMI-1640
medium, containing 10% FBS and 5 µg/ml puromycine to select for
cells expressing the transduced vector. After 48 h, a flow
cytometer was used to sort cells containing green fluorescent
protein. These processes generated five groups: MKN-45 cells,
MKN-45 cells infected with lentiviral suspension expressing NANOG
shRNA-1, MKN-45 cells infected with NANOG shRNA-2, MKN-45 cells
infected with negative control lentiviral suspension, and MKN-45
cells infected with DBC1 shRNA.
Expression vector construction
The complete open reading frame of NANOGP8 was
amplified by polymerase chain reaction (PCR) using the following
primer pair (23):
5′-CAGGCAACTCACTTTATCC-3′ and 5′-TTAGGCTCCAACCATACTC-3′.
The pcDNA3.1(+) vector (Thermo Fisher Scientific,
Inc.) was digested 2 h with KpnI and XhoI (Takara Biotechnology
Co., Ltd., Dalian, China) at 37°C and the fragments subsequent to
digestion were recycled. A total of 3 µl PCR products and 1 µ1
recycled pcDNA3.1(+) were then connected using 1 µ1 T4 DNA ligase
(Takara Biotechnology Co., Ltd.) at 16°C for 12 h. The E.
coli DH5α competent cells were placed on ice for 10 min
following the addition of 10 µ1 ligation products. Subsequently,
the suspension were heat shocked in a 42°C water bath for 90 sec
and immediately placed on ice. The bacteria solution was used to
coat LB solid medium (Beyotime Institute of Biotechnology, Haimen,
China) containing kanamycin (25 µg/ml), which was cultured for
16–20 h. Several monoclonal positive colonies were selected the
next day and transferred into 4 ml LB liquid medium containing
kanamycin (25 µg/ml), which was placed in a 37°C shaker to
cultivate the bacteria for 12–16 h. The recombinant plasmid was
extracted by E.Z.N.A. Plasmid Minikit (Omega Bio-Tek, Inc.,
Norcross, GA, USA), according to manufacturer's protocols, and
identified by electrophoresis following digestion. The digested
products were subsequently sent to Invitrogen for sequencing
identification. The functional constructs were transfected using
Lipofectamine 2000 (Invitrogen) into MKN-45-shDBC1 cells, which
were screened using 500 mg/l G418 (Gibco) for 3 weeks. This yielded
MKN-45-shDBC1+NANOGP8 cells indicating stable downregulation of
DBC1 and upregulation of NANOGP8.
Reverse transcription (RT)-PCR and
sequencing of NANOG
Total RNA was extracted from MKN-45 cells and
gastric cancer biopsy samples using TRIzol® reagent
(Life Technologies; Shanghai, China) according to the
manufacturer's protocol. Primers for NANOG, DBC1, and β-actin were
as follows: NANOG forward, 5′-CAGAAGGCCTCAGCACCTAC-3′ and reverse,
5′-ATTGTTCCAGGTCTGGTTGC-3′; DBC1 forward,
5′-ATGTCCCAGTTTAAGCGCCAG-3′ and reverse,
5′-CAACCCCAAAGTAGTCATGCAA-3′; β-actin forward,
5′-ACTGTGCCCATCTACGAGG-3′ and reverse, 5′-GAAAGGGTGTAACGCAACTA-3′.
PCR was performed with the following thermocycling conditions: 94°C
for 5 min, 94°C for 30 sec, 53°C for 30 sec, 72°C for 35 sec for 35
cycles, with a final extension step at 72°C for 2 min. Products
were analyzed by electrophoresis on a 2% agarose gel. PCR products
were subsequently cloned into the pCR-Blunt vector (Invitrogen) and
sequenced.
Western blot analysis
MKN-45 cells were washed twice for 2 min with PBS
and resuspended in radioimmunoprecipitation assay buffer (Nanjing
KeyGen Biotech Co., Ltd., Nanjing, China) at 4°C. The protein
content was quantified using a bicinchoninic acid protein assay kit
(Beyotime Institute of Biotechnology), according to manufacturer's
protocols. A total of 200 µl protein lysate was separated using 10%
SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF)
membranes (Nanjing KeyGen Biotech Co., Ltd.), which were incubated
for 1 h in TBST (TBS with 1% Tween-20) containing 5% BSA (Gibco) at
room temperature. Tween-20 is a surfactant also known as
polyethylene glycol sorbitan monolaurate. Membranes were
subsequently incubated with primary antibodies overnight at 4°C as
follows: Anti-NANOG (dilution, 1:5,000; cat. no. ab109250),
anti-DBC1 (dilution, 1:10,000; cat. no. ab128890) and anti-β-actin
(dilution, 1:1,500; cat. no. ab8226; all Abcam, Cambridge, UK) at
4°C overnight. Membranes were subsequently washed 5 min in
triplicate with TBST at room temperature, incubated with
horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (dilution, 1:3,000; cat. no. k2034; Nanjing KeyGen Biotech
Co., Ltd.) at 37°C for 1 h, and washed in triplicate with TBST for
5 min at 37°C. The A (Lumino) and B (Hydrogen peroxide) solutions
of the electrochemiluminescence detection kit (Bio-rad, Franklin
Lakes, NJ, USA) were mixed in 1:1, according to the manufacturer's
protocols. The mixture was added to a PVDF membrane and allowed to
react at room temperature for 5 min in the dark. The protein
expression levels were subsequently detected through X-ray film
(Kodak, Rochester, NY, USA). The bands were obtained with
Imagequant LAS 4000 mini software (GE Healthcare Bio-Sciences,
Pittsburgh, PA, USA) and quantified with Quantity One 4.62 software
(Bio-rad).
Cell proliferation assay
Following cell transfection with sh-NANOGP8, the
effect of NANOGP8 silencing on cell proliferation was measured
using an MTT assay according to the manufacturer's protocol.
Control and transfected cells were seeded at a density of
5×103 cells/well in a 96-well flat-bottom plate and
cultured for 6 h at 37°C. MTT reagent (20 µl, 5 mg/ml) was then
added to each well, and cells were further incubated at 37°C for 4
h. Absorbance at 490 nm was measured using a microplate reader at
0, 24, 48 and 72 h. Each experiment was performed in triplicate and
repeated three times. The proliferation rate was calculated using
the following formula: Proliferation rate=survival
rate=[(ODtest-ODnegative
control)/ODnegative control] ×100%.
Flow cytometry analysis
Annexin V-APC (Allophycocyanin; BD Biosciences,
Franklin Lakes, NJ, USA) apoptosis detection was used in accordance
with manufacturer instructions to analyze apoptosis rate. Cells
were dissociated using trypsin then centrifuged at 100 × g for 5
min at room temperature. Cells were subsequently washed twice for 2
min with PBS at room temperature and resuspended at a density of
5×105 cells/ml in binding buffer (BD Biosciences).
Subsequently, 5 µl Annexin V-APC and 5 µl 7AAD (7-Aminoactinomycin
D) were added to the cell suspension, which was then incubated at
room temperature and protected from light for 15 min. Data was
acquired using the FACS Calibur Flow Cytometer (BD Biosciences) and
results were analyzed using FlowJo software V10 platform (Tree Star
Inc., Ashland, OR, USA). Each experiment was repeated three
times.
Colony forming assay
Cells were harvested and plated at a density of 800
cells/well in six-well plates. Following incubation for two weeks
at 37°C, cells were washed twice with PBS and fixed with 100%
methyl alcohol for 15 min at room temperature. Methyl alcohol was
then removed by washing the wells twice for 2 min with PBS and the
cells were stained with Giemsadye for 20 min at room temperature
and flushed with double distilled water. Clone formation was
quantified under phase contrast microscopy (magnification, ×400). A
clone was defined as containing more than 50 cells. Each assay was
performed in triplicate.
Tumorigenecity assay in nude mice
Cells were harvested and resuspended in PBS at a
density 1×107 cells/ml. Six-week old male athymic nude
mice were subcutaneously injected in the right armpit region with
0.2 ml cell suspension. Two groups of mice were injected with
MKN-45-NC and MKN-45-shNANOGP8 stable cells (n=5/group)
respectively. Tumor size was measured using calipers every 5 days.
Tumor volume was calculated using the formula (LxW2)/2,
where L is the length and W is the width of the tumor. At 30 days
after injection, mice were sacrificed and tumor volume and weight
were measured.
DNA microarray analysis
DNA microarray analysis was performed by Kangchen
BioTechCo., Ltd. (Shanghai, China). NANOG silenced and negative
control MKN-45 cells were analyzed by Nimble Gen Human Gene
Expression Microarrays, which were comprised of 29,250 genes. The
total RNA was extracted from cells, reverse transcribed into cDNA,
and marked with Cy3 dyes and Nimble Gen Microarray hybridization.
Microarrays were washed with buffer solution I for 5 min at room
temperature and with buffer solution II for 60 sec at 37°C (Roche
Diagnostics Indianapolis, IN, USA). Subsequently to being washed in
an ozone-free environment, the slides were scanned using the Axon
Genepix4000B microarray scanner (Molecular Devices, LLC, Sunnyvale,
CA, USA). The data was collected using NimbleScan software V2.5
(Roche Diagnostics, Basel, Switzerland). The results were
normalized and input into Agilent GeneSpringGX 11.0 software
(Agilent Technologies, Inc., Santa Clara, CA, USA) for further
analysis. The selection standard for differential gene expression
was a ratio ≥0.5.
Rescue experiment
Total RNA was extracted from sh-control, sh-NANOG,
sh-DBC1 and sh-DBC1+NANOG cells. RT-quantitative PCR (RT-qPCR) was
performed with the PrimerScript RT reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China) and SYBR-Green real-time
PCR Master Mix (Takara Biotechnology Co., Ltd. Dalian, China)
according to the manufacturer's protocol. The primer sequences were
the same as those aforementioned. Relative quantifications were
obtained via Cq values, and each sample for NANOGP8, DBC1, and
β-actin was run in triplicate. The expression levels of NANOGP8 and
DBC1 in gastric cancer cells was calculated using the
2−ΔΔCq relative quantification method with β-actin as a
reference (24).
Dual-luciferase assays
The predicted promoter region of DBC1 gene was from
−2000 base pairs to +500 base pairs (data not shown). A DBC1
promoter luciferase reporter vector was constructed, designated
pGL3-DBC1. For a dual-luciferase assay, 293T cells were cultured
without antibiotics overnight and then co-transfected with
pGL3-DBC1, pcDNA3.1(+)-NANOGP8, and the reference vector pRL-TK.
After 24 h, cells were lysed using PBS, and their luciferase
activities measured using a Dual-Luciferase Reporter Assay System
kit (Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocols.
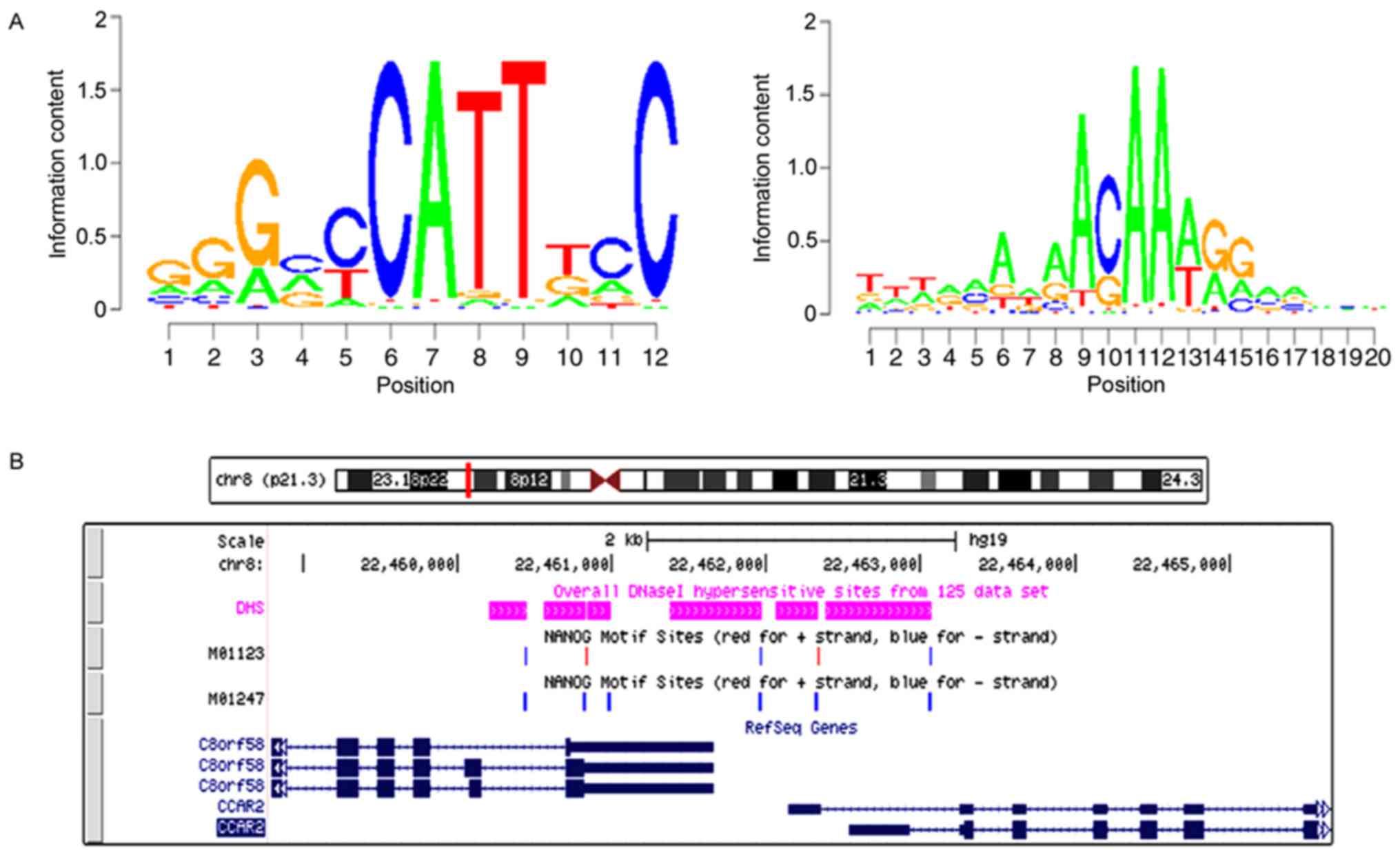
Predicting NANOG transcription factor
binding sites by MEME-chip software 5.0.1
Using MEME-chip 5.0.1 software (http://meme-suite.org/tools/meme-chip),
three known motif features (Motif) of NANOG transcription factor
binding site were identified from the transcription factor Motif
database (TRANSFAC http://www.gene-regulation.com/pub/databases.html#transfac),
and their corresponding IDs were the sequences of the three Motifs
of EN0298, M01247 and M01123. The open region of the chromosome
contained in the promoter region corresponding to the DBC1 was
extracted and Motif Screening analysis was performed on the Motif
sequences of EN0298, M01247 and M01123 in the open region of the
chromosome.
Statistical analysis
Data were analyzed using SPSS 16.0 software (SPSS,
Inc., Chicago, IL, USA). Results are depicted as the mean ±
standard deviation. A Student's t-test was used to compare means
between groups. To compare values between different groups, a
one-way analysis of variance was used and the least significant
difference test method was used as the post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
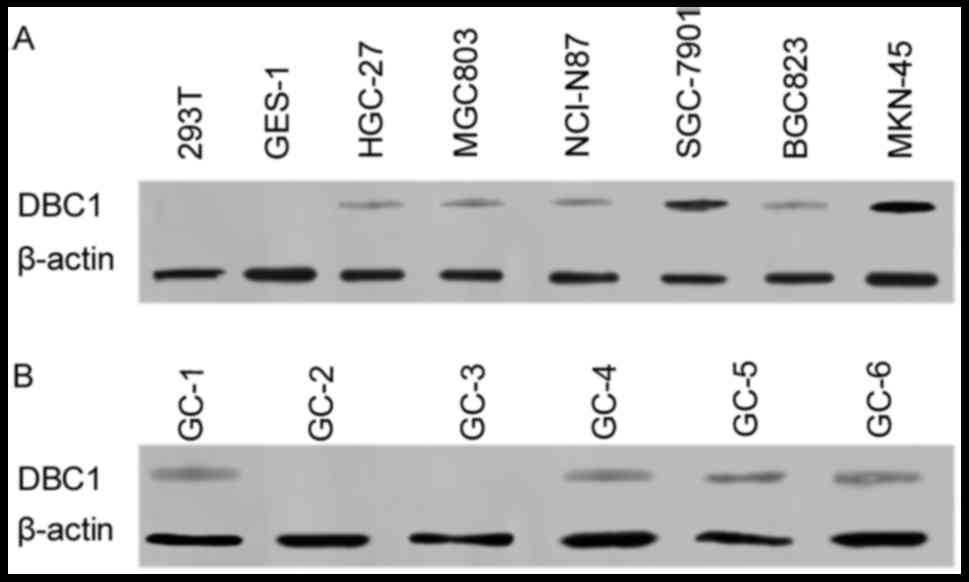
NANOG mRNA and protein expression in
gastric cancer cell lines and surgical specimens
To determine NANOG mRNA and protein expression
levels in gastric cancer, total RNA and protein was collected from
the human gastric carcinoma cells lines BGC823, SGC7901, MKN-45,
HGC-27, MGC-803 and NCI-N87, as well as from the normal human
gastric epithelial cell line GES-1. NANOG mRNA and protein
expression levels were examined by RT-PCR and western blotting,
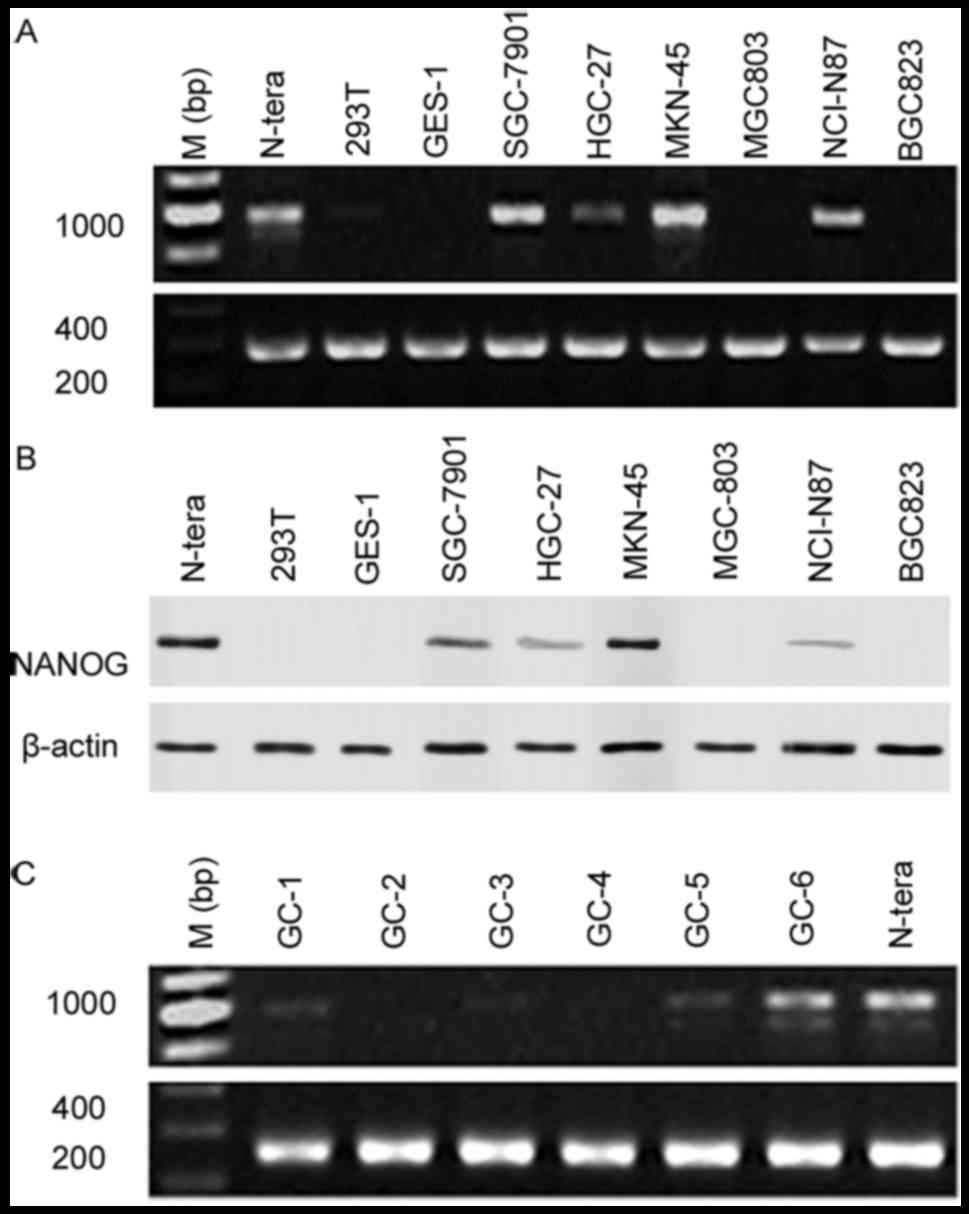
respectively. Data indicated that NANOG mRNA (SGC-7901, MKN-45,
NCI-N87) and protein (SGC-7901, HGC-27, MKN-45, NCI-N87) expression
was increased in gastric cancer cells compared with GES-1/293T
normal epithelial cells, as well as MGC803/BGC823 gastric cancer
cells (Fig. 1A and B), with NANOG
mRNA and protein expression highest in MKN-45 cells. NANOG mRNA
expression levels was also analyzed on 25 gastric cancer patient
specimens using PCR, with NANOG mRNA expression identified in 6
samples (Fig. 1C).
It was confirmed using PCR that N-tera cells
expressed NANOG1 mRNA. However, the PCR products in the MKN-45,
SGC-7901, HGC-27, and NCI-N87 cells, as well as in the gastric
cancer surgical specimens were highly homologous with the NANOGP8
gene. This suggested that the main NANOG gene expressed by gastric
cancer cell lines was NANOGP8 (Table
II). The NANOG shRNA target sequence was identical between the
NANOG and NANOGP8 genes, thus the NANOG shRNA may also be used to
silence NANOGP8 mRNA expression.
 | Table II.Sequence detection of NANOG
expression in gastric cancer cells and surgical samples. |
Table II.
Sequence detection of NANOG
expression in gastric cancer cells and surgical samples.
|
| Nucleotide |
|---|
|
|
|
|---|
| Sample | 47 | 144 | 165 | 246 | 276 | 531 | 759 | 798 |
|---|
| NANOG1
(NM_024865.2) | C | G | T | G | G | T | G | C |
| NANOGP8
(NG_004093) | A | A | T | T | G | C | C | C |
| N-tera | C | G | T | G | G | T | G | C |
| MKN-45 | A | A | T | T | G | C | C | C |
| SGC-7901 | C | A | T | T | G | C | C | C |
| HGC-27 | A | A | T | T | G | C | C | C |
| NCI-N87 | C | A | T | T | G | C | C | C |
| GC-1 | A | A | C | T | G | C | C | C |
| GC-5 | C | A | T | T | G | C | C | C |
| GC-6 | A | A | T | T | G | C | C | C |
Downregulation of NANOGP8 inhibits
cell proliferation and promotes apoptosis in human gastric
carcinoma cell lines
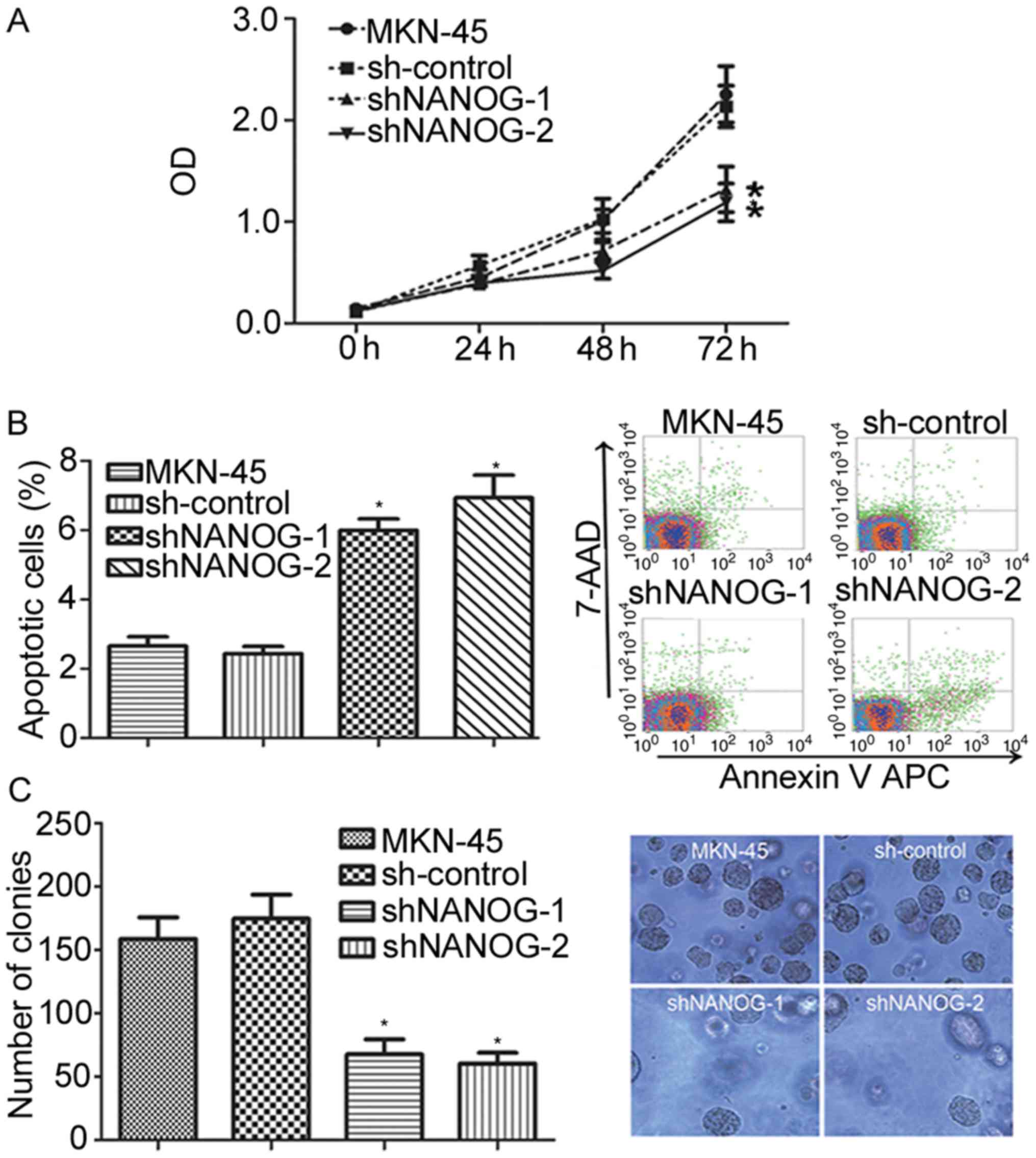
To observe the effects of NANOGP8 silencing on
MKN-45 cells, cell proliferation capacity was evaluated using MTT
and colony formation assays. Observation of cell proliferation for
72 h indicated that the proliferation rate of the sh-NANOG-1 and
sh-NANOG-2 groups was significantly inhibited compared with control
and parental cells (Fig. 2A;
P<0.05). The effects of NANOGP8 silencing on apoptosis were then
investigated using flow cytometry analysis, and it was identified
that the apoptosis rate of the sh-NANOG-1 and sh-NANOG-2 groups was
significantly increased compared to control or parental cells
(Fig. 2B). To further evaluate
proliferation ability, a colony formation assay was performed which
revealed that the sh-NANOG-1 and sh-NANOG-2 groups formed smaller
and fewer colonies than the parental or control cells (Fig. 2C). These results indicate that the
suppression of NANOGP8 expression in human gastric cancer cells
inhibits cell proliferation and promotes cell apoptosis in
vitro. In addition, the effects were greater in the shNANOG-2
group than the shNANOG-1 group. Therefore, shNANOG-2 was selected
for subsequent analyses.
Downregulation of NANOGP8 expression
in human gastric cancer cells inhibits tumor growth
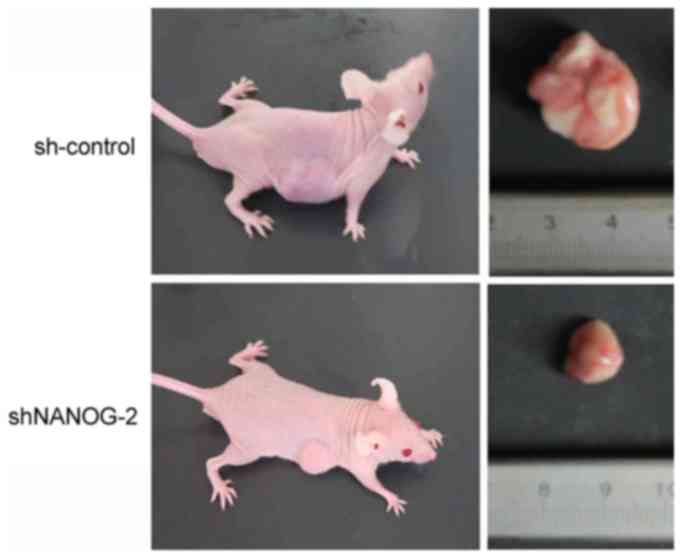
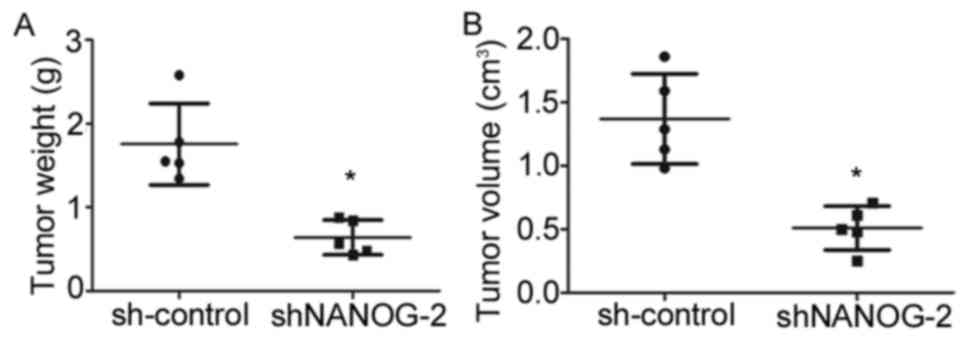
To determine whether long-term suppression of
NANOGP8 in MKN-45 cells affected tumor growth in vivo, a
tumorigenicity assay was performed in nude mice. Cell lines were
injected subcutaneously into the right armpit region of nude mice.
Tumor size and weight measurements revealed that nude mice injected
with NANOGP8-silenced cells generated smaller and lighter tumors
compared to sh-control cells (Figs. 3
and 4).
DBC1 mRNA and protein expression in
gastric cancer cell lines and surgical specimens
To identify downstream targets of NANOGP8, DNA
microarray analysis was performed. The results suggested multiple
gene expression changes following knockdown of NANOGP8 expression.
In particular, DBC1 was significantly downregulated (data not
shown). Previous research has demonstrated that DBC1 is
overexpressed and associated with poor prognosis in gastric cancer
(15). DBC1 also has influences on
gastric cancer cell proliferation and invasiveness (19). We subsequently hypothesized that DBC1
was a differentially expressed gene and has an intrinsic
association with the NANOGP8 gene to participate in the occurrence
and development of gastric cancer. Therefore, RT-PCR and western
blotting were used to detect whether DBC1 expression was altered in
gastric cancer cell lines and surgical specimens. The results, as
predicted, indicated that DBC1 mRNA expression was increased in
gastric cancer cells compared to normal gastric epithelial cells.
Furthermore, DBC1 protein expression was identified in 4/6
NANOG-expressing gastric cancer surgical specimens (Fig. 5).
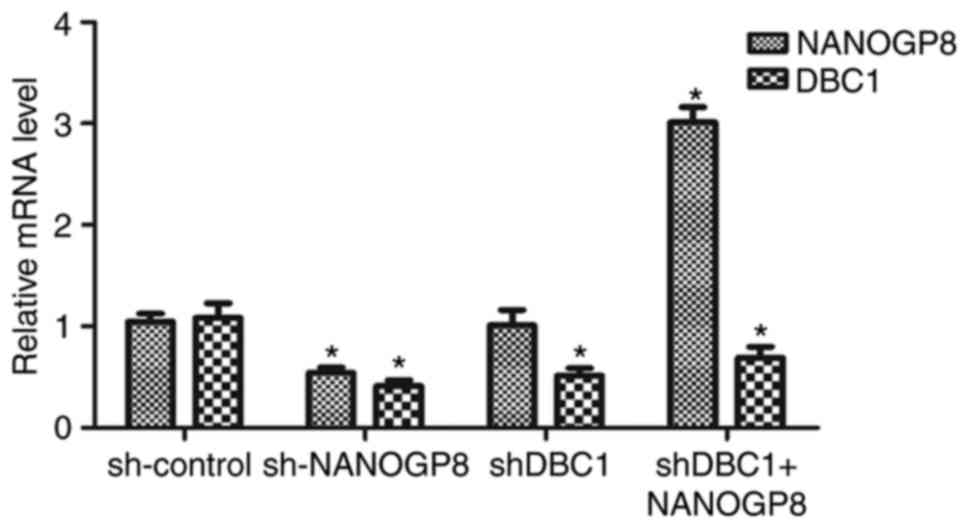
Rescue experiment
To further confirm the association between NANOGP8
and DBC1 expression, MKN-45 cells were divided into four groups as
follows: sh-control, sh-NANOGP8, sh-DBC1 and shDBC1+NANOGP8. The
results indicated that DBC1 expression was markedly downregulated
following NANOGP8 knockdown. However, downregulation of DBC1 did
not affect NANOG mRNA expression and the effect of silencing DBC1
on MKN-45 cells could be rescued by overexpression of NANOGP8,
indicating that NANOGP8 could regulate DBC1 mRNA expression
(Fig. 6).
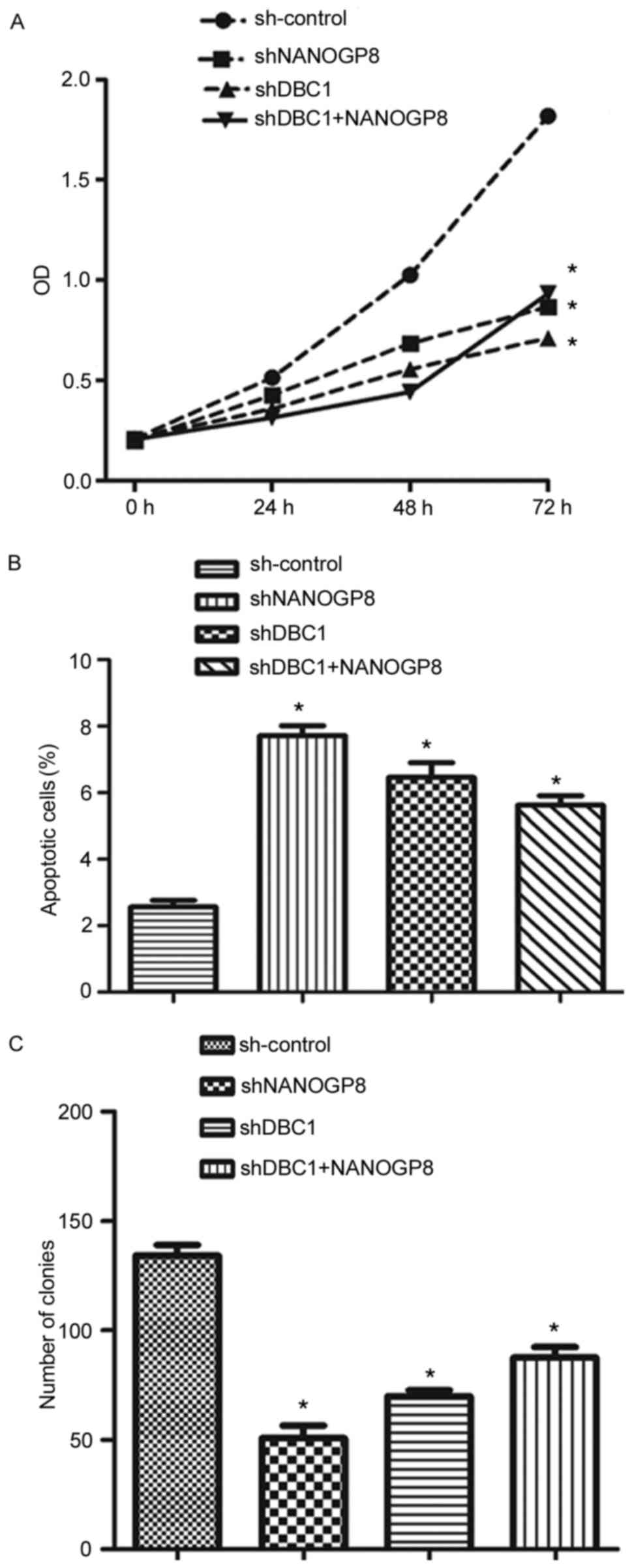
To confirm the aforementioned results, MTT, flow
cytometry and colony formation assays were performed. The results
indicated that silencing DBC1 inhibited cell proliferation and
promoted apoptosis relative to sh-control cells. Meanwhile, the
effects of DBC1 downregulation on cell proliferation and apoptosis
could be rescued by upregulation of NANOGP8 (Fig. 7). Taken together, these findings
indicated that NANOGP8 promoted gastric cancer progression by
regulating DBC1.
NANOGP8 promotes gastric cancer cell
progression by transactivating DBC1
The binding site for NANOGP8 within the DBC1
promoter region was analyzed. Based on the known motif sequence
signature of NANOGP8, motif screening analysis was performed from
M01123 Motif to M01247 Motif. The results revealed that the NANOGP8
binding site with DBC1 was in the DBC1 promoter region (Fig. 8).
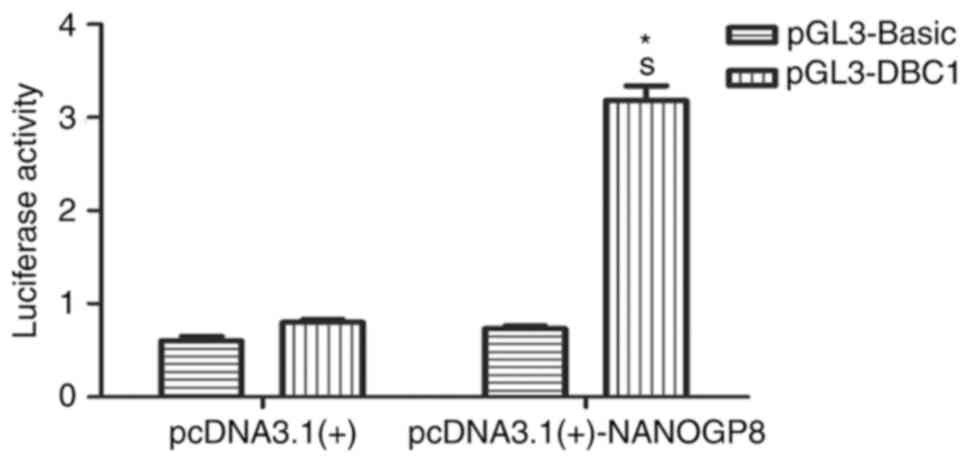
To test whether NANOG regulates DBC1 transcription,
a dual-luciferase reporter assay was performed. pGL3-DBC1 or
pGL3-Basic vectors were co-transfected with pcDNA3.1-NANOGP8 or
pcDNA3.1 vectors into 293T cells. Cellular luciferase activity was
measured at 24 h following transfection. The results revealed that
NANOGP8 transactivated the DBC1 promoter (Fig. 9; P<0.05). Taken together, these
results indicated that NANOGP8 promotes gastric cancer cell
progression by transactivating DBC1 expression.
Discussion
The objective of the present study was to determine
the effects of NANOGP8 in gastric cancer cell proliferation,
apoptosis and tumorigenicity, and resolve the underlying molecular
mechanisms. NANOG, a transcription factor expressed in primordial
germ cells and embryonic stem cells, is an important regulatory
factor for maintaining gastric cancer stem cells self-renewal and
pluripotency (25). In addition,
NANOGP8 expression regulates proliferation and migration inhuman
gastric cancer SGC-7901 cell line (26). In the present study, the
differentiation status between SGC-7901 and MKN-45 cell lines
differed. However, the effects of NANOGP8 on proliferation and
apoptosis are consistent in SGC-7901 and MKN-45 cell lines,
indicating that NANOG, serving as a promoter of gastric cancer
progression, is an independent factor in gastric cancer cell
differentiation status.
It has been reported that NANOGP8 overexpression
significantly promotes the proliferation of tumor cells in
vitro and in vivo (27,28). Chiou
et al (29) also demonstrated
that NANOG is positively associated with late stage progression and
poorer prognosis for patients with oral cancer. However, the
underlying NANOGP8-mediated mechanisms of tumor development remain
unknown. In the present study, NANOG was overexpressed in most
gastric cancer cell lines, and NANOG mRNA expression was detected
in 6/25 gastric surgical specimens. These results demonstrate that
NANOG, as a cell-fate regulatory molecule known to be important for
ESC self-renewal, may serve a novel function in gastric cancer
progression. NANOG1 is an authentic gene that encodes NANOG. NANOG1
and NANOGP8 genes responsible for NANOG, encode similar
polypeptides that differ from NANOG1 by only six nucleotides and
two amino acids (30), thus their
gene functions are similar and products are almost
indistinguishable due to high degree of homology between them. The
results of sequencing indicated that the products of gastric cancer
cells and specimens were highly homologous with regard to the
NANOGP8 gene. Therefore, it was hypothesized that NANOGP8 is likely
a primary contributor of NANOG protein expression in gastric
cancer.
The in vitro study results suggested that
downregulating NANOGP8 expression inhibited cell proliferation,
colony formation and promoted cell apoptosis in MKN-45 cells. An
in vivo tumorigenicity assay in nude mice was also employed
to verify this assumption. The gathered data indicated that NANOGP8
acted as an oncogene in gastric carcinomas. Furthermore, DBC1 has
previously been associated with human breast cancer (31) and, in the present study, identified as
a target gene of NANOG8. However, rather than being deleted, DBC1
expression is increased in certain human cancers (16–19).
Silencing DBC1 in MKN-45 cells could be rescued by overexpression
of NANOGP8. Furthermore, NANOGP8 protein was identified to directly
bind to the promoter region of DBC1. It was therefore hypothesized
that NANOGP8 may modulate tumor growth and metastasis through
directly regulating the expression of DBC1.
To conclude, the results of the present study
demonstrated that NANOGP8 serves as an oncogene to promote
proliferation and suppress apoptosis in MKN-45 human gastric cancer
cells. The role of NANOGP8 gene was observed through
transcriptional regulation by binding to the DBC1 promoter region.
Further study is required in order to examine whether the
NANOGP8/DBC1 pathway is associated with clinicopathological
characteristics of gastric cancer.
Acknowledgments
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant no. 81470303), The Jiangsu
Province Natural Science Foundation of China (grant no. BK20141140)
and natural science research projects in the universities in
Jiangsu province (grant no. 14KJB320021).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL, GJ and JZ designed the experiment. LL, SF and JC
performed the experiment. RF and QZ analyzed the data. LL, RF and
JZ wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients were required to provide written
informed consent prior to their inclusion in the present study. The
study was approved by the Ethical Committee of The Affiliated
Hospital of Xuzhou Medical University.
Patient consent for publication
All patients provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidenceand mortality worldwide: Sources, methods and
majorpatterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
3
|
Ferlay J, Shine HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wadhwa R, Taketa T, Sudo K, Blum MA and
Ajani JA: Modern oncological approaches to gastric adenocarcinoma.
Gastroenterol Clin North Am. 42:359–369. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Colmont CS, Harding KG, Piguet V and Patel
GK: Human skin cancer stem cells: A tale of mice and men. Exp
Dermatol. 21:576–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reya T, morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chambers I, Colby D, Robertson M, Nichols
J, Lee S, Tweedie S and Smith A: Functional expression cloning of
Nanog, a pluripotency sustaining factor in embryonic stem cells.
Cell. 113:643–655. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitsui K, Tokuzawa Y, Itoh H, Segawa K,
Murakami M, Takahashi K, Maruyama M, Maeda M and Yamanaka S: The
homeoprotein Nanog is required for maintenance of pluripotency in
mouse epiblast and ES cells. Cell. 113:631–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ben-Porath I, Thomson MW, Carey VJ, Ge R,
Bell GW, Regev A and Weinberg RA: An embryonic stem cell-like gene
expression signature in poorly differentiated aggressive human
tumors. Nat Genet. 40:499–507. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gu G, Yuan J, Wills M and Kasper S:
Prostate cancer cells with stem cell characteristics reconstitute
the original human tumor in vivo. Cancer Res. 67:4807–4815. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ye F, Zhou C, Cheng Q, Shen J and Chen H:
Stem-cell-abundant proteins Nanog, Nucleostemin and musashi1 are
highly expressed in malignant cervical epithelial cells. BMC
Cancer. 8:1082008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fairbanks DJ and Maughan PJ: Evolution of
the NANOG pseudogene family in the human and chimpanzee genomes.
BMC Evol Biol. 6:122006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Wang X, Chen B, Suo G, Zhao Y,
Duan Z and Dai J: Expression of Nanog gene promotes NIH3T3 cell
proliferation. Biochem Biophys Res Commun. 338:1098–1102. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hamaguchi M, Meth JL, von Klitzing C, Wei
W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC and Wigler MH:
DBC2, A candidate for a tumor suppressor gene involved in breast
cancer. Proc Natl Acad Sci USA. 99:13647–13652. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim JE, Chen J and Lou Z: p30 DBC is a
potential regulator of tumorigenesis. Cell Cycle. 8:2932–2935.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH,
Park HS, Lee H, Chung MJ, Kang MJ, Lee DG, et al: Expression of
DBC1 and SIRT1 is associated with poor prognosis of gastric
carcinoma. Clin Cancer Res. 15:4453–4459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hiraike H, Wada-Hiraike O, Nakagawa S,
Koyama S, Miyamoto Y, Sone K, Tanikawa M, Tsuruga T, Nagasaka K,
Matsumoto Y, et al: Identification of DBC1 as a transcriptional
repressor for BRCA1. Br J Cancer. 102:1061–1067. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kang Y, Jung WY, Lee H, Lee E, Kim A and
Kim BH: Expression of SIRT1 and DBC1 in Gastric Adenocarcinoma.
Korean J Pathol. 46:523–531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Gu Y, Sha S, Kong X, Zhu H, Xu B,
Li Y and Wu K: DBC1 is over-expressed and associated with poor
prognosis in colorectal cancer. Int J Clin Oncol. 19:106–112. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bae JS, Park SH, Kim KM, Kwon KS, Kim CY,
Lee HK, Park BH, Park HS, Lee H, Moon WS, et al: CK2α
phosphorylates DBC1 and is involved in the progression of gastric
carcinoma and predicts poor survival of gastric carcinoma patients.
Int J Cancer. 136:797–809. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao J, Meng FJ, Li L, Lu C, Zhou J, Cheng
H, Chen W, Chen C and Xu KL: Expression of NANOG gene in acute
lymphoblastic leukemia cells and construction of lentiviral vector
carrying NANOG specific shRNA. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
22:275–279. 2014.(In Chinese). PubMed/NCBI
|
|
22
|
Qin B, Minter-Dykhouse K, Yu J, Zhang J,
Liu T, Zhang H, Lee S, Kim J, Wang L and Lou Z: DBC1 functions as a
tumor suppressor by regulating p53 stability. Cell Rep.
10:1324–1334. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao J, Chen C, Zeng L, Li L, Li Z and Xu
K: Engineered regulatory T cells prevent graft-versus-host disease
while sparing the graft versus-leukemia effect after bone marrow
transplantation. Leuk Res. 34:1374–1382. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takaishi S, Okumura T and Wang TC: Gastric
cancer stem cells. J Clin Oncol. 26:2876–2882. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang Z, Liu Y and Wang C: Oncogenic
NANOGP8 expression regulates cell proliferation and migration
through the Akt/mTOR signaling pathway in human gastric
cancerSGC-7901cell line. Onco Targets Ther. 9:4859–4866. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang J, Wang X, Li M, Han J, Chen B, Wang
B and Dai J: NANOGP8 is a retrogene expressed in cancers. FEBS J.
273:1723–1730. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uchino K, Hirano G, Hirahashi M, Isobe T,
Shirakawa T, Kusaba H, Baba E, Tsuneyoshi M and Akashi K: Human
Nanog pseudogene8 promotes the proliferation of gastrointestinal
cancer cells. Exp Cell Res. 318:1799–1807. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ,
Tsai TH, Chou SH, Chien CS, Ku HH and Lo JF: Positive correlations
of Oct-4 and NANOG in oral cancer stem-like cells and high-grade
oral squamous cell carcinoma. Clin Cancer Res. 14:4085–4095. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Booth HA and Holland PW: Eleven daughters
of NANOG. Genomics. 84:229–238. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zannini L, Buscemi G, Kim JE, Fontanella E
and Delia D: DBC1 phosphorylation by ATM/ATR inhibits SIRT1
deacetylase in response to DNA damage. J Mol Cell Biol. 4:294–303.
2012. View Article : Google Scholar : PubMed/NCBI
|