Introduction
Osteosarcoma (OS) is one of the most frequently
diagnosed malignancies in child and adolescent populations
(1,2).
With the advance of multiple therapeutic strategies, including
surgical resection, adjuvant chemotherapy and radiotherapy, the
survival rate of OS patients has significantly increased (3,4). However,
the 5-year survival rate remains poor due to local relapse and
metastasis, especially pulmonary metastasis, after resection of the
primary OS (4,5). Therefore, research is needed to further
elucidate the complex molecular mechanisms underlying OS
tumorigenesis and progression and to identify novel early molecular
markers of diagnostic and therapeutic targets for OS.
Long non-coding RNAs (lncRNAs) are more than 200
nucleotides in length, endogenously expressed, and have important
biological functions (6–8). Increasing evidence indicates that
lncRNAs play critical roles in the modulation of cellular
biological function, such as differentiation, apoptosis and
metastasis (9–12). Several specific lncRNAs have been
reported to increase in OS and serve as a diagnostic marker for OS
(13–17). The lncRNA X-inactive specific
transcript (XIST), derived from XIST gene, was found to dysregulate
in many cancers and play an important role in cell proliferation,
differentiation and genome maintenance (18–22). For
example, lncRNA XIST was increased in aggressive tumor phenotypes
and down-regulated the expression of EZH2 via miR-101 in gastric
cancer (18). IncRNA XIST was found
to be over-expressed in non-small-cell lung carcinoma (NSCLC) and
this high level was associated with a shorter survival rate and
poorer prognosis. Knockdown of XIST impaired NSCLC cell
proliferation, migration and invasion by interacting with EZH2 to
suppress the transcription of KLF2 (19). More recently, it has been shown that
upregulation of XIST contributes to cell proliferation and invasion
by inhibiting miR-497 expression (23). However, biological roles and
underlying molecular mechanisms of XIST in OS tumorigenesis remain
largely unknown.
In our study, we explored the expression and roles
of XIST in OS progression. We found the expression level of XIST
was increased in OS tumor tissues, and high XIST expression was
associated with Enneking stage, metastasis and a short overall
survival rate. Furthermore, knockdown of XIST inhibited cell
proliferation, migration and invasion.
Materials and methods
Tissue specimens
A total of 64 pairs of OS tissues and their matched
adjacent normal bone were obtained from patients who underwent
surgery at The Xuanwu Hospital of Capital Medical University
(Beijing, China) from March 2008 to October 2009. All patients'
operation treatments were performed by the same senior doctors
according to the patients' Enneking staging. None of the patients
had received radiotherapy or chemotherapy before surgery. This
study was approved by the Research Ethics Committee of the Capital
Medical University with the permit no. 2014A083. Written informed
consent was acquired from all the patients. Tissue specimens were
immediately frozen in liquid nitrogen after surgery and stored at
−80°C until use.
Cell culture
OS cell lines (KHOS-240S, SaOS2, MG-63, SOSP-9607
and U2OS), and osteoblastic cell line (hFOB1.19) were purchased
from the Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China). All cells were cultured in Dulbecco's
modified Eagle's medium or Ham's F12/Dulbecco's modified Eagle's
medium (both GE Healthcare, Logan, UT, USA) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 100 U/ml of penicillin and 100 mg/ml of streptomycin at
37°C in a humidified atmosphere with 5% CO2.
RNA isolation and quantitative
real-time PCR (qRT-PCR)
Total RNA was extracted from the tissue samples and
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The
first-strand cDNA was reverse-transcribed from 500 ng of total RNA
using a PrimeScript™ II 1st Strand cDNA Synthesis kit
(Takara, Dalian, China). qRT-PCR was performed using the FastStart
Universal SYBR-Green Master Mixes (Roche Diagnostics Corp.,
Indianapolis, IN, USA) on a 7900 Fast Real-Time PCR system (Applied
Biosystems, Foster City, CA, USA). The following primers were used
to detect the expression of XIST and GAPDH: XIST sense,
5′-CTCTCCATTGGGTTCAC-3′ and reverse, 5′-GCGGCAGGTCTTAAGAGATGAG-3′;
GAPDH sense, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse,
5′-AGGGGCCATCCACAGTCTTC-3′. The expression of XIST was normalized
to GAPDH.
Cell transfection
The small interfering RNA (siRNA) specifically
targeting XIST was designed and commercially constructed by
GenePharma (Shanghai, China). The scrambled nucleotide was used as
the negative control (si-NC). The sequences were as follows: siRNA1
sense, 5′-GUAUCCUAUUUGCACGCUAdTdT-3′; siRNA2 sense,
5′-GCCCUUCUCUUCGAACUGUdTdT-3′; negative control siRNA sense,
5′-UUCUCCGAACGUGUCACGUdTdT-3′. Cells were transfected separately
with the siRNAs and si-NC using FuGENE 6 transfection reagent
(Roche Diagnostics) to knockdown the XIST. The interfering
efficiency was confirmed by qRT-PCR after transfection for 48
h.
Cell proliferation assay
Cell proliferation assay was used to examine the
effect of XIST knockdown on viability of OS cells. In brief,
2×103 cells transfected with MG-63 and U2OS were seeded
into 96-well plates. At 24, 48, 72, and 96 h, 10 µl Cell Counting
kit-8 (CCK-8) solution (Dojindo, Tokyo, Japan) was added into each
well and incubated for 2 h at 37°C. The absorbance was measured on
a microplate reader at 450 nm by Biotek Elx800 ELISA (BioTek,
Winooski, VT, USA).
Flow cytometry analysis
For cell apoptosis analysis, cells were harvested
and stained with FITC-Annexin V and propidium iodide (PI; BD
Biosciences, San Jose, CA, USA). The fluorescence of stained cells
was then examined using flow cytometry (BD Biosciences) according
to the manufacturer's protocol. For cell cycle analysis, cells were
harvested, washed with cold PBS, and fixed in 70% ethanol at 4°C
overnight. The cells were then stained with PI in the dark for 15
min at room temperature. The proportion of cells in the G0/G1, S,
and G2/M phases were determined by flow cytometry.
Cell migration and invasion
assays
Cell migration ability was assessed using a 24-well
plate with 8-µm pore size chamber inserts (Corning Inc., New York,
NY, USA). Cell invasion ability was assessed using a 24-well plate
with 8-µm pore size chamber inserts coated with Matrigel (BD
Biosciences). 5×104 cells were suspended in 150 µl of
serum-free medium and were placed in the upper chamber. Six hundred
µl of medium containing 10% FBS was added to the lower chamber as
the chemoattractant. Following incubation for 36 h at 37°C, the
non-invading cells were gently removed with a cotton swab. Invasive
cells located on the lower side of the chamber were fixed in 95%
ethanol and stained with 0.5% crystal violet. The numbers of cells
in five randomly selected fields were counted, and the cells were
imaged through a CKX41 inverted microscope (Olympus Corp., Tokyo,
Japan).
Western blot analysis
Total protein from cells was extracted by lysing
cells in RIPA buffer (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) containing protease and phosphatase inhibitors (Roche
Diagnostics). Equivalent protein lysates were separated by 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred to polyvinylidene fluoride membranes (EMD Millipore,
Billerica, MA, USA). The membranes were then blocked using blocking
buffer and then incubated with primary antibody overnight at 4°C.
After the membranes were incubated with secondary antibody, the
signals were visualized using a SuperSignal West Pico
Chemiluminescent Substrate kit (Pierce; Thermo Fisher Scientific,
Inc.). Antibodies against Vimentin, c-Myc, E-cadherin, and GAPDH
were purchased from Cell Signaling Technology (Danvers, MA,
USA).
Statistical analysis
All the experiments were repeated in triplicate, and
data was expressed as the mean ± standard deviation (SD).
Statistical analyses were performed using SPSS 18.0 software
package (SPSS Inc, Chicago, IL, USA). One-way or two-way analyses
of variance was performed with Dunnett's post hoc test to determine
statistical differences between >2 groups. The Chi-squared test
was used to evaluate the association between the XIST expression
and the clinicopathological features. The Kaplan-Meier method and
log-rank test were used for the survival analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
LncRNA XIST is highly expressed in OS
cell lines and tissues
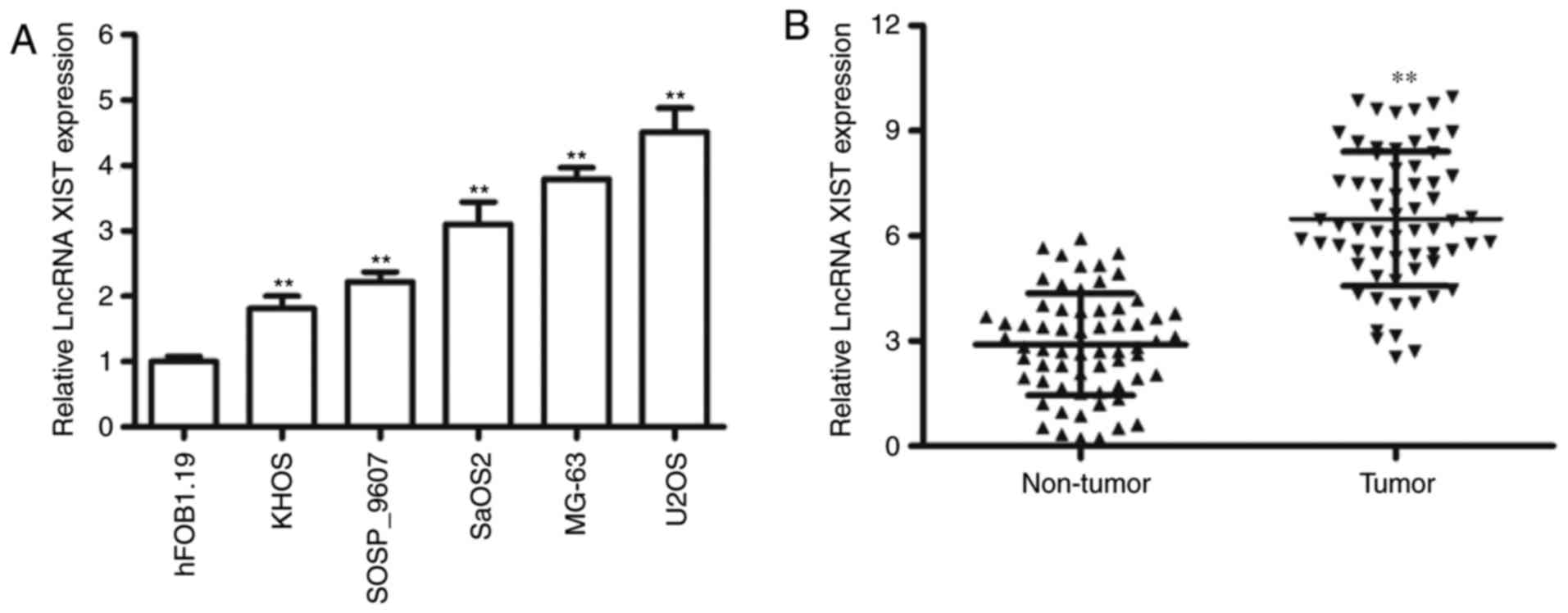
To evaluate the expression of XIST, we measured the
expression of XIST in OS cell lines and our results indicated that
the expression level of XIST was relatively high in four OS cell
lines, including KHOS-240S, SaOS2, MG-63 and U2OS, compared to the
osteoblastic cell line hFOB1.19 (Fig.
1A). Furthermore, we determined the levels of XIST expression
in 64 pairs of OS tissues and adjacent non-tumor tissues. The
results suggested that XIST was significantly increased in the
cancerous tissues compared with the adjacent normal samples
(Fig. 1B). Our results suggested that
significant upregulation of XIST was frequently observed in the
majority of the OS tumors and cell lines.
High levels of lncRNA XIST predict
poor prognosis in OS patients
To assess the clinical significance, OS tissues were
divided into two groups including the high XIST expression group (n
= 32) (higher than median value) and the low XIST expression group
(n = 32) (lower than median value) according to the median value of
XIST expression level of all samples. As shown in Table I, the high XIST expression group was
positively associated with Enneking stage and metastasis. However,
there was no significant correlation between XIST expression and
other clinicopathological features such as sex, age, tumor
location, tumor size and pathologic fracture. In addition,
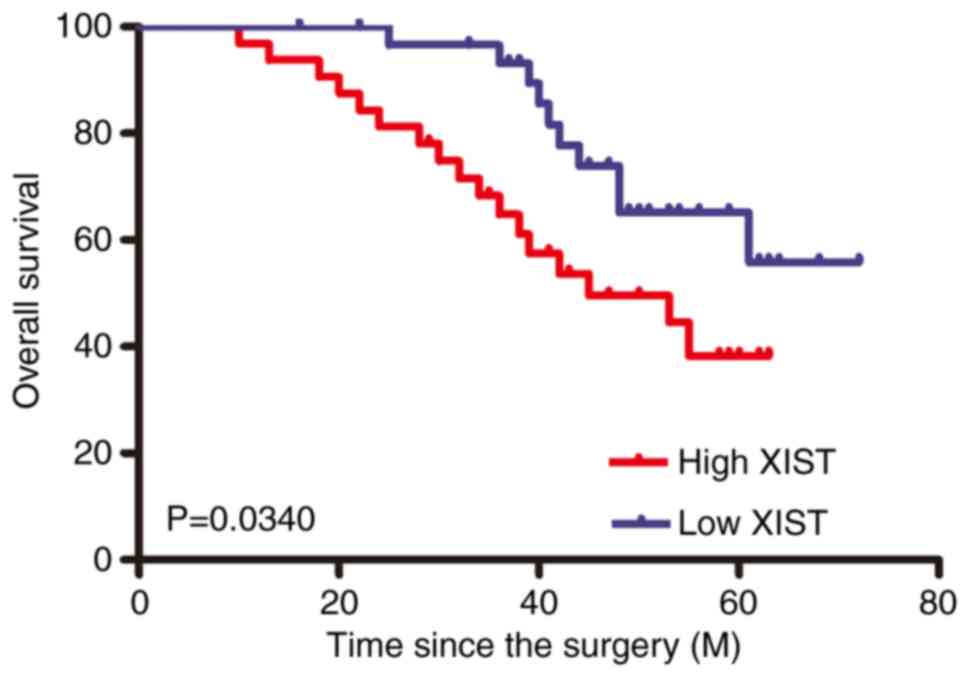
Kaplan-Meier survival curves showed that patients in the high XIST
expression group had a worse overall survival rate than those in
the low XIST expression group (Fig.
2).
 | Table I.Correlation between XIST and
clinicopathologic factors in patients with osteosarcoma. |
Table I.
Correlation between XIST and
clinicopathologic factors in patients with osteosarcoma.
|
| XIST expression |
|
|---|
|
|
|
|
|---|
| Variables | Low (n=32) | High (n=32) | P-value |
|---|
| Sex |
|
| 0.3171 |
|
Female | 14 | 19 |
|
| Male | 18 | 13 |
|
| Age, years |
|
| 0.5950 |
| ≤18 | 20 | 23 |
|
|
>18 | 12 | 9 |
|
| Enneking stage |
|
| 0.0051 |
|
IA-IIA | 24 | 12 |
|
|
IIB-III | 8 | 20 |
|
| Tumor size, cm |
|
| 0.3114 |
|
<5 | 16 | 21 |
|
| ≥5 | 16 | 11 |
|
| Pathologic
fracture |
|
| 0.7323 |
|
Absent | 28 | 26 |
|
|
Present | 4 | 6 |
|
| Location |
|
| 0.7818 |
| Upper
limbs | 7 | 8 |
|
| Lower
limbs | 19 | 20 |
|
|
Other | 6 | 4 |
|
| Metastasis |
|
| 0.0120 |
|
Absent | 29 | 22 |
|
|
Present | 3 | 12 |
|
Knockdown of LncRNA XIST reduced HCC
cell growth
Since XIST was significantly upregulated in OS
tissues, we investigated the biological function of XIST silencing
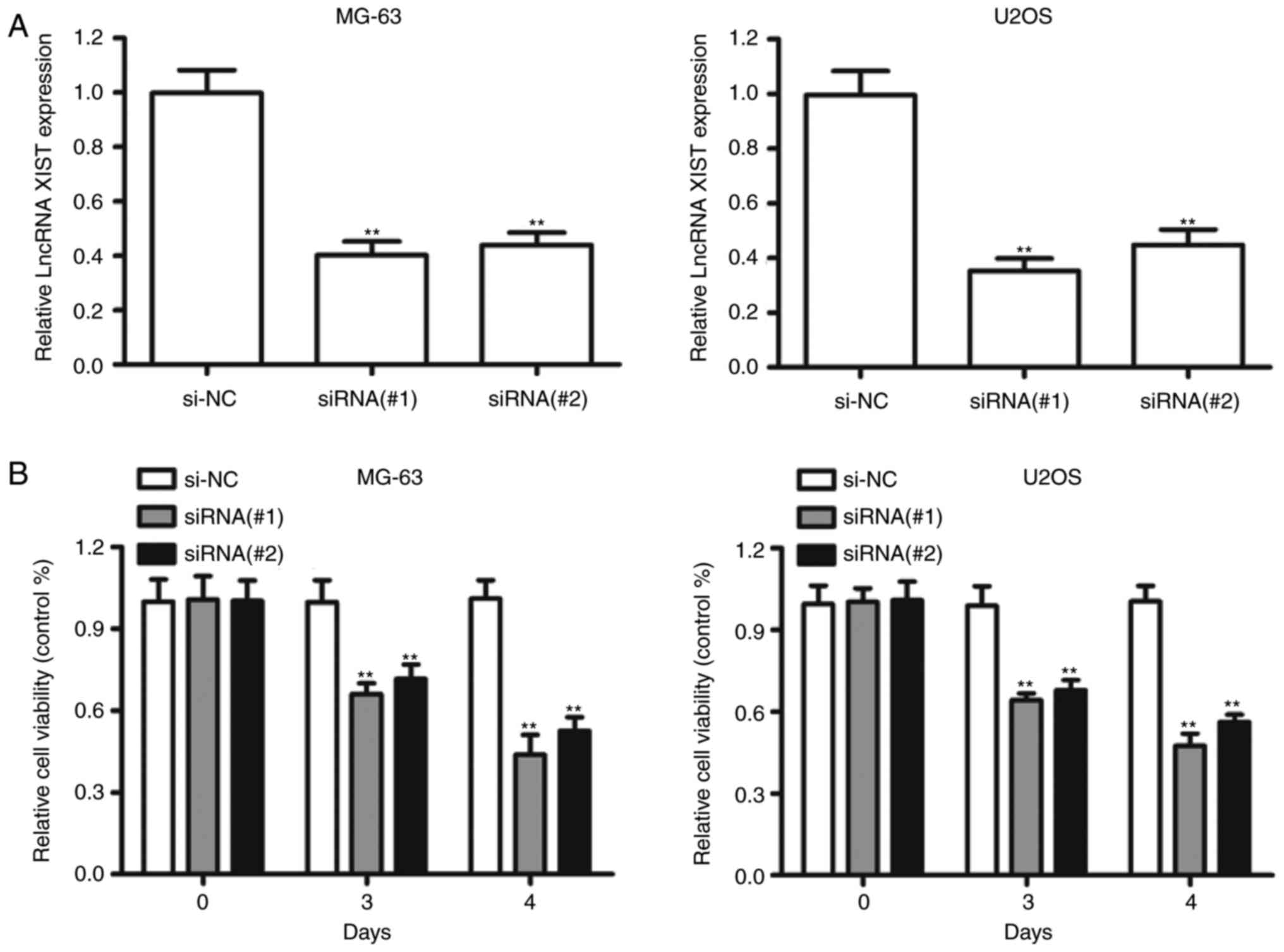
in OS cells. XIST was downregulated by transfecting the siRNAs
against XIST into the MG-63 and U2OS cell lines, which harbored the
highest expression level of XIST (Fig.
3A). Knockdown of XIST markedly impaired the growth of MG-63
and U2OS cells compared to the NC-transfected cells (Fig. 3B). The siRNA#1 against XIST had more
efficient inhibition and was chosen for the following experiments.
Furthermore, compared to si-NC, transfection with XISTsiRNA#1
resulted in a considerable increase of the apoptotic percentage in
MG-63 and U2OS cells (Fig. 3C).
Additionally, XIST silencing exhibited a significant decrease in
the percentage of cells in S phase and an increase in cells in the
G2/M phase compared with si-NC in both MG-63 and U2OS cells
(Fig. 3D). These data suggested that
XIST promoted cell proliferation by mediating cell apoptosis and
cell cycle in vitro.
Inhibition of LncRNA XIST impeded cell
migration and invasion
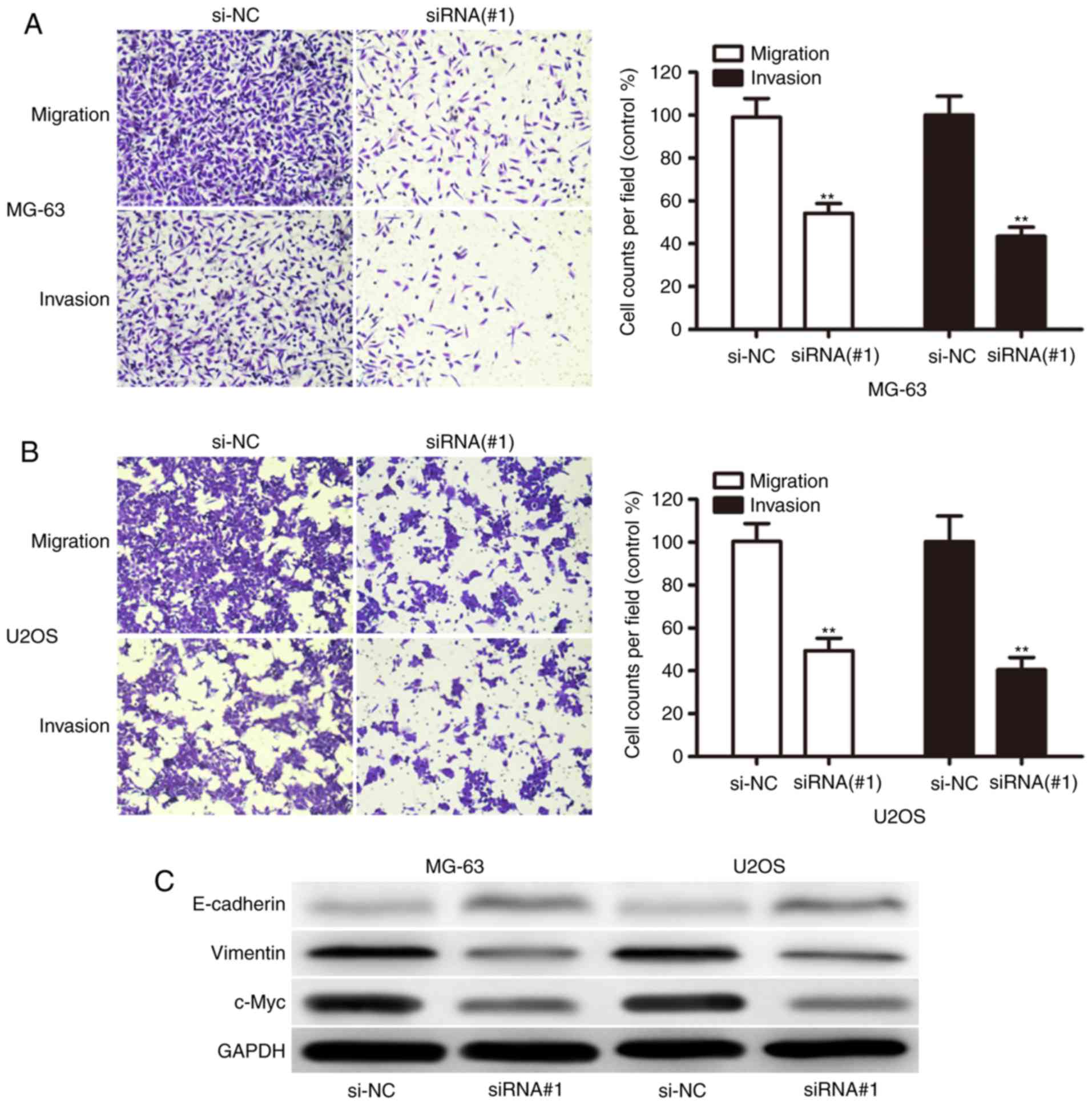
To investigate the potential role of XIST in OS
metastasis, the present study detected the effect of XIST on the
migration and invasion capacity of OS cells. We found that XIST
knockdown dramatically decreased their migration and invasion
capabilities in both MG-63 (Fig. 4A)
and U2OS (Fig. 4B) cells. We then
sought to explore whether there was some interaction between
proliferation- and metastasis-related markers and XIST expression.
Interestingly, XIST knockdown upregulated E-cadherin and repressed
Vimentin and c-Myc (Fig. 4C), which
are clearly involved in OS cell proliferation and invasion. These
results indicated that XIST functioned as an oncogene and promoted
the invasion of OS cells.
Discussion
In our present study, we found that XIST was
expressed significantly higher in OS cell lines and tissues.
Further investigation indicated that high XIST expression was
positively associated with advanced Enneking stage and metastasis,
suggesting that XIST might contribute to the progression of OS by
acting as an oncogene. Our results of the expression of XIST were
consistent with previous research demonstrating high XIST
expression in other cancers, such as NSCLC, gastric cancer,
nasopharyngeal carcinoma and OS (18–20,23,24).
In addition, Kaplan-Meier analysis showed that patients with high
XIST expression have poorer prognosis in OS. Consistent with our
results, previous research has shown that a high expression of XIST
was associated with short overall survival rates in patients with
gastric cancer and OS (18,24). However, XIST was found to be
significantly downregulated in both OS tissues and cell lines, and
overexpression of XIST inhibited cell proliferation and invasion
in vitro as well as tumor growth in vivo (25). These results suggested that XIST might
be involved in the development and progression of OS.
Several studies have emphasized the role of XIST in
tumorigenesis and progression in NSCLC and gastric cancer (18,19,23).
Knockdown of XIST repressed cell proliferation and tumorigenicity
in vitro and in vivo by suppressing KLF2 expression
in NSCLC (19). Knockdown of XIST
inhibited gastric cancer cell proliferation, migration invasion,
and tumor growth by suppressing miR-101, which increased EZH2
expression XIST (18). Deletion of
XIST inhibited cell proliferation and invasion through the
miR-320b/RAP2B axis (26). Consistent
with previous studies, our study showed that XIST silencing
significantly inhibited OS cell growth, cell migration and invasion
ability, as well as inducing cell apoptosis and cell cycle
alteration, implying that XIST played an important role in OS
progression. C-Myc, Vimentin and E-cadherin have been shown to play
critical roles in the processes of tumor cell proliferation and
invasion in OS (27–29). Furthermore, we found that the
knockdown of XIST resulted in decreased expression of c-Myc and
Vimentin and increased E-cadherin expression in OS cells. These
results indicated that XIST promoted cell proliferation and
invasion by mediating c-Myc, Vimentin and E-cadherin.
In summary, we have provided evidence demonstrating
that upregulated XIST expression in OS was significantly associated
with advanced Enneking stage and poor prognosis. Knockdown of
lncRNA XIST inhibited cell proliferation, migration and invasion
in vitro. These findings shed a light on the potential role
of XIST in OS pathogenesis and provided a valuable therapeutic
target for OS.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant no. 81541135).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WW and JH conceived and designed the study. WW, HS
and CG performed the experiments, coordinated the study and wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Capital Medical University (approval no.
2014A083). Written informed consent was acquired from all patients
prior to their inclusion in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berner K, Johannesen TB and Bruland OS:
Clinical epidemiology of low-grade and dedifferentiated
osteosarcoma in Norway during 1975 and 2009. Sarcoma.
2015:9176792015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allison DC, Carney SC, Ahlmann ER,
Hendifar A, Chawla S, Fedenko A, Angeles C and Menendez LR: A
meta-analysis of osteosarcoma outcomes in the modern medical era.
Sarcoma. 2012:7048722012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21 Suppl 7:vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA involved in canceR, neurobiology, and
DEvelopment. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu CH, Hsu CL, Lu PC, Lin WC, Juan HF and
Huang HC: Identification of lncRNA functions in lung cancer based
on associated protein-protein interaction modules. Sci Rep.
6:359392016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Niu J, Lin Y, Liu P, Yu Y, Su C and Wang
X: Microarray analysis on the lncRNA expression profile in male
hepatocelluar carcinoma patients with chronic hepatitis B virus
infection. Oncotarget. 7:76169–76180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo W, He H, Xiao W, Liu Q, Deng Z, Lu Y,
Wang Q, Zheng Q and Li Y: MALAT1 promotes osteosarcoma development
by targeting TGFA via MIR376A. Oncotarget. 7:54733–54743. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qi D, Li J, Que B, Su J, Li M, Zhang C,
Yang M, Zhou G and Ji W: Long non-coding RNA DBCCR1-003 regulate
the expression of DBCCR1 via DNMT1 in bladder cancer. Cancer Cell
Int. 16:812016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou Q, Chen F, Fei Z, Zhao J, Liang Y,
Pan W, Liu X and Zheng D: Genetic variants of lncRNA HOTAIR
contribute to the risk of osteosarcoma. Oncotarget. 7:19928–19934.
2016.PubMed/NCBI
|
|
14
|
Ma B, Li M, Zhang L, Huang M, Lei JB, Fu
GH, Liu CX, Lai QW, Chen QQ and Wang YL: Upregulation of long
non-coding RNA TUG1 correlates with poor prognosis and disease
status in osteosarcoma. Tumour Biol. 37:4445–4455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Zhao L and Wang Q: Overexpression of
long non-coding RNA HOTTIP increases chemoresistance of
osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J
Transl Res. 8:2385–2393. 2016.PubMed/NCBI
|
|
16
|
Xia WK, Lin QF, Shen D, Liu ZL, Su J and
Mao WD: Clinical implication of long noncoding RNA 91H expression
profile in osteosarcoma patients. Onco Targets Ther. 9:4645–4652.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei X, Wang C, Ma C, Sun W, Li H and Cai
Z: Long noncoding RNA ANRIL is activated by hypoxia-inducible
factor-1α and promotes osteosarcoma cell invasion and suppresses
cell apoptosis upon hypoxia. Cancer Cell Int. 16:732016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL,
Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al: Long non-coding
RNA XIST regulates gastric cancer progression by acting as a
molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin
Cancer Res. 35:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang J, Sun CC and Gong C: Long noncoding
RNA XIST acts as an oncogene in non-small cell lung cancer by
epigenetically repressing KLF2 expression. Biochem Biophys Res
Commun. 478:811–817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song P, Ye LF, Zhang C, Peng T and Zhou
XH: Long non-coding RNA XIST exerts oncogenic functions in human
nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 592:8–14.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schouten PC, Vollebergh MA, Opdam M,
Jonkers M, Loden M, Wesseling J, Hauptmann M and Linn SC: High XIST
and Low 53BP1 expression predict poor outcome after high-dose
alkylating chemotherapy in patients with a BRCA1-like breast
cancer. Mol Cancer Ther. 15:190–198. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang YS, Chang CC, Lee SS, Jou YS and
Shih HM: Xist reduction in breast cancer upregulates AKT
phosphorylation via HDAC3-mediated repression of PHLPP1 expression.
Oncotarget. 7:43256–43266. 2016.PubMed/NCBI
|
|
23
|
Ma L, Zhou Y, Luo X, Gao H, Deng X and
Jiang Y: Long non-coding RNA XIST promotes cell growth and invasion
through regulating miR-497/MACC1 axis in gastric cancer.
Oncotarget. 8:4125–4135. 2017.PubMed/NCBI
|
|
24
|
Li GL, Wu YX, Li YM and Li J: High
expression of long non-coding RNA XIST in osteosarcoma is
associated with cell proliferation and poor prognosis. Eur Rev Med
Pharmacol Sci. 21:2829–2834. 2017.PubMed/NCBI
|
|
25
|
Zhang R and Xia T: Long non-coding RNA
XIST regulates PDCD4 expression by interacting with miR-21-5p and
inhibits osteosarcoma cell growth and metastasis. Int J Oncol.
51:1460–1470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lv GY, Miao J and Zhang XL: Long
non-coding RNA XIST promotes osteosarcoma progression by targeting
ras-related protein RAP2B via miR-320b. Oncol Res. Apr
12–2017.(Epub ahead of print).
|
|
27
|
Baker EK, Taylor S, Gupte A, Sharp PP,
Walia M, Walsh NC, Zannettino AC, Chalk AM, Burns CJ and Walkley
CR: BET inhibitors induce apoptosis through a MYC independent
mechanism and synergise with CDK inhibitors to kill osteosarcoma
cells. Sci Rep. 5:101202015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng Q, Ren C, Wang L, Zhao Y and Wang S:
Knockdown of ST6Gal-I inhibits the growth and invasion of
osteosarcoma MG-63 cells. Biomed Pharmacother. 72:172–178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin DS, Cai LY, Ding J and Gao WY:
Correlation between E-cadherin-regulated cell adhesion and human
osteosarcoma MG-63 cell anoikis. Asian Pac J Cancer Prev.
15:8203–8207. 2014. View Article : Google Scholar : PubMed/NCBI
|