Introduction
Renal cell carcinoma (RCC) comprises 90% of all
kidney tumors and its incidence is increasing worldwide (1). Worldwide, approximately 30% of patients
with RCC were diagnosed with metastatic disease, which decreased
the 5-year survival rate to only 10% as reported in 2006 (2,3).
Conventional radical nephrectomy is not able to cure metastatic
RCC, which is resistant to radiotherapy and chemotherapy (4). In the past decade, tyrosine kinase
inhibitors, also known as targeted therapies, were introduced and
demonstrated relatively high rates of response but the 5-year
survival rate of patients with metastatic RCC remains low (5). More efficient targets are urgently
needed to improve long term survival of patients with advanced
RCC.
FOXF1 adjacent non-coding developmental regulatory
RNA (FENDRR), a long noncoding RNA lacking an open reading frame of
significant length, was first identified as a gene that is
specifically expressed in the posterior mesoderm of mouse embryos
and is essential for heart and body wall development (6). FENDRR is a conserved transcript of ~3
kilobases, located on human chromosome 16 (6). Further characterization revealed that it
is downregulated in patients with gastric and lung cancer (7,8). Further
analysis has demonstrated that histone deacetylation is involved in
downregulation of FENDRR and may serve as prognostic factor for
human gastric cancer (7).
Although the key functions of FENDRR in
embryogenesis and development have been well elucidated (6,9,10), the biological functions of FENDRR and
whether aberrant expression of FENDRR occurs in RCC remains to be
determined. In the present study, it was identified that FENDRR was
frequently downregulated in RCC and other types of human cancer.
Overexpression of FENDRR attenuates RCC cells proliferation,
migration, invasion and colony formation capabilities, suggesting
pivotal functions of FENDRR in RCC progression. Mechanistically,
FENDRR physically associates with Polycomb Repressive Complex 2
(PRC2) and lysin methyltransferase 2A (MLL) histone modifying
complexes. Importantly, downregulation of FENDRR predicts poor
prognosis of RCC.
Materials and methods
Tissue samples
Samples of fresh frozen RCC cancer tissues, together
with normal adjacent tissues (3 cm away from the tumor as in
previously published studies) (11),
were obtained during radical nephrectomy from Sun Yat-sen Memorial
Hospital of Sun Yat-sen University (Guangzhou, China). All samples
were snap frozen using liquid nitrogen and transferred to −80°C
until processed. Tissue samples were collected with written
informed consent from patients, and approved by the Ethic Review
Committees of Sun-Yat Sen Memorial Hospital. All samples had their
diagnoses confirmed pathologically. The nuclear grade was assessed
with the Fuhrman grade system (12).
Cell lines and transfection
Human RCC cell lines (769-P, 786-O, ACHN) used in
the study were obtained from the American Type Culture Collection
(Manassas, VA, USA). 769-P and 786-O cells were cultured in
RPMI-1640 medium (Hyclone; GE Healthcare, Logan, UT, USA). ACHN
cells were cultured in Dulbecco's modified Eagle's medium (Hyclone;
GE Healthcare, Logan, UT, USA). All media were supplemented with
10% fetal bovine serum (FBS; Hyclone; GE Healthcare) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Cells were cultured in a humidified air
atmosphere of 5% CO2 at 37°C. Routine tests for
mycoplasm infection were negative. Cell transfection was performed
using Lipofectamine 2000 or RNAiMAX (Life Technologies; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol
for overexpression or knockdown experiments, respectively.
Sequences of siRNAs were as follows: siFENDRR#1,
5′-CCAGCCAUGUGAUUCCAAATT-3′; siFENDRR#2,
5′-GCGAUUGACUGUCUUAUAATT-3′; siControl,
5′-UUCUCCGAACGUGUCACGUTT-3′.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cells was isolated using
TRIzol® reagent (Life Technologies; Thermo Fisher
Scientific, Inc.) and treated with an RNase-free DNase set (Qiagen
China Co., Ltd., Shanghai, China) according to the manufacturer's
protocols. RNA electrophoresis on a denaturing agarose gel at 5–6
V/cm was performed to inspect RNA integrity. Intact total RNA run
on a denaturing gel has sharp 28S and 18S rRNA bands, and the 28S
rRNA band is approximately twice as intense as the 18S rRNA band.
RT was performed using M-MLV reverse transcriptase (Life
Technologies; Thermo Fisher Scientific, Inc.) and qPCR was
performed with SYBR-Green master mix (Roche Diagnostics, Basel,
Switzerland). Relative expression values were calculated using the
2−ΔΔCq method with GAPDH as a normalizer (13). The primer sequences used were as
follows: FENDRR: Forward, 5′-AGAGTGCTTCCACTGCCCTA-3′ and reverse,
5′-CCCATTTGCAAAGGCTACAT-3′; GAPDH, forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse,
5′-GAAGATGGTGATGGGATTTC-3′.
Cell proliferation, motility, invasion
and colony formation assays
Cell proliferation, motility, invasion and colony
formation assays were performed as previously described (14). For the cell proliferation assay,
2x103 cells were plated in quadruplicates in 96-well
plates and incubated in a humidified air atmosphere of 5%
CO2 at 37°C. Cell number was calculated using an MTT
assay kit (Roche Diagnostics). The cell motility assay was
performed using Transwell chambers (BD Falcon; BD Biosciences, San
Jose, CA, USA). A total of 1x104 cells were plated in
Transwell chambers and incubated in complete medium in a humidified
air atmosphere of 5% CO2 at 37°C for 8–24 h. Cells were
stained with 0.1% crystal violet. For the colony formation assay,
100–500 transfected cells were placed in a fresh six-well plate and
maintained for two weeks and colonies were fixed with methanol and
stained with 0.1% crystal violet and counted. An in vitro
cell invasion assay was performed using the BD BioCoat Matrigel
Invasion Chamber (BD Falcon; BD Biosciences) according to
manufacturer's instructions. A total of 1x104 cells were
plated in BD BioCoat Matrigel Invasion Chambers and incubated in
complete medium in a humidified air atmosphere of 5% CO2
at 37°C for 12–36 h. Subsequently, crystal violet on the Transwell
membrane was dissolved in 33% acetic acid solution and measured the
absorbance of crystal violet (OD570 nm). Cells were visualized with
inverted microscope (magnification, ×200; Nikon Corporation, Tokyo,
Japan). All experiments were performed in triplicate.
5′-Ethynyl-2′-deoxyuridine (EdU)
incorporation assay
786-O and 769-P cells in S-phase were measured using
an EdU assay kit (Guangzhou Ribobio Co., Ltd., Guanzhou, China),
according to the manufacturer's protocol. Briefly, cells were
incubated with 5 µM EdU in culture media supplemented with 10% FBS
for 2 h at 37°C. Then cells were fixed with 4% formaldehyde for 30
min at room temperature and treated with 0.5% Triton X-100 for 20
min for permeabilization. Subsequent to washing with PBS for 30
seconds at room temperature for three times, the cells were
incubated with 1× Apollo® reaction cocktail (Guangzhou
RiboBio Co., Ltd.) at 37°C for 30 min. Subsequently, the DNA
contents were stained with Hoechst 33342 (5 µg/ml) for 30 min at
37°C and visualized under a fluorescent microscope (magnification,
×100; Nikon Corporation, Tokyo, Japan).
RNA immunoprecipitation assay
RNA immunoprecipitation was performed using the
EZ-MagnaRIP kit (EMD Millipore, Billerica, MA, USA) according to
manufacturer's protocols. Briefly, 5x106 cells were
harvested and lysed with radioimmunoprecipitation assay lysis
buffer with one freeze-thaw cycle. Cell extracts were
co-immunoprecipitated with anti-enhancer of zeste 2 polycomb
repressive complex 2 (EZH2; 1:500 dilution; cat. no. ab195409, ChIP
grade), suppressor of zeste 12 protein homolog (SUZ12; 1:500
dilution; cat. no. ab12073, ChIP grade) and WD repeat domain 5
(WDR5; 1:500 dilution; cat. no. ab56919; ChIP grade; all Abcam,
Cambridge, UK), and incubated with agitation overnight at 4°C. The
retrieved RNA was subjected to RT-qPCR analysis. Anti-snRNP70
(1:500 dilution; cat. no. CS203216; EMD Millipore) was used as a
technical positive control, and normal mouse Immunoglobulin G was
used as a negative control. For RT-qPCR analysis, U1 splicesome RNA
was used as a non-specific control. Experiments were performed in
triplicates.
Bioinformatic analysis
Data from The Cancer Genome Atlas (TCGA) were
obtained from The Atlas of Noncoding RNAs in Cancer website
(http://ibl.mdanderson.org/tanric/_design/basic/resources.html).
Data from the Gene Expression Omnibus (GEO) were downloaded from
the GEO website (https://www.ncbi.nlm.nih.gov/geo/).
Statistical analysis
All quantitative data were presented as the mean ±
standard deviation from at least three independent experiments. The
differences between two independent groups were analyzed using
independent t-tests, and the differences between paired tumor and
normal adjacent tissues were analyzed with paired t-tests. The
differences between multiple groups were assessed using one-way
analysis of variance followed by Dunnett's multiple-comparison
test. The association between FENDRR and clinical characteristics
was analyzed using Pearson Chi-square test. All tests performed are
two sided. Statistical tests were performed using SPSS software
(version 10.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
FENDRR is downregulated in multiple
human cancers
To explore the biological roles of FENDRR in human
cancer, publically available databases were searched. As presented
in Fig. 1, FENDRR is downregulated in
multiple types of human cancer, including bladder and lung cancer,
and pancreatic ductal adenocarcinoma. Interestingly, in the
observation of multistep pancreatic carcinogenesis, the expression
levels of FENDRR were downregulated in intraductal papillary
mucinous adenoma, intraductal papillary mucinous carcinoma and
invasive cancer of intraductal papillary mucinous neoplasm. More
importantly, the expression levels of FENDRR were downregulated in
the metastatic site compared with the primary tumor in colorectal
cancer and prostate cancer.
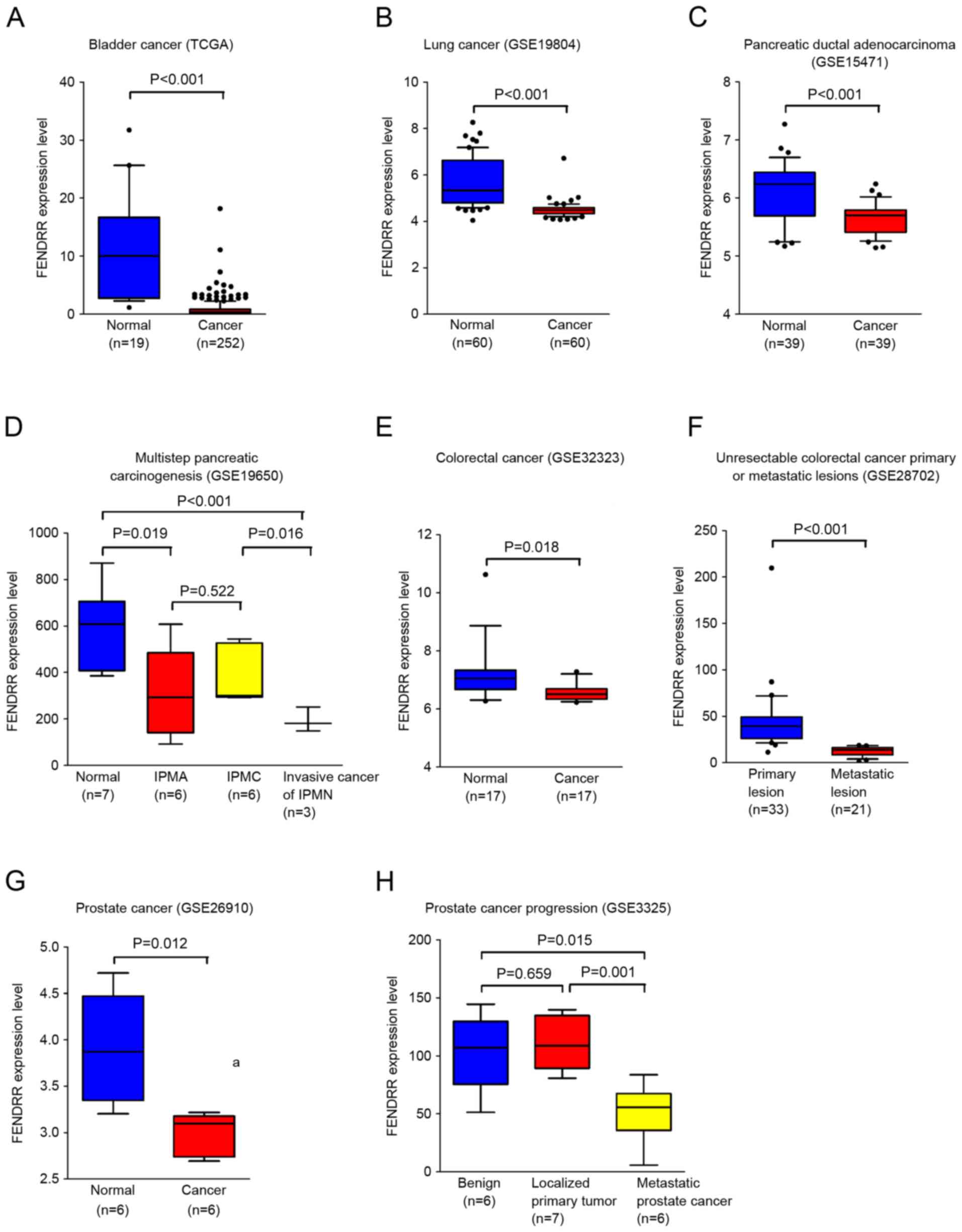 | Figure 1.FENDRR is downregulated in multiple
human cancers. (A) Analysis of data from TCGA revealed that FENDRR
is downregulated in bladder cancer. Data are expressed as
box-and-whisker diagrams depicting the smallest observation, 10–90%
percentile, median, and largest observation. Data from GEO
demonstrates that FENDRR is downregulated in (B) lung cancer, (C)
pancreatic ductal adenocarcinoma, (D) different stages of
pancreatic carcinogenesis, (E) colorectal cancer, (F) unresectable
colorectal primary or metastatic lesions, (G) prostate cancer and
(H) process of prostate cancer progression. Statistical
significance was assessed using two-tailed Student's t-tests (A, B,
C, E, F and G) and a one-way analysis of variance followed by
Dunnett's tests for multiple comparisons (D and H). FENDRR, FOXF1
adjacent non-coding developmental regulatory RNA; TCGA, the cancer
genome association; IPMA, intraductal papillary-mucinous adenoma;
IPMC, intraductal papillary-mucinous carcinoma; IPMN, intraductal
papillary-mucinous neoplasm. |
Collectively, these data demonstrated that FENDRR
serves important functions in carcinogenesis and cancer metastasis
in multiple types of human cancer.
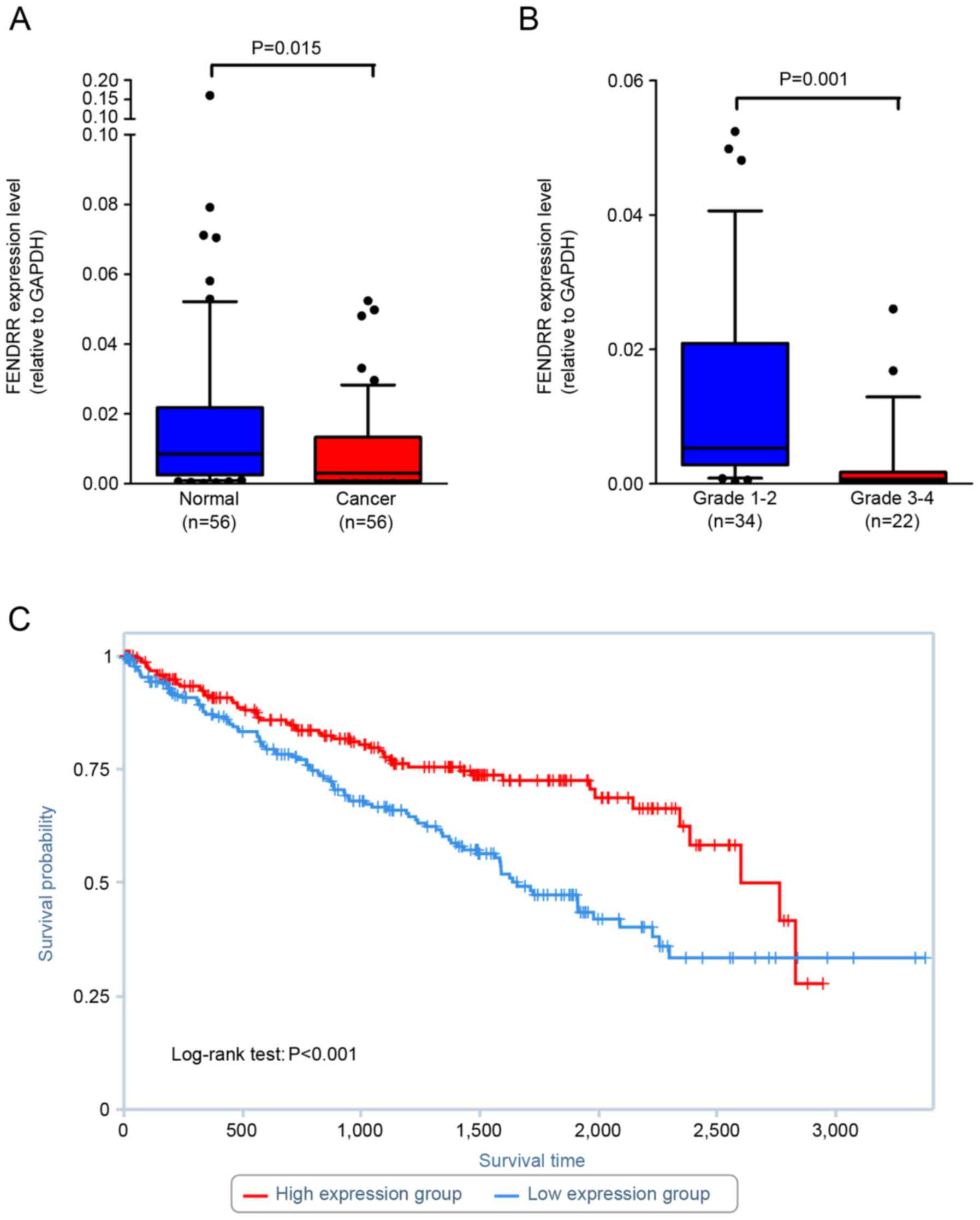
FENDRR is downregulated in renal cell
carcinoma and associates with poor prognosis of RCC
In order to determine whether dysregulation of
FENDRR expression indeed occurs in renal cell carcinoma, the RNA
level of FENDRR was quantified in renal cell carcinoma tissues and
paired normal adjacent cortex tissues using RT-qPCR. Fold changes
of <1.0 were designated as downregulated in cancer tissues
compared with normal adjacent tissues. The results revealed that
FENDRR was downregulated in 73.2% of renal cell carcinoma tissues
(Table I; Fig. 2A). Statistical analysis revealed that
lower FENDRR expression was associated with Fuhrman grade (Fig. 2B). As a result of the limited number
of cases and the follow up period, TCGA database was searched to
identify whether FENDRR expression associates with the prognosis of
patients with renal cell carcinoma. The results demonstrated that
lower FENDRR expression associates with shorter overall survival
(P=0.00025; Fig. 2C).
 | Table I.Characteristics of renal cell
carcinoma patients. |
Table I.
Characteristics of renal cell
carcinoma patients.
|
|
| FENDRR |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Patient frequency
(%) | Low | High | Pearson
χ2 | P-value |
|---|
| Total | 56 | 41 (73.2%) | 15 (26.8%) |
|
|
| Sex |
|
|
|
|
|
| Male | 39 (69.6) | 30 | 9 | 0.386 | 0.535 |
|
Female | 17 (30.4) | 11 | 6 |
|
|
| Age, years |
|
|
|
|
|
|
<60 | 32 (57.1) | 25 | 7 | 0.918 | 0.338 |
|
≥60 | 24 (42.9) | 16 | 8 |
|
|
| Tumor stage |
|
|
|
|
|
| T1 | 38 (67.9) | 26 | 12 | 0.730 | 0.39 |
|
T2-4 | 18 (32.1) | 15 | 3 |
|
|
| Fuhrman grade |
|
|
|
|
|
|
1–2 | 34 (60.7) | 21 | 13 | 5.785 | 0.016 |
|
3–4 | 22 (39.3) | 20 | 2 |
|
|
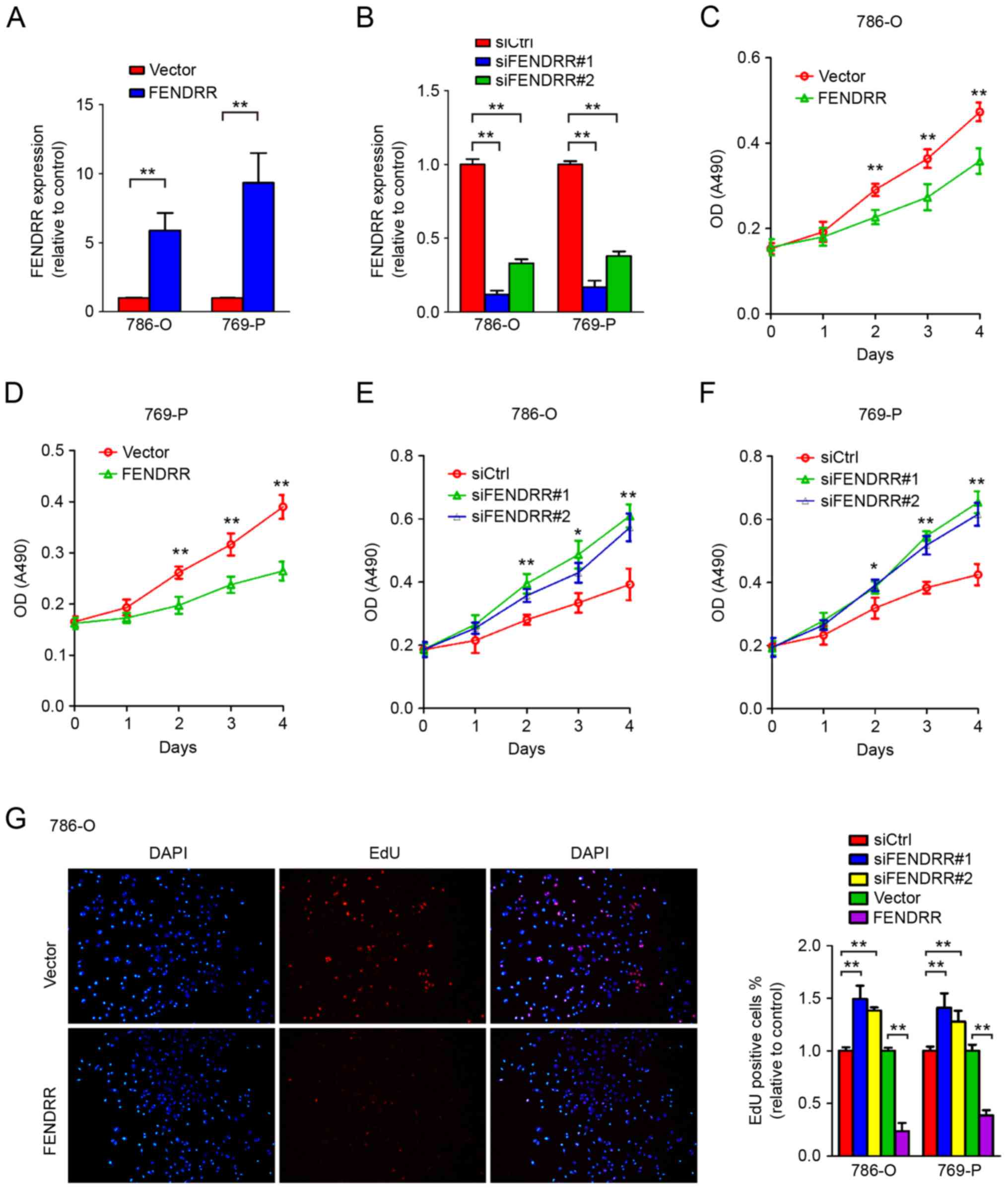
FENDRR downregulation results in
accelerated cell proliferation of RCC cells
Unlimited proliferation ability comprises one of the
most important characteristics of malignant tumor. Reports have
identified that downregulation of FENDRR promotes tumor growth and
associates with poor prognosis in gastric cancer (7). To further determine the role of FENDRR
in regulating RCC cell proliferation, FENDRR was overexpressed
using expression vectors or depleted in RCC cell lines using
siRNAs. As presented in Fig. 3A-D,
knockdown of endogenous FENDRR accelerated cell proliferation in
786-O and 769-P RCC cells, compared with the non-specific control
siRNA that shared no homology with the human genome. Conversely,
overexpression of FENDRR significantly inhibited proliferation of
RCC cells (Fig. 3E-F). These data
suggested a function of FENDRR in regulating RCC cell
proliferation.
To further validate the role of FENDRR in cell cycle
modulation, an EdU incorporation assay was performed. EdU is a
nucleotide analog of thymidine and is incorporated into DNA during
DNA synthesis. Thus EdU positive cells indicate the cell is in S
phase. Markedly increased EdU positive cells were observed
following FENDRR depletion in 786-O and 769-P RCC cells (Fig. 3G), and overexpression of FENDRR
decreased the proportion of cells in S phase, further indicating
that FENDRR expression inhibited cell proliferation of RCC
cells.
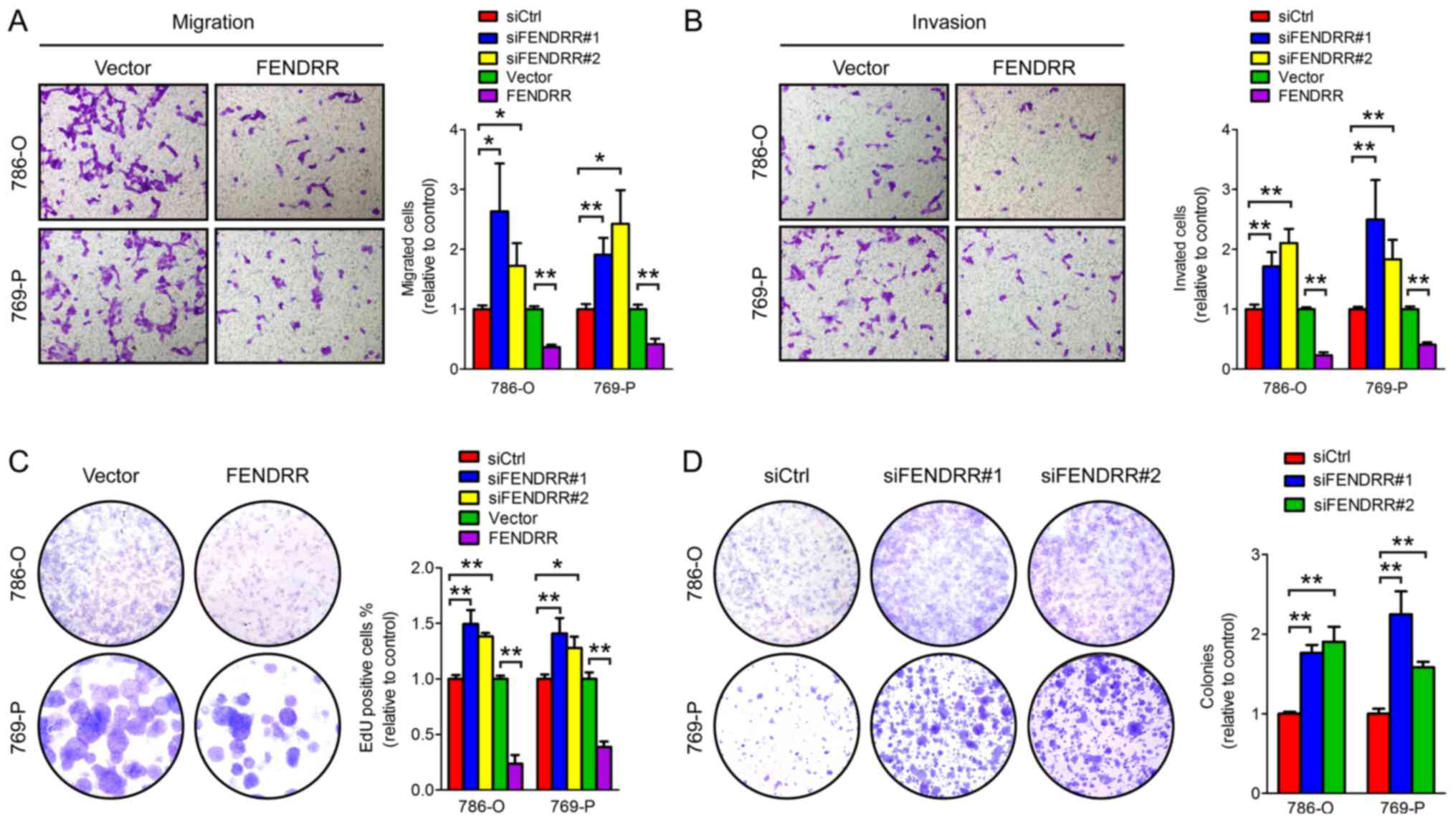
FENDRR overexpression inhibits the
migration, invasion and colony formation abilities of RCC
cells
The ability to invade the basement membrane and
migrate from the primary site to a distant site is critical for
malignant cancer cells to form metastatic lesions. Cell migration
ability was evaluated using Transwell chambers and cell invasion
was assessed using a Matrigel invasion assay which mimics the
process of tumor cells invading the basement membrane. As presented
in Fig. 4A-B, FENDRR knockdown
promoted, while FENDRR overexpression inhibited, cell migration and
invasion of RCC cells, suggesting that FENDRR serves a regulatory
function in RCC cell migration and invasion.
Colony formation ability demonstrates the
cell-population dependent proliferation capacity of cancer cells.
Knockdown of endogenous FENDRR markedly promoted, and FENDRR
overexpression inhibited, colony formation both in 786-O and 769-P
cells (Fig. 4C-D). Together, these
results indicate the critical function of FENDRR in the process of
metastasis of renal cell carcinoma.
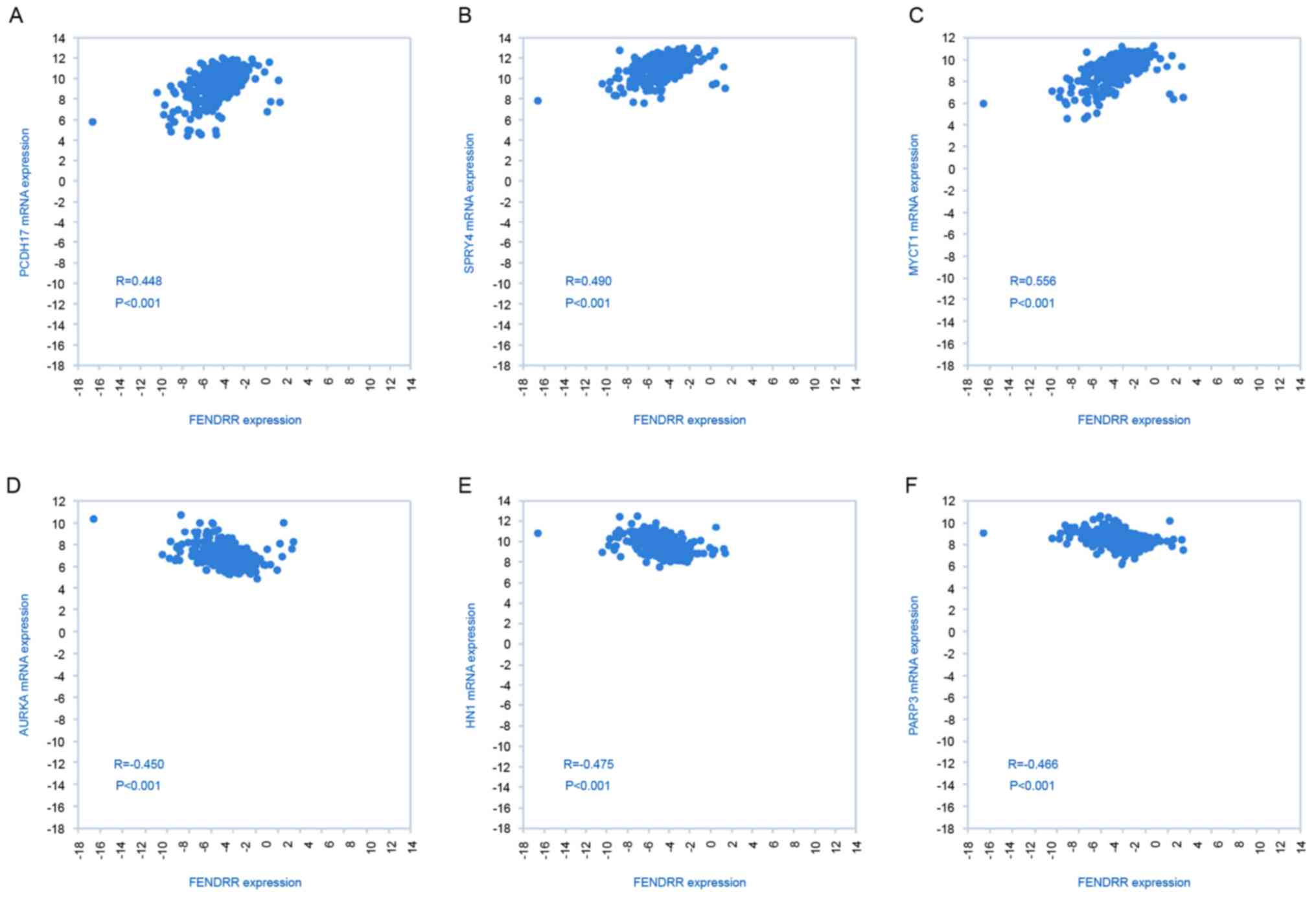
Potential targets of FENDRR in
RCC
To identify potential target genes of FENDRR
involved in the metastasis of RCC, an analysis of FENDRR and mRNA
expression was performed using TCGA database. As presented in
Fig. 5, a positive association was
identified between FENDRR expression and tumor suppressors
protocadherin 17, sprouty RTK signaling antagonist 4, MYC target 1
and a negative association was identified between FENDRR and
oncogene aurora kinase A, jupiter microtubule associated homolog 1
and poly(ADP-ribose) polymerase family member 3, suggesting that
FENDRR may serve as a tumor suppressor by activating important
tumor suppressor genes and inhibiting expression of oncogenes.
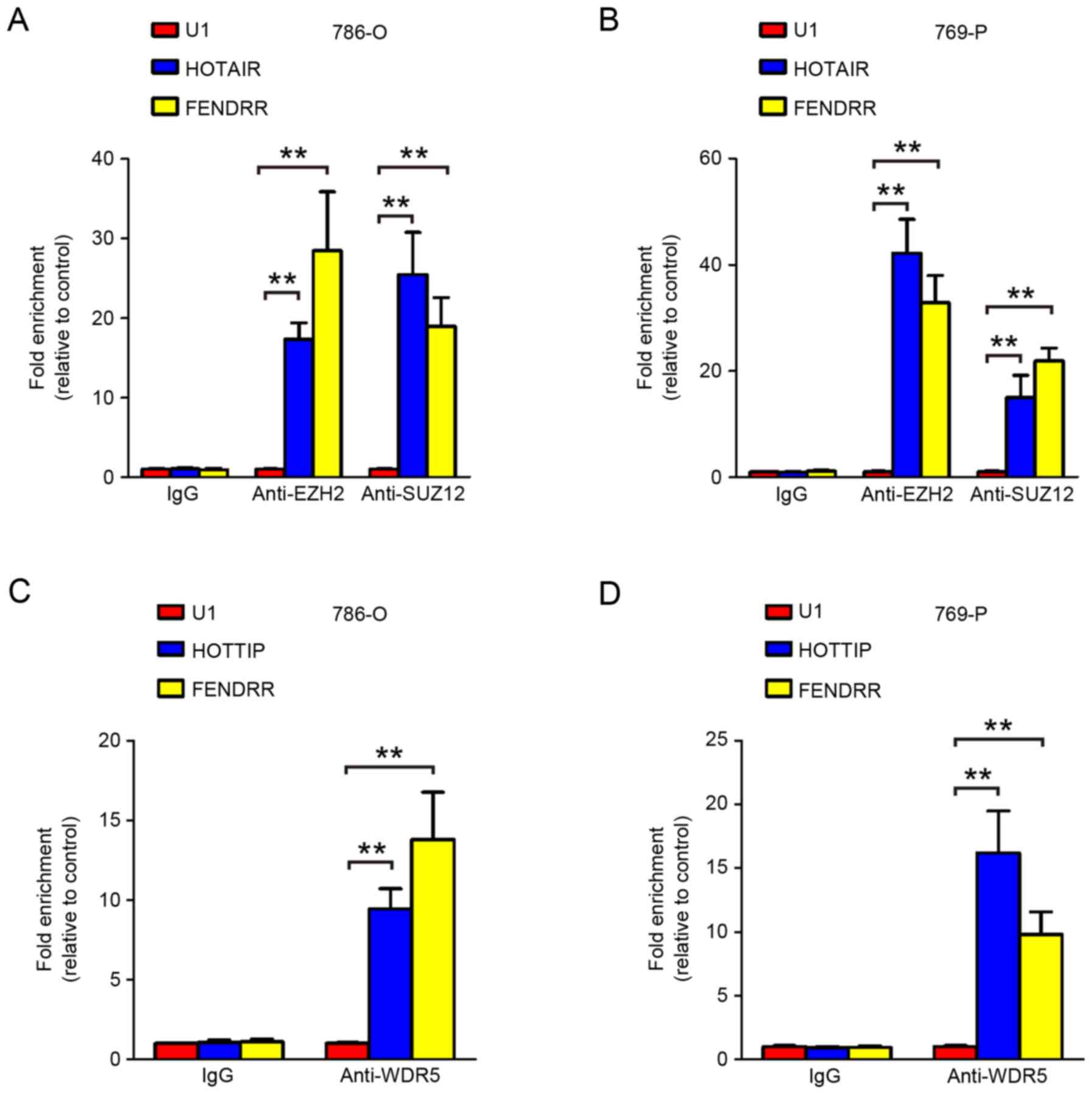
FENDRR physically associates with PRC2
complex and MLL complex
To further elucidate how FENDRR regulates
down-stream target genes, we next performed RNA immunoprecipitation
to identify FENDRR interacting proteins. Our RIP assay confirmed
that FENDRR physically associates with EZH2, SUZ12 and WDR5 in RCC
cells (Fig. 6), suggesting that
FENDRR may exert its function at least partially by interacting
with PRC2 complex and MLL complex.
Discussion
FENDRR has been suggested to serve important
functions in vertebral development and is downregulated in numerous
types of cancer (7–9,15). A low
level of FENDRR expression promotes local invasion and lymphatic
metastasis in gastric cancer, which are key steps for cancer
progression (7). To interpret how
this transcript exerts its biological function, potential
mechanisms were proposed, including controlling epigenetic
modifications of promoters of target genes critical for embryonic
development (6), and promoting cancer
invasion through regulating expression of matrix metalloproteinase
(MMP)2/MMP9 (7). In the present
study, it was identified that FENDRR downregulation occurs in the
process of carcinogenesis and cancer metastasis through searching a
publicly available database. Though a tumor suppressing role was
identified in multiple types of human cancer, functional
characterization of FENDRR in RCC remains undocumented. Consistent
with observations in other cancers, the present data revealed that
FENDRR downregulation is very common in RCC and associates with
poor prognosis of patients with RCC. Importantly, overexpression of
FENDRR significantly inhibited cell proliferation and invasion,
pointing to the possibility of FENDRR as a therapeutic target for
cancer treatment.
Dysregulation of chromatin modifications including
H3K4 and H3K27 methylation is implicated in the carcinogenesis of
RCC (16). The MLL complex is
responsible for H3K4 methylation (17) and polycomb group proteins (PRC1 and
PRC2) are epigenetic silencers implicated in cancer development
(18). EZH2, a core component of
PRC2, directly controls the histone H3 lysine 27 trimethylation and
interacts with DNA methyltransferases promoting DNA methylation,
and EZH2 is critical for these processes (19,20) and
predicts cancer specific survival of RCC (16). Studies have uncovered that numerous
long intergenic noncoding RNAs (lincRNAs) associate with epigenetic
modifying complexes, and >20% of lincRNAs interact with PRC2
complex (21). In the present study,
the gathered data suggested that FENDRR is able to physically
associate with WDR5, EZH2 and SUZ12 in renal carcinoma cells. It is
of note that certain studies provide evidence that EZH2 promotes
the progression of RCC by modulating vascular endothelial growth
factor and E-cadherin expression (22,23).
However, a more precise mechanism detailing how FENDRR affects the
targeting specificity of PRC2 complex and MLL complex to promote
carcinogenesis and cancer metastasis remains unresolved and
warrants further investigation.
It is a common phenomenon that genes associated with
developmental process also serve important functions in cancer
progression. Recent studies have demonstrated that long noncoding
RNAs serve important functions in the process of limb development
and lymph node metastasis of multiple types of cancer (HOXA
transcript at the distal tip) (24,25),
skeletal development and cancer metastasis (HOX transcript
antisense RNA) (24), muscle
differentiation and cancer metastasis (H19) (24,26).
Specifically, FENDRR is dispensable for lung and heart development,
and others and the present experiments suggested that FENDRR binds
to PRC2 and MLL complexes (6,9). Markedly, the present study identified
that FENDRR is not only critical for RCC but also serves a function
in the development of multiple cancers. However, the precise
molecular mechanisms by which FENDRR affects cellular phenotypic
alterations, including cell proliferation and migration, is largely
unexplored, which requires further investigation in future
studies.
To conclude, the present study reports that the long
noncoding RNA FENDRR acts as a tumor suppressor in RCC. It was
identified that FENDRR is frequently downregulated in cancerous
tissues of RCC, promoting cell migration and invasion. Furthermore,
overexpression of FENDRR decelerates cell migration, invasion and
colony formation of RCC cells. These findings suggest that
targeting FENDRR may help controlling RCC progression and
metastasis.
Acknowledgements
The authors would like to thank Dr. Changhao Chen
(Sun Yat-sen University) for helpful discussion of the article. Mr.
Wang He is a recipient of Yat-sen Scholarship for Young
Scientists.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant nos. 81472381, 81572514,
U1301221, 81472384, 81402106, 81372729, 81272808, 81172431), the
Science and Technology Program of Guangzhou (grant nos.
201707010116, 201604020156, 201604020177), the National Natural
Science Foundation of Guangdong (grant nos. 2016A030313321,
2015A030311011, 2015A030310122, S2013020012671, 07117336,
10151008901000024), the Three Big Construction funds of Sun Yat-sen
University, the Specialized Research Fund for the Doctoral Program
of Higher Education (grant no. 20130171110073), the Fundamental
Research Funds for the Central Universities, the Guangdong Province
Higher Vocational Colleges & Schools Pearl River Scholar Funded
Scheme, the Elite Young Scholars Program of Sun Yat-Sen Memorial
Hospital (grant no. J201401), the National Clinical Key Specialty
Construction Project for Department of Urology and Department of
Oncology, the Key Laboratory of Malignant Tumor Gene Regulation and
Target Therapy of Guangdong Higher Education Institutes,
Sun-Yat-Sen University (grant no. KLB09001) and the Key Laboratory
of Malignant Tumor Molecular Mechanism and Translational Medicine
of Guangzhou Bureau of Science and Information Technology [grant
no. (2013)163].
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the TCGA repository, http://ibl.mdanderson.org/tanric/_design/basic/index.html.
Authors' contributions
WH and JH participated in the study design. GZ, PW
and NJ carried out in vitro cell experiments and data
analysis. PW and CJ performed the PCR experiments and data
analysis. WH conceived the study and wrote the manuscript. All the
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Tissue samples were collected with written informed
consent from patients, and approved by the Ethics Review Committee
of Sun Yat-sen Memorial Hospital.
Patient consent for publication
All samples were collected with written informed
consent from patients. No identifying information, including names,
initials, date of birth or hospital numbers, images or statements
were included in the manuscript.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jonasch E, Gao J and Rathmell WK: Renal
cell carcinoma. BMJ. 349:g47972014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hollingsworth JM, Miller DC, Daignault S
and Hollenbeck BK: Five-year survival after surgical treatment for
kidney cancer: A population-based competing risk analysis. Cancer.
109:1763–1768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mourad WF, Dutcher J and Ennis RD:
State-of-the-art management of renal cell carcinoma. Am J Clin
Oncol. 37:498–505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Escudier B, Eisen T, Stadler WM, Szczylik
C, Oudard S, Staehler M, Negrier S, Chevreau C, Desai AA, Rolland
F, et al: Sorafenib for treatment of renal cell carcinoma: Final
efficacy and safety results of the phase III treatment approaches
in renal cancer global evaluation trial. J Clin Oncol.
27:3312–3318. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grote P, Wittler L, Hendrix D, Koch F,
Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M and
Herrmann BG: The tissue-specific lncRNA Fendrr is an essential
regulator of heart and body wall development in the mouse. Dev
Cell. 24:206–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin
L, Chen WM, Han L, Zhang EB, Kong R, De W, et al: Decreased
expression of the long non-coding RNA FENDRR is associated with
poor prognosis in gastric cancer and FENDRR regulates gastric
cancer cell metastasis by affecting fibronectin1 expression. J
Hematol Oncol. 7:632014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen R, Li WX, Sun Y, Duan Y, Li Q, Zhang
AX, Hu JL, Wang YM and Gao YD: Comprehensive analysis of lncRNA and
mRNA expression profiles in lung cancer. Clin Lab. 63:313–320.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sauvageau M, Goff LA, Lodato S, Bonev B,
Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E,
Spence M, et al: Multiple knockout mouse models reveal lincRNAs are
required for life and brain development. Elife. 2:e017492013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grote P and Herrmann BG: The long
non-coding RNA Fendrr links epigenetic control mechanisms to gene
regulatory networks in mammalian embryogenesis. RNA Biol.
10:1579–1585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H and Men CP: Correlation of
increased expression of MicroRNA-155 in bladder cancer and
prognosis. Lab Med. 46:118–122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He W, Cai Q, Sun F, Zhong G, Wang P, Liu
H, Luo J, Yu H, Huang J and Lin T: linc-UBC1 physically associates
with polycomb repressive complex 2 (PRC2) and acts as a negative
prognostic factor for lymph node metastasis and survival in bladder
cancer. Biochim Biophy Acta. 1832:1528–1537. 2013. View Article : Google Scholar
|
|
15
|
Li Q, Wu C, Song G, Zhang H, Shan B, Duan
Y and Wang Y: Genome-wide analysis of long noncoding RNA expression
profiles in human Xuanwei lung cancer. Clin Lab. 61:1515–1523.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vieira-Coimbra M, Henrique R and Jeronimo
C: New insights on chromatin modifiers and histone
post-translational modifications in renal cell tumours. Eur J Clin
Invest 45 (Suppl). 1:S16–S24. 2015. View Article : Google Scholar
|
|
17
|
Schuettengruber B, Chourrout D, Vervoort
M, Leblanc B and Cavalli G: Genome regulation by polycomb and
trithorax proteins. Cell. 128:735–745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schlesinger Y, Straussman R, Keshet I,
Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E,
Reubinoff BE, et al: Polycomb-mediated methylation on Lys27 of
histone H3 pre-marks genes for de novo methylation in cancer. Nat
Genetics. 39:232–236. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Chen Y, Geng H, Qi C, Liu Y and
Yue D: Overexpression of YB1 and EZH2 are associated with cancer
metastasis and poor prognosis in renal cell carcinomas. Tumour
Biol. 36:7159–7166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu L, Xu Z, Zhong L, Wang H, Jiang S,
Long Q, Xu J and Guo J: EZH2 promotes tumor cell migration and
invasion via epigenetic repression of E-cadherin in renal cell
carcinoma. BJU Int. 117:351–362. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu ZQ, Zhang L, Gao BS, Wan YG, Zhang XH,
Chen B, Wang YT, Sun N and Fu YW: EZH2 promotes tumor progression
by increasing VEGF expression in clear cell renal cell carcinoma.
Clin Transl Oncol. 17:41–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Z, He A, Wang D, Liu Y and Huang W:
Long noncoding RNA HOTTIP as a novel predictor of lymph node
metastasis and survival in human cancer: A systematic review and
meta-analysis. Oncotarget. 8:14126–14132. 2017.PubMed/NCBI
|
|
26
|
Jing W, Zhu M, Zhang XW, Pan ZY, Gao SS,
Zhou H, Qiu SL, Liang CZ and Tu JC: The significance of long
noncoding RNA H19 in lredicting progression and metastasis of
cancers: A Meta-Analysis. Biomed Res Int. 2016:59026782016.
View Article : Google Scholar : PubMed/NCBI
|