Introduction
Cancer is a major global public health problem and a
leading cause of mortality. Digestive system cancer types (DSCs)
are the most common malignant tumors affecting the organs and
glands of the digestive tract. DSCs include esophageal, stomach,
colon, rectum, liver, gallbladder and pancreatic carcinomas
(1). Colorectal cancer (CRC) is the
third most commonly diagnosed cancer type, whilst liver cancer and
gastric cancer (GC) were second and third to lung cancer as the
most frequent causes of cancer-associated cases of mortality
worldwide in 2012 (2). Pancreatic
cancer (PC) is usually detected at an advanced stage with extensive
invasion and lymphatic metastasis, when the estimated 5-year
survival rate is well below 5% (3).
Early detection of DSCs is limited by the absence of specific
symptoms prior to metastasis, which leads to poor prognosis.
Therefore, it is of great importance to identify novel predictive
and prognostic biomarkers, including genetic and epigenetic
alterations at early stages that can detect and monitor tumor
dynamics.
A number of studies have identified that microRNAs
(miRNAs) are aberrantly expressed in human cancer types due to
genomic or epigenetic alterations (4,5). miRNAs
are a class of single-stranded, small RNAs approximately 22
nucleotides in length. miRNAs serve as essential
post-transcriptional regulators by binding to the 3′-untranslated
region (3′-UTR), 5′-UTR or coding sequences of target mRNAs
(6). Notably, miRNAs are thought to
modulate almost 30% of human genes (5,7). The
extensive regulatory functions of miRNAs are not only associated
with developmental timing, cell proliferation and apoptosis, but
also play a critical role in oncogenesis and tumor suppression
(7). Numerous miRNAs have been
demonstrated to interact with their target genes and
tumor-associated pathways through subtly regulated networks
(7,8).
The current review summarizes the latest findings regarding the
crucial roles of miR-126 in major DSCs. Furthermore, the potential
clinical value of miR-126 as a novel diagnostic and prognostic
biomarker and therapeutic target in DSCs is discussed. The current
review aims to improve understanding regarding the significance of
the miR-126 regulatory mechanisms in human cancer types.
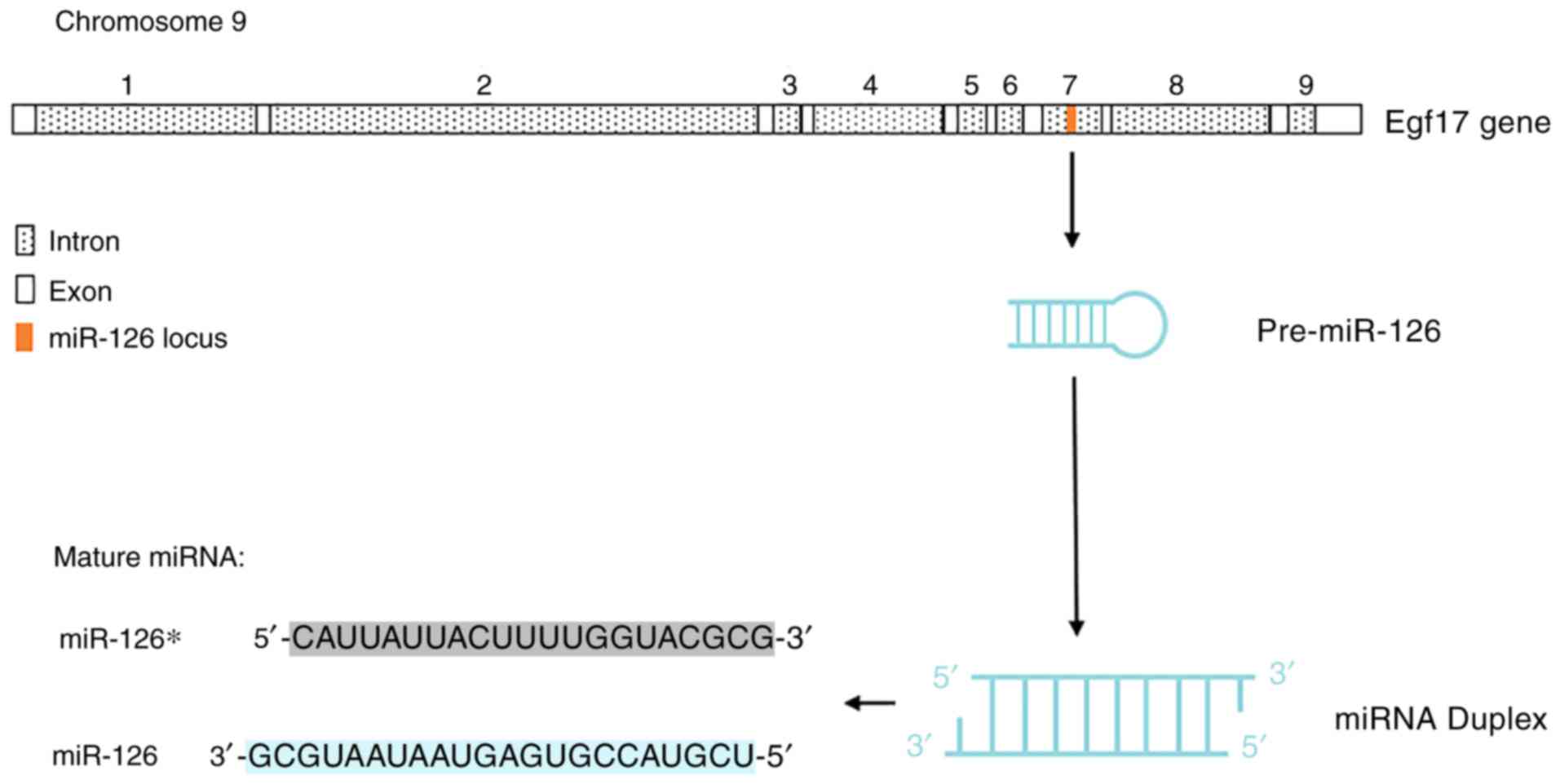
Structure and function of miR-126
miR-126 and its passenger strand, miR-126*, are two
endothelial cell-specific miRNAs that mediate angiogenesis
(9). Both miR-126 and miR126* are
encoded by the epidermal growth factor-like domain 7 (EGFL7) gene
(10). miR-126 usually refers to the
3′ region of the transcript, also termed miR-126-3p, located within
the 7th intron of EGFL7. Whereas, miR-126* refers to the 5′ region
of the transcript, also termed miR-123 or miR-126-5p, which binds
to the main miR-126 transcript at the stem loop structure of the
precursor miRNA (10,11) (Fig. 1).
EGFL7 is located on chromosome 9 in humans and encodes an
endothelial cell-derived factor involved in the repression of
smooth muscle cell migration and the regulation of angiogenesis
(12,13). EGFL7 is highly expressed in the
vasculature during embryonic development and at a relatively lower
level in adult blood vessels (13,14).
However, an aberrant upregulation of EGFL7 can be observed during
pathological angiogenesis in malignant tumors (15).
To the best of our knowledge, miR-126 is the only
miRNA with specific expression in the endothelial cell lineage and
hematopoietic progenitor cells (11).
Targeted deletion of miR-126 results in a loss of vascular
integrity, which leads to ruptured vessels and impaired endothelial
cell migration (9,10). Functionally, miR-126 is significantly
associated with angiogenesis and inflammation. miR-126 suppresses
sprouty-related EVH1 domain-containing protein 1 (SPRED1) and
phosphoinositide-3-kinase regulatory subunit 2 (PI3KR2), both of
which inhibit receptor tyrosine kinase-induced signaling via the
mitogen-activated protein kinase (MAPK) and
phosphinositide-3-kinase (PI3K) signaling pathways, thereby
promoting vascular endothelial growth factor (VEGF) signaling and
angiogenesis (9). Although miR-126
expression is usually altered in parallel with the expression of
EGFL7 protein, miR-126 can downregulate EGFL7, leading to reduced
angiogenesis and cell proliferation via a negative feedback loop
mechanism (9,16). In addition, endogenous miR-126 has
been revealed to inhibit the expression of vascular cell adhesion
molecule 1 (VCAM-1) and prevent leukocyte adhesion and infiltration
to further control vascular inflammation (17). Furthermore, miR-126 can suppress
inflammation and reactive oxygen species production in endothelial
cells under hyperglycemic conditions by post-transcriptionally
inhibiting the expression of high-mobility group box 1 (18). The functional properties of miR-126*
are less well understood. The significant role of miR-126 as a
tumor suppressor involved in various signaling cascades has been
demonstrated in previous studies (11,19).
Aberrant expression of miR-126 has been identified in numerous
human cancer types, including melanoma, osteosarcoma, leukemia and
carcinomas of the digestive system, endocrine glands, urogenital
system and respiratory system (20).
Aberrant expression of miR-126 in DSCs
Numerous miRNAs have been revealed to be abnormally
expressed in multiple cancer types (21). Both miR-126 and miR-126* are highly
expressed in endothelial cells in vivo (9). In addition, miR-126 is overexpressed in
highly vascularized organs, including the heart, liver and lungs
(22,23). Table I
lists previous studies that have identified that miR-126 expression
is significantly decreased in human DSC tissues compared with
adjacent non-cancerous tissues (24–58).
Notably, miR-126 is involved in angiogenesis and has significantly
higher expression levels in hepatocellular carcinoma (HCC) tissues
treated with transcatheter arterial chemoembolization (TACE) and
surgery, compared with tissues only subjected to surgical resection
(53). Another study identified that
HCC tissues exhibit an elevated level of miR-126 compared with
adjacent non-cancerous tissues, although miR-126 may inhibit tumor
angiogenesis and proliferation (16).
In addition, miR-126 is aberrantly expressed in certain GC tissues
(33,38).
 | Table I.Aberrant expression, target genes and
direct functions of miR-126 in various types of digestive system
cancer. |
Table I.
Aberrant expression, target genes and
direct functions of miR-126 in various types of digestive system
cancer.
| A, Esophageal
cancer |
|---|
|
|---|
| Expression | Sample(s) | Target gene(s) | Direct
function(s) | (Refs.) |
|---|
| Down | Tissues | – | – | (24,25) |
| Down | Tissues/cell
lines | DNMT1 and
ADAM9 | Forming
DNMT1-miR-126 loop and suppressing ADAM9 expression | (26) |
| Down | Tissues/cell
lines | IRS-1 and
GOLPH3 | Downregulating the
expression of IRS-1 and GOLPH3 protein | (27) |
| Down | Tissues/cell
lines | SOX2 | Inhibiting SOX2
expression by targeting its mRNA 3′-UTR | (28) |
| Down | Tissues/cell
lines | PI3KR2 | Inhibiting PI3K/AKT
signaling pathway | (29) |
| Down | Tissues/cell
lines | VEGF-A | Negatively
regulating VEGF-A expression | (30) |
|
| B, Gastric
cancer |
|
|
Expression |
Sample(s) | Target
gene(s) | Direct
function(s) | (Refs.) |
|
| Down | Tissues/cell
lines | CRK | Downregulating the
expression of CRK | (19,31,32) |
|
Up/downa | Cell lines | SOX2 | Repressing SOX2
expression by targeting its mRNA 3′-UTR | (33) |
| Down | Tissues/cell
lines | CRKL | Negatively
regulating CRKL expression by targeting its mRNA 3′-UTR | (34) |
| Down | Tissues/cell
lines | VEGF-A | Reducing the
expression of VEGF-A by binding to its mRNA 3′-UTR | (35) |
| Down | Tissues/cell
lines | PLK2, PI3KR2 and
CRK | Synergistically
reducing the expression of PLK2, PI3KR2 and CRK | (36) |
| Down | Tissues/cell
lines | LAT-1 | Negatively
regulating the expression of LAT-1 | (37) |
| Up | Cell lines | CADM1 | Downregulating
CADM1 expression by targeting its mRNA 3′-UTR | (38) |
| Down | Cell lines | EZH2 | Decreasing EZH2
expression to regulate chemotherapy resistant | (39) |
| Down | Cell lines | ADAM9 | Downregulating
ADAM9 expression by binding to its mRNA 3′-UTR | (40) |
| Down | Tissues/cell
lines | RGS3 |
Post-transcriptionally downregulating the
expression of RGS3 | (41) |
|
| C, Colorectal
cancer |
|
|
Expression |
Sample(s) | Target
gene(s) | Direct
function(s) | (Refs.) |
|
| Down | Cell lines | p85β (PI3KR2) | Reducing the level
of p85β and phospho-AKT | (42,43) |
| – | Cell lines | VCAM-1 | The expression
level of miR-126 inversely correlates with VCAM-1 | (42) |
| Down | Tissues | VEGFR-2 | Elevated miR-126 is
associated with high VEGFR-2 expression | (44) |
| Down | Tissues/cell
lines | VEGF | Suppressing the
expression of VEGF by binding to its mRNA 3′-UTR | (45) |
| Down | Tissues/cell
lines | CXCR4 | Negatively
regulating CXCR4 expression | (46,47) |
| Down | Tissues/cell
lines | RhoA/ROCK | Inhibiting
RhoA/ROCK signaling pathway | (46,48) |
| Down | Cell lines | IRS-1 | Inhibiting IRS-1
expression by binding to its mRNA 3′-UTR | (49) |
| Down | Tissues/cell
lines | IRS-1, SLC7A5 and
TOM1 | Reducing the
expression of IRS1, SLC7A5 and TOM1 | (50) |
|
| D,
Hepatocellular carcinoma |
|
|
Expression |
Sample(s) | Target
gene(s) | Direct
function(s) | (Refs.) |
|
| Down | Tissues/cell
lines | LRP6 and
PI3KR2 | Negatively
regulating LRP6 and PI3KR2/phospho-AKT expression | (51) |
| Down | Cell lines | SOX2 | Inhibiting SOX2
expression by binding to its mRNA 3′-UTR | (52) |
| Down | Tissues | SPRED1 and
VEGF | Inhibiting the
expression of SPRED1 and VEGF | (53) |
| Down | Tissues/cell
lines | EGFL7 | Suppressing the
expression of EGFL7 | (54) |
| Up | Tissues | EGFL7 | Downregulating
EGFL7 expression to inhibit proliferation and angiogenesis | (16) |
| Down | Tissues/cell
lines | ADAM9 | Downregulating the
expression of ADAM9 | (55) |
|
| E, Pancreatic
cancer |
|
|
Expression |
Sample(s) | Target
gene(s) | Direct
function(s) | (Refs.) |
|
| Down | Tissues/cell
lines | ADAM9 | Inhibiting ADAM9
expression by binding to its mRNA 3′-UTR | (56) |
| Down | Tissues/cell
lines | KRAS and CRK | Reducing the
expression of KRAS and CRK | (57) |
| Down | Tissues/cell
lines | VEGF-A and
SOX2 | Downregulating the
expression of VEGF-A and SOX2 | (58) |
miR-126 expression levels are downregulated in
almost all established esophageal cancer (EC), CRC, HCC and PC cell
lines and certain GC cell lines (Table
I). Restoring or upregulating miR-126 expression can inhibit
cancer cell proliferation, migration and invasion in vitro,
indicating a potential tumor suppressive function or miR-126 in
DSCs (19,49,51,56).
However, Otsubo et al (33)
have revealed that five GC cell lines (HSC43, HSC58, NUGC3, NUGC4
and GCIY) exhibit high levels of miR-126. Similarly, miR-126 has
also been identified to be significantly upregulated in the AGS,
HGC-27, BGC-823, SGC-7901 and MKN-7 cell lines of GC (38), indicating miR-126 may exhibit a
potential oncogenic role in GC (33).
Genetic and epigenetic changes lead to alterations
in tumor-associated gene expression. Aberrant changes in gene
sequence, dicer abundance and epigenetic modifications, including
DNA methylation, histone modifications and nucleosome positioning,
predominantly account for miR-126 dysregulation in cancer (20). For example, the downregulation of
miR-126 in CRC is partly due to promoter methylation of its host
gene EGFL7 (45). Pagano et al
(59) have identified the existence
of epigenetic-miRNA loops in hematopoietic cells, which can either
modulate or be modulated by epigenetic factors and therefore
regulate the gene expression profile. Subsequently, Liu et
al (26) identified a DNA
(cytosine-5)-methyltransferase 1 (DNMT1)-miR-126 epigenetic circuit
in esophageal squamous cell carcinoma (ESCC) cells. DNMT1 is
aberrantly upregulated in ESCC cells, which leads to the
hypermethylation of EGFL7 and downregulation of miR-126, whilst
upregulation of miR-126 markedly reduces the expression of DNMT1
(26). In addition, overexpressed HOX
antisense intergenic RNA (HOTAIR) in GC serves as a competing
endogenous RNA and directly associates with miR-126 to dysregulate
the expression of miR-126 (60).
Finally, the tumor suppressor nasopharyngeal carcinoma-associated
gene 6 has been revealed to upregulate miR-126 in colon cancer
(61).
Targets of miR-126 in DSCs
miRNAs modulate their target gene expression by
binding to the 3′-UTR of the corresponding mRNA. Numerous studies
have identified that miR-126 regulates key processes in different
cancer types by targeting various genes, including insulin receptor
substrate 1, v-crk sarcoma virus CT10 oncogene homolog (CRK), CXC
chemokine receptor type 4 (CXCR4), disintegrin and
metalloproteinase domain-containing protein 9 (ADAM9) and
sex-determining region Y-box 2 (SOX2). As presented in Table I, the majority of these target genes
are overexpressed in DSCs and function as oncogenes involved in
cell proliferation, cell cycle, cell senescence, differentiation,
apoptosis and metastasis at both genetic and epigenetic levels.
Therefore, miR-126 is an effective regulator of tumorigenesis and
progression. VCAM-1 and PI3KR2 are associated with the
anti-inflammatory and anti-neoplastic functions of pomegranate
polyphenols in colon cancer (42).
Golgi phosphoprotein 3 (GOLPH3), the target of miR-126 in ESCC,
contributes to tumorigenicity and tumor cell proliferation
(27,62). Several studies have demonstrated that
miR-126 targets CXCR4 and suppresses its expression, which inhibits
tumor cell proliferation, migration, invasion and cell apoptosis,
and arrests the cell cycle at the G0/G1
transition (46,47). VEGF overexpression and
neo-angiogenesis in CRC is associated with miR-126 downregulation
(45). The oncogenic targets of
miR-126 associated with HCC include the pro-angiogenic PI3KR2
(51) and EGFL7 (16,54), in
addition to the pro-metastatic low-density lipoprotein
receptor-related protein 6 (LRP6) (51). Abnormally high expression of EGFL7 in
HCC is also associated with increased proliferation and decreased
apoptosis in HCC cells (54). ADAM9
overexpression in PC (61), regulator
of G protein signaling 3 (RGS3) overexpression in GC (41,63) and
aberrantly high levels of SOX2 in pancreatic ductal adenocarcinoma
(PDAC) (64) are all positively
associated with cellular migration, invasion and
epithelial-mesenchymal transition (EMT) (56).
The dysregulation of these targets not only promotes
the initiation and development of DSCs, but may also be useful as
novel markers for diagnosing, monitoring and predicting the
prognosis of DSCs. As targets of miR-126 in GC, CRK and CRK-like
(CRKL) have been revealed to facilitate adhesion, invasion and
migration of cancer cells. Furthermore, overexpression of CRK and
CRKL has been associated with more aggressive clinicopathological
features, including a larger tumor size, a higher number of lymph
node metastases, deeper local invasion and higher
tumor-node-metastasis (TNM) staging (19,34).
Overexpression of GOLPH3 may be associated with a poor outcome for
patients with ESCC (62). Similarly,
the expression level of ADAM9 is a confirmed prognostic factor for
HCC (55) and a VEGF polymorphism is
associated with higher susceptibility to GC (65). In summary, the aforementioned markers
may assist in screening populations at high risk and preventing
progression of tumorigenesis.
While the anti-tumor function of miR-126 is well
established and is known to depend on targeting and dysregulating
the aforementioned oncogenes, there are certain reports that
suggest miR-126 serves an oncogenic role in DSCs by targeting
certain tumor-suppressor genes. Cell adhesion molecule 1 (CADM1)
and SOX2 are suppressed upon miR-126 overexpression in GC cells,
leading to carcinogenesis, migration and invasion (33,38).
Furthermore, a pro-angiogenic role of miR-126 has been identified
in HCC tissues, with SPRED1 inhibition as the underlying mechanism
(53). Hansen et al (44) also revealed that a higher expression
of miR-126 in CRC tissues is associated with increased expression
of VEGFR-2 and a denser microvessel density (MVD).
The targets of miR-126 in DSCs can also be
negatively modulated by miR-126 in other tumors. For instance, CRK
is targeted by miR-126 in non-small cell lung cancer cells
(66) and solute carrier family 7
member 5 has been identified as an miR-126 target in colon cancer
and thyroid cancer (50,67). In addition, miR-126 can regulate the
same or different targets in DSCs by cooperating with other miRNAs.
For instance, miR-145 exerts its role of inhibiting tumor growth in
colon cancer by targeting and regulating IRS-1 (68). Additionally, miR-126 and miR-34a can
synergistically exert a tumor suppressive effect by targeting
VEGF-A, SOX2, cyclin D1, E2F1 and B-cell lymphoma 2 in pancreatic
adenocarcinoma (PAC) (58).
Notably, several targets of miR-126 in DSCs are
exclusively regulated by miR-126 in specific cancer types. Studies
indicate that PI3KR2 and VEGF-A are regulated by miR-126 solely or
together in EC (29,30), GC (35,36), CRC
(42), HCC (51) and PAC (58). It has also been demonstrated that
miR-126 targets IRS-1 only in ESCC (27) and CRC (50). KRAS has been confirmed as a target of
miR-126 in PDAC and in contrast to the typical miRNA-target gene
interaction, miR-126 binds to KRAS at an uncommon site in its
3′-UTR (57). However, KRAS
expression is not influenced by miR-126 in CRC (69). Furthermore, certain genes that are
targeted by miR-126 in several DSCs serve different roles in
different cancer types. SOX2, an inhibitor of GC growth (70), promotes tumor cell proliferation and
invasion in EC (28). A feedback
regulatory loop may also exist between miR-126 and its target
genes, as identified with DNMT1 in ESCC, which increases the
intricacy of the regulation network of the miRNA (26).
Signaling cascades modulated by miR-126 in
DSCs
In addition to numerous cancer-associated genes,
miR-126 also interacts with the signaling pathways involved in
DSCs. Various signaling cascades involved in the regulation of
angiogenesis and vascular integrity, as well as cell proliferation,
migration and invasion are modulated by miR-126 (Fig. 2).
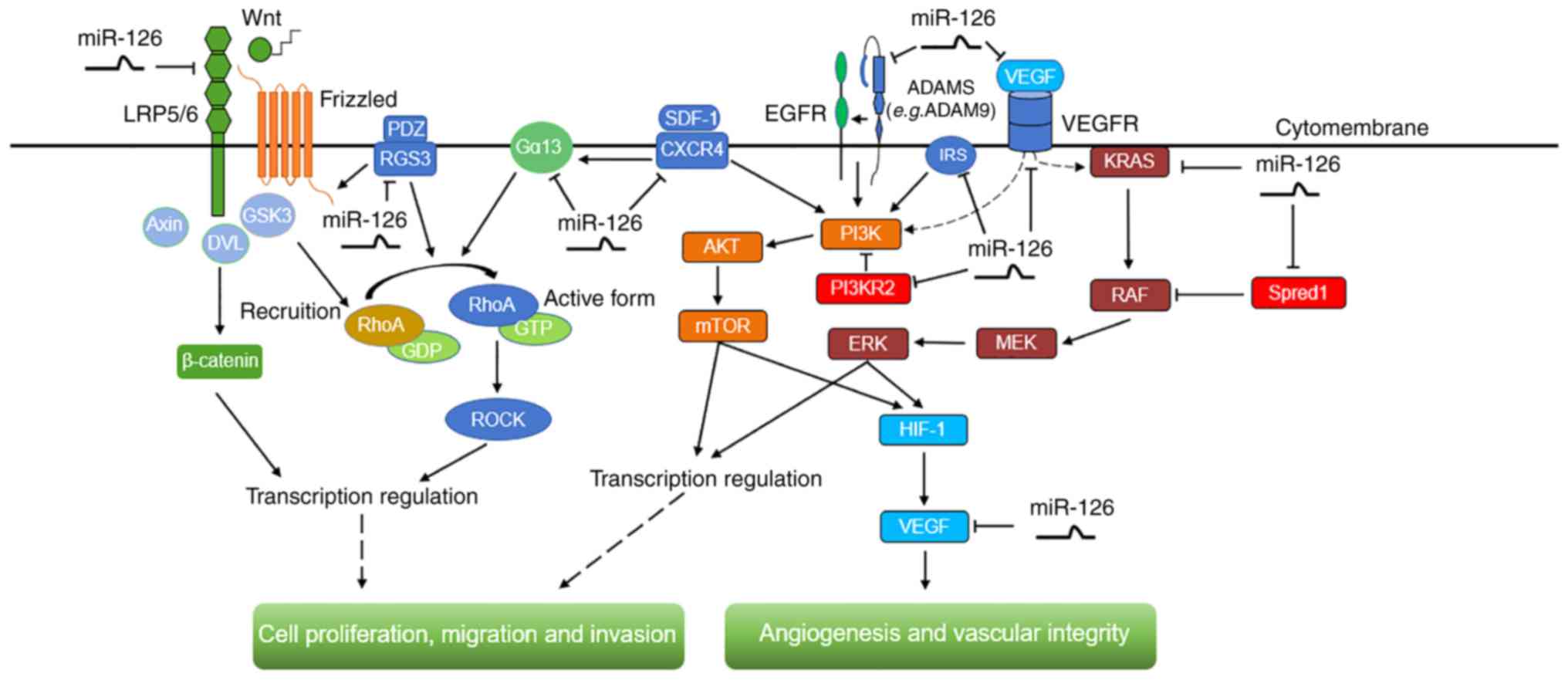 | Figure 2.The signaling pathways, including
Ras/ERK, PI3K/AKT/mTOR, ADAM9/EGFR/AKT, RhoA/ROCK and
Wnt/β-catenin, modulated by miR-126 in digestive system cancer
types. These signaling cascades are involved in the regulation of
angiogenesis and vascular integrity, as well as cell proliferation,
migration and invasion. miR-126, microRNA-126; ERK, extracellular
signal-regulated kinase; PI3K, phosphatidylinositol-3-kinase; mTOR,
mammalian target of rapamycin; ADAM9, disintegrin and
metalloproteinase domain-containing protein 9; EGFR, epidermal
growth factor receptor; RhoA, ras homology gene family, member A;
ROCK, Rho-associated protein kinase; LRP5/6, low density
lipoprotein receptor related protein 5/6; GSK3, glycogen synthase
kinase 3; DVL, dishevelled; RGS3, regulator of G protein signaling
3; Gα13, G protein α13; GTP, guanosine 5′-triphosphate; GDP,
guanosine 5′-diphosphate; SDF-1, stromal cell-derived factor-1;
CXCR4, CXC chemokine receptor type 4; PI3KR2,
phosphoinositide-3-kinase regulatory subunit 2; IRS-1, insulin
receptor substrate 1; VEGF, vascular endothelial growth factor;
VEGFR, VEGF receptor; SPRED1, sprouty-related EVH1
domain-containing protein 1; MEK, mitogen-activated protein kinase;
HIF1, hypoxia-inducible factor 1. |
miR-126 regulates the expression of VEGFR-2 and VEGF
in DSCs via the MAPK/ERK and PI3K/AKT pathways (44,45), and
can impede the growth of cancer cells by targeting p85β (also
termed PI3KR2), leading to decreased levels of phosphorylated AKT,
the active form of the kinase (29).
AKT phosphorylates a wide range of proteins involved in cell
survival, motility and proliferation (71). Myelin transcription factor 1, which is
phosphorylated by AKT to modulate the G2/M-phase transition
(72), is also upregulated by miR-126
(29). Zhou et al (49) identified that miR-126 suppresses the
proliferation, migration, invasion and cell cycle progression of
cancer cells by targeting IRS-1 via the AKT and ERK1/2 signaling
pathways. The anti-angiogenic role of miR-126 in cancer tissues is
exerted by targeting VEGF-A, possibly via the MAPK/ERK and
AKT/mammalian target of rapamycin (mTOR) signaling pathways
(16,35,53,54), which
reduces the MVD in cancer tissues (51). In addition, miR-126 exhibits a
suppressive role in the proliferation, apoptosis and angiogenesis
of tumor cells and tissues by targeting EGFL7, through the
inactivation of the ERK signaling pathway (73). HOTAIR, a long non-coding RNA that is
increased in GC and is responsible for drug-resistance, potentially
acts by binding to and dysregulating miR-126, leading to the
upregulation of miR126-targeted oncogenes, including VEGF-A and
PI3KR2, and activation of the PI3K/AKT/multidrug
resistance-associated protein 1 pathway (60).
Furthermore, miR-126 inhibits the activity and
expression of the Ras homology gene family, member A
(RhoA)/Rho-associated protein kinase (ROCK) signaling pathway
mediators that are aberrantly upregulated in cancer cells. The
expression of these mediators positively correlates with clinical
stage and lymph node metastasis but negatively correlates with
overall survival of patients with cancer (46). CXCR4 is a putative mediator between
miR-126 and the RhoA/ROCK signaling pathway (46,48). It
has been identified that miR-126 exerts a tumor suppressive effect
by targeting CXCR4 and involving the stromal cell-derived factor
1/CXCR4 axis, which possibly promotes MAPK p42/44 phosphorylation
and activates the PI3K/AKT pathway (74,75).
Furthermore, miR-126 was revealed to be a tumor suppressor in CRC
through the AKT and ERK1/2 signaling pathways, by dysregulating
CXCR4 (47). Additionally, Yuan et
al (46) observed that CXCR4
could prevent the inhibitory action of miR-126 on the RhoA
signaling pathway by coupling with Gα13, therefore validating that
miR-126 acts as a tumor suppressor by inactivating RhoA signaling
via the CXCR4/Gα13/RhoA axis in DSCs.
It has been hypothesized that upregulating miR-126
can weaken the metastatic ability of HCC by suppressing the
expression of LRP6, a Wnt co-receptor, and its downstream mediator
β-catenin (51). This may be
consistent with a study demonstrating that the activation of the
Wnt/β-catenin signaling pathway results in spontaneous HCC in
farnesoide X receptor-knockout mice (76). In addition, activation of the
Wnt/β-catenin signaling pathway is involved in promoting EMT of GC
cells (77) and numerous studies have
confirmed that various miRNAs, including miR-544a, modulate EMT in
GC via Wnt/β-catenin signaling (78).
Upregulation of RGS3 can enhance the pro-tumorigenic signals
generated by the Wnt/β-catenin pathway and induce EMT (41,63).
Therefore, miR-126 directly targeting and downregulating RGS3 is
possibly involved in its inhibition of EMT via the Wnt/β-catenin
pathway (41). Furthermore, miR-126
functions as a tumor suppressor in ESCC via the ADAM9-EGFR-AKT axis
and the downregulation of ADAM9 suppresses the activation of the
EGFR-AKT pathway by restricting phosphorylation (26). In addition to the anti-tumor effects
of miR-126 through the aforementioned signaling pathways, miR-126
also exhibits a pro-tumorigenic effect in DSCs. A pathway has been
proposed in which miR-126 dysregulates SOX2, leading to
overexpression of placenta-specific 1, a tumor suppressor for
gastric adenocarcinoma; this pathway may serve a positive role in
gastric carcinogenesis (33).
It is noteworthy that VEGF, VEGFR, EGFR and CXCR4
are pro-inflammatory factors that are frequently upregulated in the
tumor microenvironment, possibly leading to oncogenic events
(79). As aforementioned, the
interactions between miR-126 and these cytokines or their receptors
exert modulatory effects on several dysregulated pathways,
including MAPK/ERK, AKT/mTOR and RhoA signaling pathways, which
indicates a complicated cross-talk among cytokines, miRNAs and
signaling pathways in tumorigenesis and tumor progression. Other
miRNAs have also been reported to interact with cytokines in DSCs.
For example, Ma et al (80)
have demonstrated that interleukin (IL)-1β-induced nuclear factor
κ-light-chain-enhancer of activated B cells activation results in
increased miR-425 transcription, which further facilitates GC cell
proliferation by suppressing phosphatase and tensin homolog.
miR-126 serves as a useful biomarker in
DSCs
miRNAs exhibit several essential features of
biomarkers, including an average length of 22 nucleotides, stable
expression and ease of detection (81). Due to their ubiquitous and abnormal
expression in human cancer types, they exhibit great potential as
diagnostic and prognostic factors for cancer (82). As aforementioned, the dysregulation of
several target genes due to aberrant expression of miR-126 is
involved in DSCs, indicating the potential of miR-126 as a
biomarker for this group of cancer types.
Previous studies have revealed that miR-126 is
highly expressed in normal tissues compared with cancer tissues and
DSC cell lines (Table I). Liu et
al (25) identified 60 miRNAs,
including miR-126, that were expressed differentially between
matched EC and paracancerous normal tissues. Lower expression of
miR-126 has been observed in both HCC tissues and liver cancer
cells compared with normal controls (16,51). In
addition, higher levels of miR-126 can be used to discriminate HCC
from metastatic adenocarcinoma in the liver along with other miRNAs
(83). Hamada et al (56) demonstrated that miR-126 is
significantly downregulated in invasive ductal adenocarcinoma (IDA)
compared with normal pancreatic tissues, intraductal papillary
mucinous adenoma and intraductal papillary mucinous carcinoma
tissues. Furthermore, patients with IDA were revealed to exhibit
more adverse outcomes compared with patients with intraductal
papillary mucinous adenoma or intraductal papillary mucinous
carcinoma due to a higher invasive potential (56). Similarly, other studies consider
miR-126 as a representative of 21 dysregulated miRNAs when
comparing PDAC with serous microcystic adenoma (57). Therefore, detecting the levels of
miR-126 in tissues or cells may promote promising new strategies to
diagnose DSCs.
Overwhelming evidence has indicated that miR-126 is
associated with metastasis, cancer stage, tumor size or other
clinicopathological characteristics of DSCs, in which metastasis is
a leading cause of cancer-associated cases of mortality (84). Low expression of miR-126 is observed
in metastatic CRC cell lines and miR-126 levels are significantly
lower in patients with numerous metastatic sites compared with
those with only one metastatic location (48,85).
Furthermore, the expression of miR-126 is negatively correlated
with TNM stage and metastasis in patients with CRC, indicating that
lower expression levels of miR-126 may predict a poorer prognosis
(46,47). Low levels of miR-126 in tumors are
similarly associated with lymph node metastasis, local invasion and
TNM stage in GC, ESCC and other DSCs (19,27,29). In
addition, miR-126 may serve as an independent prognostic indicator
for patients with ESCC (26). Li
et al (86) established a
seven-miRNA signature, with miR-126 designated as protective, to
predict the relapse-free survival and overall survival of patients
with GC. Similarly, miR-126 was identified as a predictor of venous
metastases and survival of patients with HCC in a 20-miRNA
prognostic signature (87).
Dysregulation of miR-126 is inversely correlated with the
clinicopathological parameters of HCC, including tumor size, tumor
weight and alpha-fetoprotein (AFP) level, and is involved with
tumor angiogenesis, microvascular invasion, tumor metastasis and
early recurrence (16,51,55). The
gene targets of miR-126 may also exhibit a potential prognostic
function. For example, downregulation of miR-126 and upregulation
of CRK are synergistically correlated with tumor progression of GC
and can therefore be a combined unfavorable prognostic factor for
patients with GC (88). Consequently,
timely detection of miR-126 and CRK levels may be of great
importance in predicting the overall survival of patients with
advanced GC (32).
In addition to the tissues, circulating miRNAs are
also detected in plasma, which has been demonstrated to be a
feasible method for diagnosing human cancer types (89). Previous studies have identified that
the serum levels of miR-126 are altered in patients with CRC
compared with healthy controls (90),
and between patients with CRC with liver metastasis and those with
localized CRC (89). Lower levels of
miR-126 were also observed in the plasma of patients with PC
compared with healthy individuals (91). Since serum miR-126 levels are
significantly different between patients with HCC with hepatitis B
virus (HBV-HCC) and their non-HCC counterparts, it is necessary to
evaluate the plasma levels of miR-126 and AFP rather than AFP alone
for diagnosing HBV-HCC (92).
However, several studies demonstrate ambiguous
results regarding the role of miR-126 in DSC diagnosis. miR-126 has
been identified as both a benign and an unfavorable prognostic
factor. Overexpression of miR-126 accompanied with downregulation
of CADM1 may function as an adverse factor for patients with stage
I GC (38). While miR-126
demonstrates no association with the prognosis of patients with EC
(24), it can act as an independent
predictor of the outcome of ESCC (26).
Therapeutic implications of miR-126 in
DSCs
miRNAs are promising therapeutic targets for various
cancer types on account of their post-transcriptional regulation of
numerous cancer-associated target genes (7). Therefore artificially modulating the
expression levels of miRNAs and thereby their targets is a rational
therapeutic approach. Accordingly, the inhibitory role of miR-126
in tumorigenesis and the development of DSCs could be exploited for
treatment of these cancer types. Restoring the expression of
miR-126 to influence different targets and pathways could be a
novel therapeutic strategy for DSCs. Ectopic expression of miR-126
in CRC (47), GC (19,31), EC
(29), HCC (51,52) and
PAC (58) attenuates cell-cycle
progression, suppresses tumor proliferation, oncogenesis, colony
formation, invasion and metastasis, and simultaneously promotes
tumor cell apoptosis. Increasing the expression of miR-126 in
pancreatic benign cystic tumors prevents them developing into PC
(57).
miRNAs have also been implicated in chemoresistance
of cancer cells and failed chemotherapy; the miRNAs associated with
apoptosis are usually responsible for this phenomenon (93). An association between miR-126 and
chemoresistance in DSCs has been reported. A low level of miR-126
in primary CRC tumors is associated with lower sensitivity to
oxaliplatin in metastatic CRC (85).
In another study, upregulation of miR-126 significantly improved
the sensitivity of colon cancer cells to oxaliplatin (50). Multiple studies have revealed lower
expression of miR-126 in drug-resistant GC cell lines and
re-sensitization to vincristine and adriamycin following ectopic
expression of miR-126 (39). miR-126
may also affect the sensitivity of GC cells to cisplatin, which is
one of the major chemotherapeutic options in treating patients with
advanced GC (60). A previous study
identified that miR-126 is downregulated along with a number of
other miRNAs in hydroxycamptothecin-resistant GC cells (94). Therefore, another feasible therapeutic
strategy is using synthetic miR-126 as a drug-sensitivity mediator
in DSCs.
However, there are a number of limitations in the
clinical application of miR-126, including delivery of the effector
molecule to target sites, which is considered to be the major
challenge for miRNA-based treatment (95). Stable nucleic acid lipid particles,
polyethylene glycol or virus-like particles have been tested as
carriers of miR-126 (95,96). Additionally, another study
investigated the possibility of using carcinoma cells targeting
oncolytic adenovirus for delivery of miR-126 (58). Despite the generally high efficacy of
viral vectors, there are apprehensions regarding their toxicity and
immunogenicity, which limit their clinical usage (97).
Recent studies have demonstrated that the
dysregulation of miRNAs in cancer is associated with DNA
methylation. Therefore, altering the methylation status of
tumor-associated genes is a promising epigenetic therapy (98). Zhang et al (45) revealed that deficiency of miR-126 in
CRC cells may be a result of aberrant DNA methylation, since
miR-126 expression was restored following demethylation of CRC
cells with 5-aza-2′-deoxycytidine. Similarly, miR-126 expression is
regulated by the DNMT1-miR-126 epigenetic circuit in ESCC cells via
hypermethylation of the EGFL7 promoter. Therefore,
miR-126-associated epigenetic modification exhibits significant
therapeutic potential in ESCC (26).
Since miRNAs exert their oncotherapeutic effects by
regulating target genes, the direct regulation of target genes
should be validated. Suppressing CRKL in GC cells had the same
effects as overexpressing miR-126, including inducing cell cycle
arrest at G0/G1 phase and inhibiting
proliferation (34). Similarly, the
therapeutic effect of inhibiting ESCC cell proliferation and
migration can be acquired not only by the re-introduction of
miR-126 but also dysregulation of ADAM9 (26). Furthermore, the miR-126/ADAM9 axis can
be a therapeutic target in PC in terms of suppressing migration of
tumor cells (56).
The heterogeneity of the miRNA regulation network
makes the therapeutic role of miR-126 in DSCs controversial. It has
been reported that decreasing but not increasing the expression of
miR-126 may inhibit migration and invasion of GC cells (38). Another study hypothesized that a
miR-126 inhibitor combined with TACE may achieve an improved
therapeutic effect in HCC (53). In
addition, the combination of miR-126 with other miRNAs, including
miR-34a, targeting different genes could achieve improved results
in PAC (58). Notably, overexpression
of miR-126 destroyed cell viability, arrested cell cycle, inhibited
clonogenicity and blocked tumorigenicity by targeting
KRAS-associated genes in multiple KRAS-mutant CRC cells but not in
KRAS-WT cancer cells (69).
Conclusions
The current review has summarized that miR-126 is a
tumor suppressor and its low levels in DSC tissues are associated
with tumor development and progression. Therefore, restoring or
increasing miR-126 levels is a promising therapeutic strategy for
DSCs. However, the difficulty in transporting miR-126 to target
sites may be a limitation until safe and effective vectors are
developed. In addition, the aberrant serum levels of miR-126 in
patients with DSCs may serve as predictive or diagnostic
biomarkers.
miR-126 can also act as an oncogene in specific
DSCs. Regardless of its function, it exerts its modulatory effects
via different target genes and regulatory pathways with associated
functions. Considering the significance and complexity of miR-126
and its regulatory network in DSCs, it is essential to further
investigate the role of miR-126 in the initiation and progression
of different cancer types to identify potential clinical
applications.
Acknowledgements
The authors thank Dr Qiang Liu for assisting with
the writing of the manuscript and Dr Tao Li for valuable
discussions (both Department of Gastroenterology of the First
Affiliated Hospital of Nanchang University, Nanchang, China).
Funding
The review was supported by funding from the
National Science Foundation Grants of China (nos. 81160307 and
81560395), the Jiangxi Science & Technology Pillar Program and
the Science Foundation for Young Scholars of Jiangxi Province
(grant no. 2007GQY1167).
Availability of data and materials
Not applicable.
Authors' contributions
MH wrote the manuscript. SX made the tables and
diagrams. QC, and SZ checked and revised the manuscript; XZ put
forward the concept, and was responsible for the organization,
revision and submission of this manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
et al: MicroRNAs exhibit high frequency genomic alterations in
human cancer. Proc Natl Acad Sci USA. 103:9136–9141. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moretti F, Thermann R and Hentze MW:
Mechanism of translational regulation by miR-2 from sites in the 5′
untranslated region or the open reading frame. RNA. 16:2493–2502.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Z, Li Z, Li Y and Zang A: MicroRNA
and signaling pathways in gastric cancer. Cancer Gene Ther.
21:305–316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fish JE, Santoro MM, Morton SU, Yu S, Yeh
RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY and Srivastava D:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Aurora AB, Johnson BA, Qi X,
McAnally J, Hill JA, Richardson JA, Bassel-Duby R and Olson EN: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meister J and Schmidt MHH: miR-126 and
miR-126* New players in cancer. ScientificWorldJournal.
10:2090–2100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Soncin F, Mattot V, Lionneton F, Spruyt N,
Lepretre F, Begue A and Stehelin D: VE-statin, an endothelial
repressor of smooth muscle cell migration. EMBO J. 22:5700–5711.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parker LH, Schmidt M, Jin SW, Gray AM,
Beis D, Pham T, Frantz G, Palmieri S, Hillan K, Stainier DY, et al:
The endothelial-cell-derived secreted factor Egfl7 regulates
vascular tube formation. Nature. 428:754–758. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Campagnolo L, Leahy A, Chitnis S,
Koschnick S, Fitch MJ, Fallon JT, Loskutoff D, Taubman MB and
Stuhlmann H: EGFL7 is a chemoattractant for endothelial cells and
is up-regulated in angiogenesis and arterial injury. Am J Pathol.
167:275–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morgenbesser SD, Dufault MR, Martin TB,
Lim E, Callahan M, Weber W, Winter SF, McLaren RP, Richards B, Cook
BP, et al: Characterization of EGFL7 expression and function in
tumorigenesis and angiogenesis. Cancer Res. 65 Suppl
9:S11032005.
|
|
16
|
Hu MH, Ma CY, Wang XM, Ye CD, Zhang GX,
Chen L and Wang JG: MicroRNA-126 inhibits tumor proliferation and
angiogenesis of hepatocellular carcinoma by down-regulating EGFL7
expression. Oncotarget. 7:66922–66934. 2016.PubMed/NCBI
|
|
17
|
Harris TA, Yamakuchi M, Ferlito M, Mendell
JT and Lowenstein CJ: MicroRNA-126 regulates endothelial expression
of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA.
105:1516–1521. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang ST, Wang F, Shao M, Wang Y and Zhu
HQ: MicroRNA-126 suppresses inflammation in endothelial cells under
hyperglycemic condition by targeting HMGB1. Vascul Pharmacol.
88:48–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feng R, Chen X, Yu Y, Su L, Yu B, Li J,
Cai Q, Yan M, Liu B and Zhu Z: miR-126 functions as a tumour
suppressor in human gastric cancer. Cancer Lett. 298:50–63. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ebrahimi F, Gopalan V, Smith RA and Lam
AK: miR-126 in human cancers: Clinical roles and current
perspectives. Exp Mol Pathol. 96:98–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Y, Correa AM, Hoque A, Guan B, Ye F,
Huang J, Swisher SG, Wu TT, Ajani JA and Xu XC: Prognostic
significance of differentially expressed miRNAs in esophageal
cancer. Int J Cancer. 128:132–143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu R, Gu J, Jiang P, Zheng Y, Liu X,
Jiang X, Huang E, Xiong S, Xu F, Liu G, et al: DNMT1-microRNA126
epigenetic circuit contributes to esophageal squamous cell
carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res.
21:854–863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Meng F, Ma J, Yu Y, Hua X, Qin J and
Li Y: Insulin receptor substrate-1 and Golgi phosphoprotein 3 are
downstream targets of miR-126 in esophageal squamous cell
carcinoma. Oncol Rep. 32:1225–1233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nie ZC, Weng WH, Li J, Xu YT and Li Z:
Down-regulation of miR-126 in esophageal carcinoma tissues and its
inhibition effect on proliferation and migration of esophageal
carcinoma cell line EC109. Tumor. 35:55–64. 2015.
|
|
29
|
Nie ZC, Weng WH, Shang YS, Long Y, Li J,
Xu YT and Li Z: MicroRNA-126 is down-regulated in human esophageal
squamous cell carcinoma and inhibits the proliferation and
migration in EC109 cell via PI3K/AKT signaling pathway. Int J Clin
Exp Pathol. 8:4745–4754. 2015.PubMed/NCBI
|
|
30
|
Kong R, Ma Y, Feng J, Li S, Zhang W, Jiang
J, Zhang J, Qiao Z, Yang X and Zhou B: The crucial role of miR-126
on suppressing progression of esophageal cancer by targeting
VEGF-A. Cell Mol Biol Lett. 21:32016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li X, Wang F and Qi Y: miR-126 inhibits
the invasion of gastric cancer cell in part by targeting Crk. Eur
Rev Med Pharmacol Sci. 18:2031–2037. 2014.PubMed/NCBI
|
|
32
|
Yue S, Shi H, Han J, Zhang T, Zhu W and
Zhang D: Prognostic value of microRNA-126 and CRK expression in
gastric cancer. Onco Targets Ther. 9:6127–6135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Otsubo T, Akiyama Y, Hashimoto Y, Shimada
S, Goto K and Yuasa Y: MicroRNA-126 inhibits SOX2 expression and
contributes to gastric carcinogenesis. PLoS One. 6:e166172011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Chen X, Li P, Su L, Yu B, Cai Q,
Li J, Yu Y, Liu B and Zhu Z: CRKL promotes cell proliferation in
gastric cancer and is negatively regulated by miR-126. Chem Biol
Interact. 206:230–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N,
Zhou X and Chen C: Reduced miR-126 expression facilitates
angiogenesis of gastric cancer through its regulation on VEGF-A.
Oncotarget. 5:11873–11885. 2014.PubMed/NCBI
|
|
36
|
Liu LY, Wang W, Zhao LY, Guo B, Yang J,
Zhao XG, Hou N, Ni L, Wang AY, Song TS, et al: Mir-126 inhibits
growth of SGC-7901 cells by synergistically targeting the oncogenes
PI3KR2 and Crk, and the tumor suppressor PLK2. Int J Oncol.
45:1257–1265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Chen X, Su L, Li P, Cai Q, Liu B,
Wu W and Zhu Z: MicroRNA-126 inhibits cell proliferation in gastric
cancer by targeting LAT-1. Biomed Pharmacother. 72:66–73. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang Z, Wang R, Zhang T and Dong X:
MicroRNA-126 regulates migration and invasion of gastric cancer by
targeting CADM1. Int J Clin Exp Pathol. 8:8869–8880.
2015.PubMed/NCBI
|
|
39
|
Wang P, Li Z, Liu H, Zhou D, Fu A and
Zhang E: MicroRNA-126 increases chemosensitivity in drug-resistant
gastric cancer cells by targeting EZH2. Biochem Biophys Res Commun.
479:91–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Zhou Y, Fei X, Chen X, Yan J, Liu
B and Zhu Z: ADAM9 functions as a promoter of gastric cancer growth
which is negatively and post-transcriptionally regulated by
miR-126. Oncol Rep. 37:2033–2040. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Zhou Y, Fei X, Chen X and Zhu Z:
Regulator of G-protein signaling 3 targeted by miR-126 correlates
with poor prognosis in gastric cancer patients. Anticancer Drugs.
28:161–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Banerjee N, Kim H, Talcott S and
Mertens-Talcott S: Pomegranate polyphenolics suppressed
azoxymethane-induced colorectal aberrant crypt foci and
inflammation: Possible role of miR-126/VCAM-1 and
miR-126/PI3K/AKT/mTOR. Carcinogenesis. 34:2814–2822. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guo C, Sah JF, Beard L, Willson JK,
Markowitz SD and Guda K: The noncoding RNA, miR-126, suppresses the
growth of neoplastic cells by targeting phosphatidylinositol
3-kinase signaling and is frequently lost in colon cancers. Genes
Chromosomes Cancer. 47:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hansen TF, Andersen CL, Nielsen BS,
Spindler KL, Sørensen FB, Lindebjerg J, Brandslund I and Jakobsen
A: Elevated microRNA-126 is associated with high vascular
endothelial growth factor receptor 2 expression levels and high
microvessel density in colorectal cancer. Oncol Lett. 2:1101–1106.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Y, Wang X, Xu B, Wang B, Wang Z,
Liang Y, Zhou J, Hu J and Jiang B: Epigenetic silencing of miR-126
contributes to tumor invasion and angiogenesis in colorectal
cancer. Oncol Rep. 30:1976–1984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yuan W, Guo YQ, Li XY, Deng MZ, Shen ZH,
Bo CB, Dai YF, Huang MY, Yang ZY, Quan YS, et al: MicroRNA-126
inhibits colon cancer cell proliferation and invasion by targeting
the chemokine (C-X-C motif) receptor 4 and Ras homolog gene family,
member A, signaling pathway. Oncotarget. 7:60230–60244. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu Y, Zhou Y, Feng X, An P, Quan X, Wang
H, Ye S, Yu C, He Y and Luo H: MicroRNA-126 functions as a tumor
suppressor in colorectal cancer cells by targeting CXCR4 via the
AKT and ERK1/2 signaling pathways. Int J Oncol. 44:203–210. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li N, Tang A, Huang S, Li Z, Li X, Shen S,
Ma J and Wang X: miR-126 suppresses colon cancer cell proliferation
and invasion via inhibiting RhoA/ROCK signaling pathway. Mol Cell
Biochem. 380:107–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou Y, Feng X, Liu YL, Ye SC, Wang H, Tan
WK, Tian T, Qiu YM and Luo HS: Down-regulation of miR-126 is
associated with colorectal cancer cells proliferation, migration
and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling
pathways. PLoS One. 8:e812032013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li N, Li X, Huang S, Shen S and Wang X:
miR-126 inhibits colon cancer proliferation and invasion through
targeting IRS1, SLC7A5 and TOM1 gene. Zhong Nan Da Xue Xue Bao Yi
Xue Ban. 38:809–817. 2013.(In Chinese). PubMed/NCBI
|
|
51
|
Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie
H, Zhou L, Wu J and Zheng S: miR-126-3p suppresses tumor metastasis
and angiogenesis of hepatocellular carcinoma by targeting LRP6 and
PIK3R2. J Transl Med. 12:2592014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao C, Li Y, Zhang M, Yang Y and Chang L:
miR-126 inhibits cell proliferation and induces cell apoptosis of
hepatocellular carcinoma cells partially by targeting Sox2. Hum
Cell. 28:91–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ji JS, Xu M, Song JJ, Zhao ZW, Chen MJ,
Chen WQ, Tu JF and Yang XM: Inhibition of microRNA-126 promotes the
expression of Spred1 to inhibit angiogenesis in hepatocellular
carcinoma after transcatheter arterial chemoembolization: In vivo
study. Onco Targets Ther. 9:4357–4367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gong C, Fang J, Li G, Liu HH and Liu ZS:
Effects of microRNA-126 on cell proliferation, apoptosis and tumor
angiogenesis via the down-regulating ERK signaling pathway by
targeting EGFL7 in hepatocellular carcinoma. Oncotarget.
8:52527–52542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xiang LY, Ou HH, Liu XC, Chen ZJ, Li XH,
Huang Y and Yang DH: Loss of tumor suppressor miR-126 contributes
to the development of hepatitis B virus-related hepatocellular
carcinoma metastasis through the upregulation of ADAM9. Tumor Biol.
39:10104283177091282017. View Article : Google Scholar
|
|
56
|
Hamada S, Satoh K, Fujibuchi W, Hirota M,
Kanno A, Unno J, Masamune A, Kikuta K, Kume K and Shimosegawa T:
miR-126 acts as a tumor suppressor in pancreatic cancer cells via
the regulation of ADAM9. Mol Cancer Res. 10:3–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jiao LR, Frampton AE, Jacob J, Pellegrino
L, Krell J, Giamas G, Tsim N, Vlavianos P, Cohen P, Ahmad R, et al:
MicroRNAs targeting oncogenes are down-regulated in pancreatic
malignant transformation from benign tumors. PLoS One.
7:e320682012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Feng SD, Mao Z, Liu C, Nie YS, Sun B, Guo
M and Su C: Simultaneous overexpression of miR-126 and miR-34a
induces a superior antitumor efficacy in pancreatic adenocarcinoma.
Onco Targets Ther. 10:5591–5604. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Pagano F, De Marinis E, Grignani F and
Nervi C: Epigenetic role of miRNAs in normal and leukemic
hematopoiesis. Epigenomics. 5:539–552. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yan J, Dang Y, Liu S, Zhang Y and Zhang G:
LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by
targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biol.
37:16345–16355. 2016. View Article : Google Scholar
|
|
61
|
Wang XY, Wu MH, Liu F, Li Y, Li N, Li GY
and Shen SR: Differential miRNA expression and their target genes
between NGX6-positive and negative colon cancer cells. Mol Cell
Biochem. 345:283–290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang JH, Chen XT, Wen ZS, Zheng M, Deng
JM, Wang MZ, Lin HX, Chen K, Li J, Yun JP, et al: High expression
of GOLPH3 in esophageal squamous cell carcinoma correlates with
poor prognosis. PLoS One. 7:e456222012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shi CS, Huang NN and Kehrl JH: Regulator
of G-protein signaling 3 isoform 1 (PDZ-RGS3) enhances canonical
Wnt signaling and promotes epithelial mesenchymal transition. J
Biol Chem. 287:33480–33487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Singh SK, Chen NM, Hessmann E, Siveke J,
Lahmann M, Singh G, Voelker N, Vogt S, Esposito I, Schmidt A, et
al: Antithetical NFATc1-Sox2 and p53-miR200 signaling networks
govern pancreatic cancer cell plasticity. EMBO J. 34:517–530. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu H, Wang S and Huang C: VEGFA+936C/T
and −634G/C polymorphisms and gastric cancer risk: A meta-analysis.
Asian Pac J Cancer Prev. 12:1979–1983. 2011.PubMed/NCBI
|
|
66
|
Crawford M, Brawner E, Batte K, Yu L,
Hunter MG, Otterson GA, Nuovo G, Marsh CB and Nana-Sinkam SP:
MicroRNA-126 inhibits invasion in non-small cell lung carcinoma
cell lines. Biochem Biophys Res Commun. 373:607–612. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xiong Y, Kotian S, Zeiger MA, Zhang L and
Kebebew E: miR-126-3p inhibits thyroid cancer cell growth and
metastasis, and is associated with aggressive thyroid cancer. PLoS
One. 10:e01304962015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: Micro RNA 145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hara T, Jones MF, Subramanian M, Li XL, Ou
O, Zhu Y, Yang Y, Wakefield LM, Hussain SP, Gaedcke J, et al:
Selective targeting of KRAS-mutant cells by miR-126 through
repression of multiple genes essential for the survival of
KRAS-mutant cells. Oncotarget. 5:7635–7650. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Otsubo T, Akiyama Y, Yanagihara K and
Yuasa Y: SOX2 is frequently downregulated in gastric cancers and
inhibits cell growth through cell-cycle arrest and apoptosis. Br J
Cancer. 98:824–831. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Osaki M, Oshimura M and Ito H: PI3K-Akt
pathway: Its functions and alterations in human cancer. Apoptosis.
9:667–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Okumura E, Fukuhara T, Yoshida H, Hanada
Si S, Kozutsumi R, Mori M, Tachibana K and Kishimoto T: Akt
inhibits Myt1 in the signalling pathway that leads to meiotic
G2/M-phase transition. Nat Cell Biol. 4:111–116. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xu WH, Zhang JB, Dang Z, Li X, Zhou T, Liu
J, Wang DS, Song WJ and Dou KF: Long non-coding RNA URHC regulates
cell proliferation and apoptosis via ZAK through the ERK/MAPK
signaling pathway in hepatocellular carcinoma. Int J Biol Sci.
10:664–676. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang SC, Lin JK, Wang HS, Yang SH, Li AF
and Chang SC: Nuclear expression of CXCR4 is associated with
advanced colorectal cancer. Int J Colorectal Dis. 25:1185–1191.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li Z, Li N, Wu M, Li X, Luo Z and Wang X:
Expression of miR-126 suppresses migration and invasion of colon
cancer cells by targeting CXCR4. Mol Cell Biochem. 381:233–242.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wolfe A, Thomas A, Edwards G, Jaseja R,
Guo GL and Apte U: Increased activation of the Wnt/β-catenin
pathway in spontaneous hepatocellular carcinoma observed in
farnesoid X receptor knockout mice. J Pharmacol Exp Ther.
338:12–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Huang J, Xiao D, Li G, Ma J, Chen P, Yuan
W, Hou F, Ge J, Zhong M, Tang Y, et al: EphA2 promotes
epithelial-mesenchymal transition through the Wnt/β-catenin pathway
in gastric cancer cells. Oncogene. 33:2737–2747. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yanaka Y, Muramatsu T, Uetake H, Kozaki K
and Inazawa J: miR-544a induces epithelial-mesenchymal transition
through the activation of WNT signaling pathway in gastric cancer.
Carcinogenesis. 36:1363–1371. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Germano G, Allavena P and Mantovani A:
Cytokines as a key component of cancer-related inflammation.
Cytokine. 43:374–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ma J, Liu J, Wang Z, Gu X, Fan Y, Zhang W,
Xu L, Zhang J and Cai D: NF-kappaB-dependent microRNA-425
upregulation promotes gastric cancer cell growth by targeting PTEN
upon IL-1β induction. Mol Cancer. 13:402014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai
C, Lu S, Han Q and Zhao RC: MicroRNA-1246 enhances migration and
invasion through CADM1 in hepatocellular carcinoma. BMC Cancer.
14:6162014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang JL, Hu Y, Kong X, Wang ZH, Chen HY,
Xu J and Fang JY: Candidate microRNA biomarkers in human gastric
cancer: A systematic review and validation study. PLoS One.
8:e736832013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Barshack I, Meiri E, Rosenwald S, Lebanony
D, Bronfeld M, Aviel-Ronen S, Rosenblatt K, Polak-Charcon S,
Leizerman I, Ezagouri M, et al: Differential diagnosis of
hepatocellular carcinoma from metastatic tumors in the liver using
microRNA expression. Int J Biochem Cell Biol. 42:1355–1362. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Fidler IJ: The pathogenesis of cancer
metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hansen TF, Sørensen FB, Lindebjerg J and
Jakobsen A: The predictive value of microRNA-126 in relation to
first line treatment with capecitabine and oxaliplatin in patients
with metastatic colorectal cancer. BMC Cancer. 12:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Li X, Zhang Y, Zhang Y, Ding J, Wu K and
Fan D: Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Feng R, Sah BK, Beeharry MK, Yuan F, Su L,
Jin X, Yan M, Liu B, Li C and Zhu Z: Dysregulation of miR-126/Crk
protein axis predicts poor prognosis in gastric cancer patients.
Cancer Biomark. 21:335–343. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yin J, Bai Z, Song J, Yang Y, Wang J, Han
W, Zhang J, Meng H, Ma X, Yang Y, et al: Differential expression of
serum miR-126, miR-141 and miR-21 as novel biomarkers for early
detection of liver metastasis in colorectal cancer. Chin J Cancer
Res. 26:95–103. 2014.PubMed/NCBI
|
|
90
|
Li H, Zhang H, Lu G, Li Q, Gu J, Song Y,
Gao S and Ding Y: Mechanism analysis of colorectal cancer according
to the microRNA expression profile. Oncol Lett. 12:2329–2336. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Imamura T, Komatsu S, Ichikawa D, Miyamae
M, Okajima W, Ohashi T, Kiuchi J, Nishibeppu K, Konishi H, Shiozaki
A, et al: Depleted tumor suppressor miR-107 in plasma relates to
tumor progression and is a novel therapeutic target in pancreatic
cancer. Sci Rep. 7:57082017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ghosh A, Ghosh A, Datta S, Dasgupta D, Das
S, Ray S, Gupta S, Datta S, Chowdhury A, Chatterjee R, et al:
Hepatic miR-126 is a potential plasma biomarker for detection of
hepatitis B virus infected hepatocellular carcinoma. Int J Cancer.
138:2732–2744. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhou W, Yang W, Ma J, Zhang H, Li Z, Zhang
L, Liu J, Han Z, Wang H and Hong L: Role of miR-483 in digestive
tract cancers: From basic research to clinical value. J Cancer.
9:407–414. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wu XM, Shao XQ, Meng XX, Zhang XN, Zhu L,
Liu SX, Lin J and Xiao HS: Genome-wide analysis of microRNA and
mRNA expression signatures in hydroxycamptothecin-resistant gastric
cancer cells. Acta Pharmacol Sin. 32:259–269. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Liang G, Zhu Y, Jing A, Wang J, Hu F, Feng
W, Xiao Z and Chen B: Cationic microRNA-delivering nanocarriers for
efficient treatment of colon carcinoma in xenograft model. Gene
Ther. 23:829–838. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang Y, Wang Z and Gemeinhart RA:
Progress in microRNA delivery. J Control Release. 172:962–974.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Li XM, Wang AM, Zhang J and Yi H:
Down-regulation of miR-126 expression in colorectal cancer and its
clinical significance. Med Oncol. 28:1054–1057. 2011. View Article : Google Scholar : PubMed/NCBI
|