Introduction
Pancreatic cancer (PC) is one of the most lethal
malignancy types, with a 5-year survival rate of ~8% (1). The low survival rate is partly due to
more than 50% of patients with PC being diagnosed at advanced
stages (1). Therefore, understanding
the mechanisms underlying the initiation and progression of PC may
assist the development of novel therapeutic strategies.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
that participate in diverse cellular processes and negatively
regulate gene expression at the post-transcriptional level by
binding with 3′-untranslated regions (3′-UTRs) (2–4). A number
of studies have demonstrated that altered expression of miRs serves
critical roles in human cancers by directly regulating cell
behaviors (5–7). miR-205 expression in humans was
validated by Landgraf et al (8), however, its role in tumor progression is
contradictory (9–12). Zhang et al (9) revealed that expression of miR-205 was
significantly decreased in radioresistant subpopulations of breast
cancer cells and loss of miR-205 expression was associated with
poor distant relapse-free survival in patients with breast cancer.
Furthermore, the authors identified that miR-205 mimics could
sensitize the tumor to radiation in a xenograft model. By contrast,
miR-205 expression has been identified to be significantly
increased in several human cancer types, including ovarian cancer,
endometrial cancer and laryngeal squamous cell carcinoma, in which
it was identified to function as an oncoprotein (10,11,13).
Runt-related transcription factor 2 (RUNX2), a
member of the RUNX family, functions as a critical regulator for
osteoblast differentiation (14). In
addition, Kayed et al (15)
demonstrated that RUNX2 was overexpressed in PC and could be
regulated by certain cytokines, including transforming growth
factor β1 and bone morphogenetic protein 2. However, to the best of
our knowledge, the miRs that regulate RUNX2 expression in tumors
are unknown. The current study demonstrated that miR-205 was a
tumor suppressor in PC and a regulator of RUNX2 expression. In
addition, the results revealed that miR-205-induced downregulation
of RUNX2 was associated with the inhibition of PC cell
proliferation and migration.
Materials and methods
Tissue samples
A total of 48 paired fresh frozen PC tumor tissues
and matched normal pancreatic tissues were obtained from patients
who underwent treatment at the Changhai Hospital, Second Military
Medical University (Shanghai, China) between January 2010 and
December 2011. Written informed consent was obtained from all
enrolled patients (25 female and 23 male; age, 36–74 years). No
patients had ever received preoperative chemotherapy or
embolization. The experimental protocols were approved by the
Ethics Committee of Changhai Hospital, Second Military Medical
University and conducted according to The Declaration of
Helsinki.
Cell culture
Human PC cell lines (CFPAC-1 and PANC-1) were
obtained from American Type Culture Collection (Manassas, VA, USA)
and grown in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS) (both from Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The immortalized human
pancreatic ductal epithelial cell line (HPC-Y5) was obtained from
the Cell Bank of Type Culture Collection, Chinese Academy of
Sciences (Shanghai, China) and cultured in DMEM supplemented with
10% FBS. Cultured cells were maintained in a humidified 5%
CO2 atmosphere at a temperature of 37°C.
Transfection procedure
miR-205 mimic (5′-UCCUUCAUUCCACCGGAGUCUG-3′) and
control (miR-con, 5′-GGUCCGUCCGUAAUUAUCCUCC-3′) oligonucleotides
were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
RUNX2 small interfering RNA (siRNA, 5′-AAGGACAGAGTCAGATTACAG-3′)
and control (si-con, 5′-ATAAGGTATCGAGACCAGAGA-3′) oligonucleotides
were also purchased from Guangzhou RiboBio Co., Ltd. The RUNX2 open
reading frame cloned into pcDNA3.1 vector and the empty vector
pcDNA3.1 were purchased from GenScript (Nanjing, China).
Transfection was conducted with Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol (100 nM miRNAs and siRNAs, 5 µg RUNX2 construct and empty
vector). Following transfection for 48 h, the cells were used for
subsequent experiments.
Dual-luciferase 3′-UTR reporter
assay
Using the online prediction algorithm (TargetScan
version 7.2; http://www.targetscan.org/vert_72/), it was identified
that the 3′-UTR of RUNX2 contains a putative binding sequence of
miR-205. The wild-type (wt) RUNX2 3′-UTR or mutant (mut) RUNX2
3′-UTR sequences were cloned into a pmirGLO control vector (Promega
Corporation, Madison, WI, USA). Cells were co-transfected with
either wt RUNX2 3′-UTR or mut RUNX2 3′-UTR and miR-205 mimic or
miR-con using Lipofectamine 2000 according to the manufacturer's
protocol. At 48 h following transfection, cells were harvested and
luciferase activity relative to the Renilla luciferase
activity was measured using a Dual Luciferase Reporter system
(Promega Corporation) according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from PC tumor tissue,
matched normal pancreatic tissue and the cell lines CFPAC-1, PANC-1
and HPC-Y5 using TRIzol reagent (Beyotime Institute of
Biotechnology, Haimen, China) according to the manufacturer's
protocol. For measurement of miRNA expression levels, 100 ng total
RNA was reverse transcribed into cDNA using PrimeScript miRNA cDNA
Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR
was subsequently performed using SYBR Premix Ex TaqII (Takara
Biotechnology Co., Ltd.) with an Applied Biosystems 7500 Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The expression of miRNA was normalized to the expression of the
control U6 snRNA. The primers used were as follows: miR-205
forward, 5′-GCTCCTTCATTCCACCGG-3′ and reverse,
5′-CAGTGCAGGGTCCGAGGT-3′; and U6 snRNA forward,
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′. The following thermocycling protocol
was used: Denaturation at 95°C for 10 min, followed by 45 cycles of
denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec, and
extension at 72°C for 1 min. Relative expression levels were
determined using the 2−ΔΔCq method (16).
Western blot assay
Total protein was extracted from frozen tissues and
cell lines using RIPA lysis buffer (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. The
protein concentration was measured using a bicinchoninic acid
protein concentration determination kit (Beyotime Institute of
Biotechnology). Protein extracts (50 µg) loaded to each lane were
then separated by SDS-PAGE (10% gels) and transferred onto
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with fat-free milk at room
temperature for 4 h. Then, the membranes were incubated with
primary anti-RUNX2 (1:5,000; cat. no. ab23981) or anti-GAPDH
(1:5,000; cat. no. ab181602) (both from Abcam, Cambridge, UK) at
4°C overnight. Subsequently, the membranes were incubated with
horseradish peroxidase conjugated goat anti-rabbit secondary
antibody (1:1,000; cat. no. ab205718; Abcam) at room temperature
for 1 h. Bands were visualized using an enhanced chemiluminescence
kit (Beyotime Institute of Biotechnology). GAPDH was used as a
loading control.
Cell proliferation
The rate of cell proliferation was measured using
Cell Counting Kit-8 (Beyotime Institute of Biotechnology) according
to the manufacturer's protocol. Cells were seeded onto 96-well
plates at a density of 4×103 cells per well. CCK-8
reagent (10 µl) was added to each well at indicated time points
(days 0, 1, 2 and 3). The absorbance was measured and recorded at
450 nm using a microplate reader (Thermo Fisher Scientific,
Inc.).
Cellular migration
The rate of cellular migration was measured using a
wound-healing assay. A scratch was created using a micropipette tip
on a monolayer surface of cultured cells. The initial gap length (0
h) and residual gap length at 24 h following wounding were
calculated from photomicrographs.
Statistical analysis
Data are presented as the mean ± standard deviation.
Student's t-test was used to analyze differences between two
groups. One-way analysis of variance and Tukey's test were used to
analyze differences among three or more groups. Kaplan-Meier curves
were used to establish overall survival and the survival
differences were analyzed using a log-rank test. The correlation
between miR-205 and RUNX2 expression in PC tissues was calculated
using Spearman's correlation coefficient. Data analysis was
performed using SPSS statistical software (version 17.0; SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-205 expression is downregulated in
PC tissues and cell lines
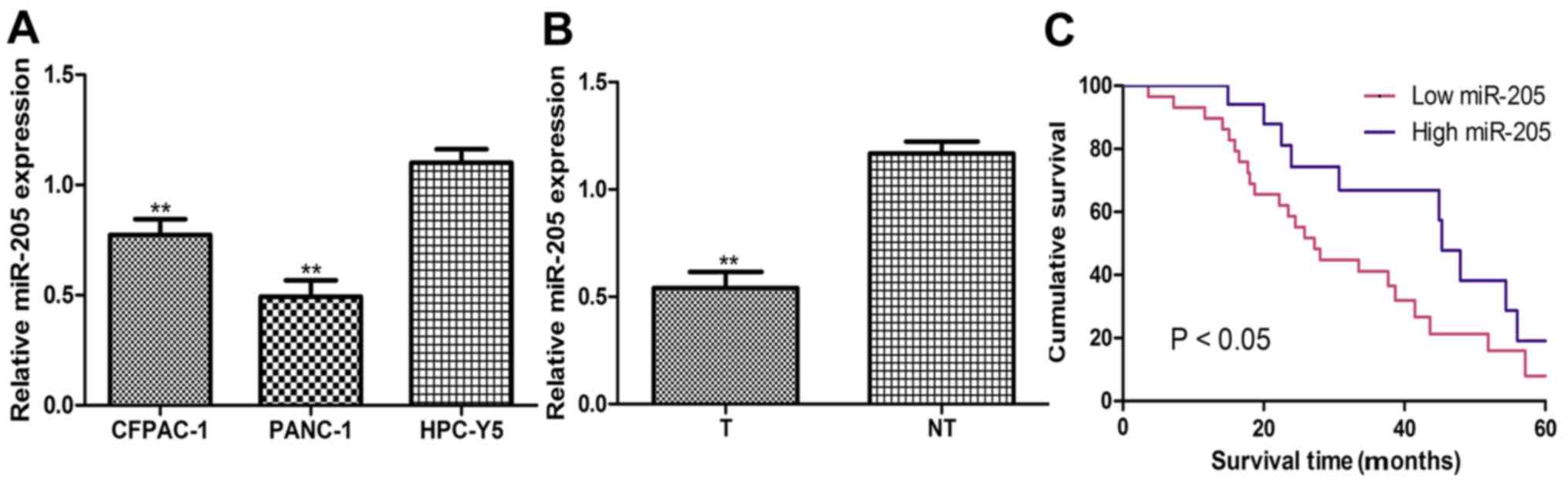
The current study identified that miR-205 expression
levels were significantly downregulated in PC cell lines (CFPAC-1
and PANC-1) compared with the HPC-Y5 cell line (Fig. 1A). It was also revealed that miR-205
expression levels were downregulated in PC tumor tissues compared
with their matched noncancerous tissues (Fig. 1B). miR-205 expression levels in the
tumor tissues were used to stratify the patients into low or high
miR-205 expression groups (cut-off value, 0.55). Results from
Kaplan-Meier survival analysis demonstrated that patients with low
levels of miR-205 expression had a significantly shorter overall
survival compared with those with high miR-205 expression levels
(Fig. 1C).
miR-205 inhibits the expression of
RUNX2 in PC
Next, it was identified that RUNX2 protein
expression levels were significantly increased in PC tissues and
cell lines compared with matched noncancerous tissues and the
HPC-Y5 cell line, respectively (Fig. 2A
and B). Furthermore, transfection with miR-205 mimic
significantly increased miR-205 expression but decreased RUNX2
protein expression levels in PC cell lines compared with miR-con
transfection (Fig. 2C and D). The
online prediction algorithm TargetScan was used to analyze whether
miR-205 could bind to the 3′-UTR of RUNX2. It was identified that
the 3′-UTR of RUNX2 contains a putative binding sequence of miR-205
(Fig. 2E). Luciferase reporter assay
revealed that transfection with miR-205 mimic significantly
decreased the luciferase activity of the wt RUNX2 3′-UTR but had no
effect on the luciferase activity of mut RUNX2 3′-UTR (Fig. 2F). Notably, an inverse correlation
between miR-205 and RUNX2 expression levels was identified in PC
tumor tissues (Fig. 2G).
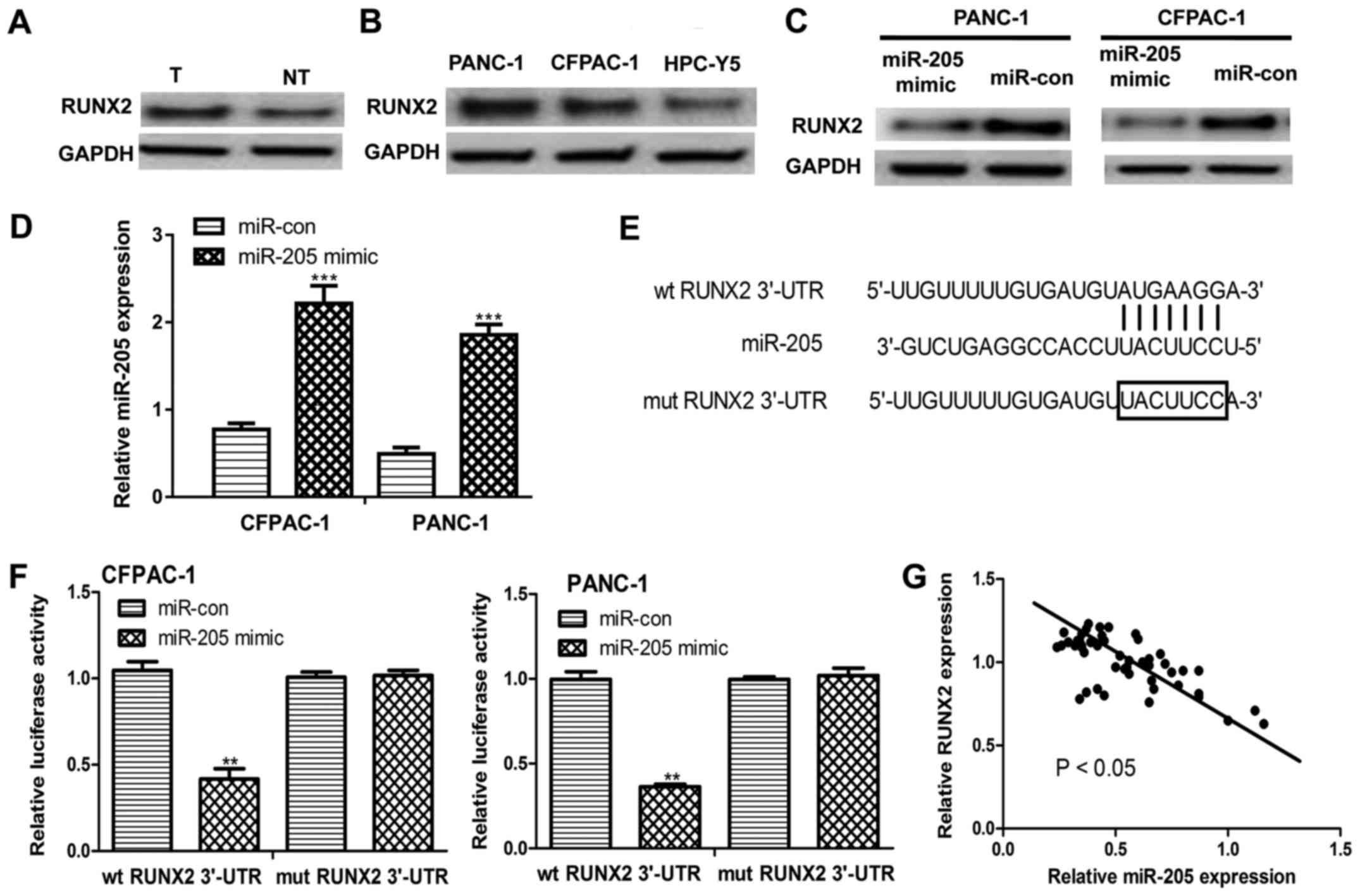 | Figure 2.miR-205 inhibits RUNX2 expression in
pancreatic cancer. (A) RUNX2 expression was evaluated by western
blot analysis in pancreatic cancer tissues and matched non-tumor
tissues. (B) RUNX2 expression was evaluated by western blot
analysis in human pancreatic cancer cell lines (CFPAC-1 and PANC-1)
and the immortalized human pancreatic ductal epithelial cell line,
HPC-Y5. (C) RUNX2 expression levels were evaluated by western blot
analysis in CFPAC-1 and PANC-1 cell lines following transfection
with miR-205 mimic or miR-con. (D) miR-205 expression levels were
analyzed by RT-qPCR in CFPAC-1 and PANC-1 cell lines following
transfection with miR-205 mimic or miR-con. (E) The predicted
miR-205 binding sequence within the 3′-UTR of RUNX2. (F) Relative
luciferase activity in human pancreatic cancer cell lines (CFPAC-1
and PANC-1) co-transfected with either wt RUNX2 3′-UTR or mut RUNX2
3′-UTR and miR-205 mimic or miR-con. (G) Inverse correlation
between miR-205 and RUNX2 expression levels in patients with
pancreatic cancer (r=−0.49). **P<0.01, ***P<0.001 vs.
miR-con. miR-205, microRNA-205; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; T, tumor; NT,
non-tumor; wt, wild-type; mut, mutant; miR-con, microRNA control;
UTR, untranslated region; RUNX2, runt-related transcription factor
2. |
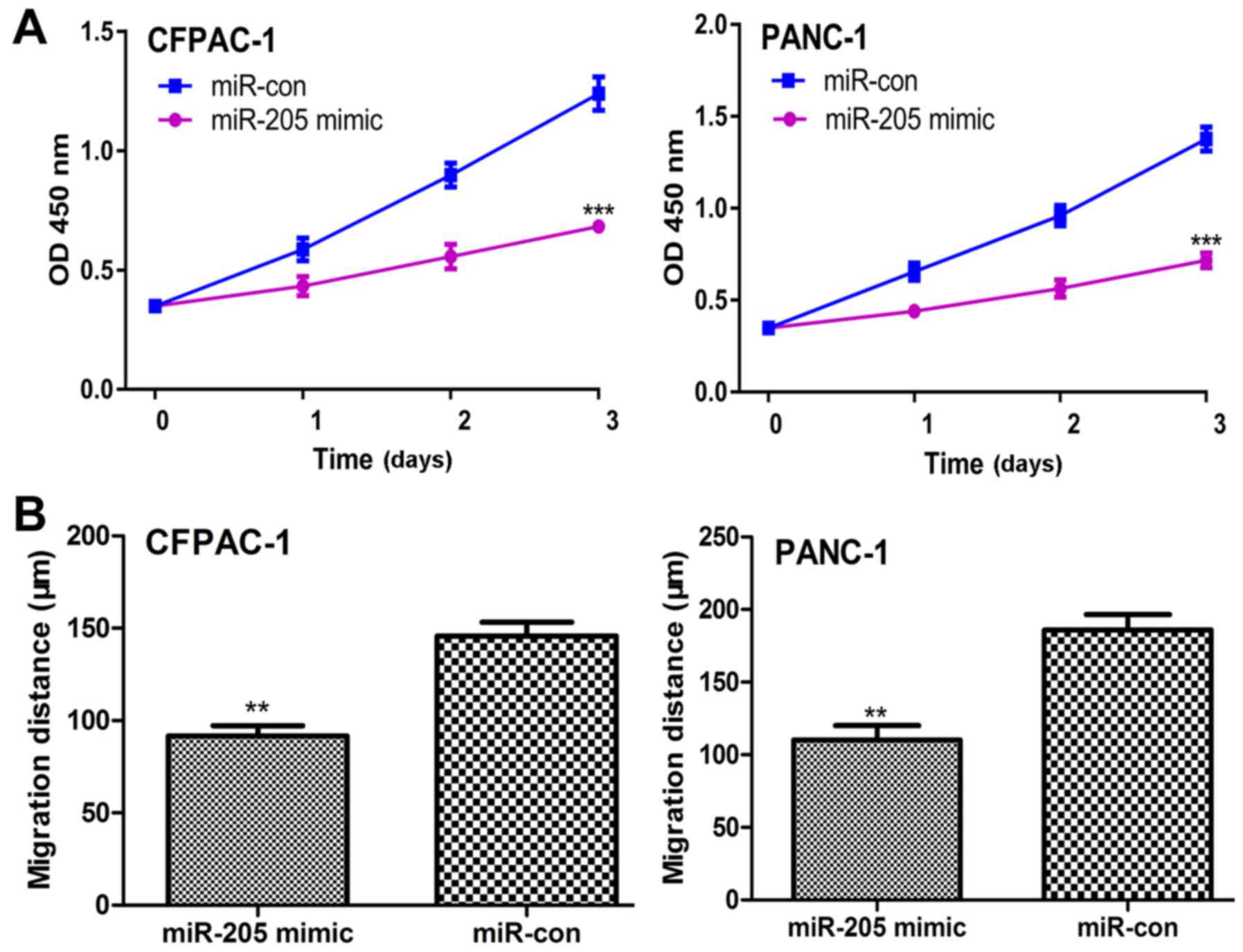
Elevated expression of miR-205
inhibits PC cell proliferation and migration in vitro
The effects of miR-205 expression on PC cell
proliferation and migration were assessed by CCK-8 and wound
healing assays, respectively. It was identified that the rate of
cellular proliferation in PC cells transfected with miR-205 mimic
was significantly lower compared with those transfected with
miR-con (Fig. 3A). A significant
decrease was also identified in the rate of cell migration in
miR-205 mimic transfected PC cells compared with those transfected
with miR-con (Fig. 3B).
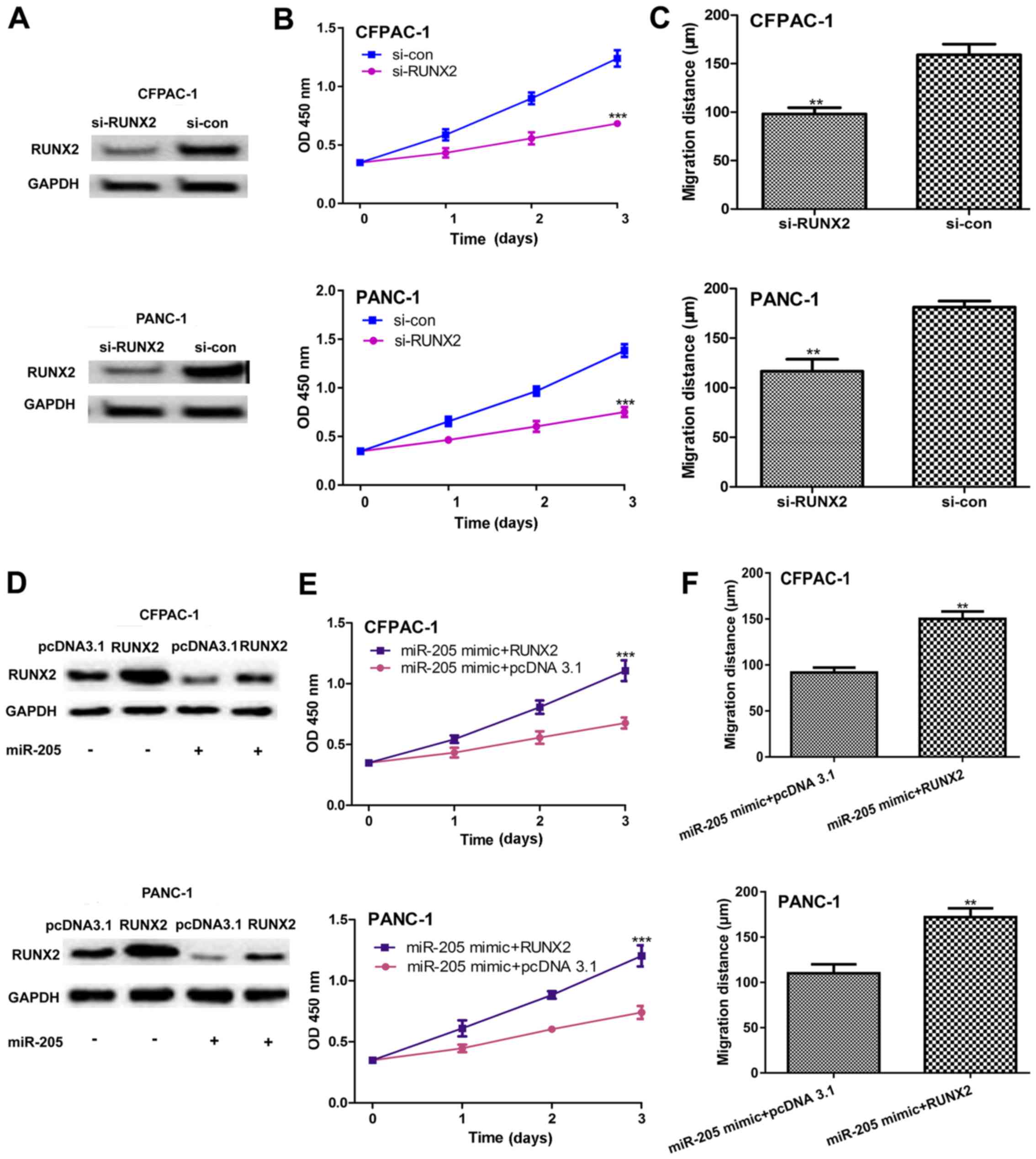
miR-205 inhibits PC cell proliferation
and migration by targeting RUNX2 in vitro
It was then investigated whether miR-205 targets
RUNX2 to regulate PC cell proliferation and migration. It was first
demonstrated that RUNX2 protein expression levels were decreased by
si-RUNX2 transfection in PC cell lines (Fig. 4A). In addition, it was identified that
the downregulation of RUNX2 significantly decreased PC cell
proliferation (Fig. 4B) and migration
(Fig. 4C) in vitro.
Furthermore, it was demonstrated that RUNX2 protein expression
levels were higher in PC cell lines co-transfected with miR-205
mimic and RUNX2 expression plasmid compared with those
co-transfected with miR-205 mimic and pcDNA3.1 (Fig. 4D). In vitro functional assays
revealed that RUNX2 restoration significantly decreased the
inhibition effect of miR-205 on cell proliferation (Fig. 4E) and migration (Fig. 4F).
Discussion
PC presents a serious public health challenge due to
its poor overall survival rate and increasing incidence rate in
China (17,18). The association between miRNA
expression and cancer development was first established in 2002 as
miR-15 and miR-16-1 were identified to be aberrantly expressed in
69% of patients with chronic lymphocytic leukemia (19). Therefore, targeting miRNAs was
recognized as a novel approach for the treatment of tumors
(20,21). Notably, a miRNA mimic termed MRX34 has
entered into a phase I clinical trial for cancer therapy (22).
miR-205 expression levels vary in humans to function
as either tumor suppressors or promoters (23). However, to the best of our knowledge,
the precise functions of miR-205 in PC have not been fully
understood. In the current study, miR-205 expression levels were
revealed to be significantly lower in PC tumor tissues compared
with noncancerous tissues and low miR-205 expression levels were
associated with poor 5-year overall survival. Therefore, it is of
interest to investigate the biological role of miR-205 in PC
development. In vitro functional assays demonstrated that
elevated miR-205 levels inhibited PC cell proliferation and
migration. A recent study demonstrated that miR-205 overexpression
inhibited PC stem cell proliferation (24). These results suggest that miR-205
functions as a tumor suppressor in PC.
It has been recognized that miRNAs exert their
biological function through regulating the expression of a number
of target genes (3). For example,
zinc finger E-box binding homeobox 1 was identified as a target of
miR-205 in epithelial ovarian cancer and breast cancer (9,10). Using
online bioinformatics analysis, the current study identified that
RUNX2 may be a target of miR-205. This prediction was further
supported by luciferase reporter and western blot analyses.
Notably, it was revealed that miR-205 and RUNX2 expression were
inversely correlated in PC tumor tissues. In addition, it was
identified that RUNX2 overexpression reversed the inhibitory
effects of miR-205 mimic on PC cell proliferation and migration
in vitro.
In summary, the current results demonstrated that
miR-205 may serve a critical role in the development and
progression of PC. miR-205 was validated as a tumor suppressor in
PC through the regulation of RUNX2 expression. The current study
provided critical insights into the use of miR-205 as a potential
therapeutic miRNA for PC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ participated in the experimental design,
coordinated the experimental work, interpreted the results, wrote
the manuscript and contributed to the critical revision. LZ, JG and
YY performed experiments and analyzed the results. ZL designed the
research plan, interpreted the results and wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Changhai Hospital, Second Military Medical University (Shanghai,
China). Written informed consent was obtained from all the
participating patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gregory RI, Chendrimada TP, Cooch N and
Shiekhattar R: Human RISC couples microRNA biogenesis and
posttranscriptional gene silencing. Cell. 123:631–640. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao X, Zhao H, Diao C, Wang X, Xie Y, Liu
Y, Han J and Zhang M: miR-455-3p serves as prognostic factor and
regulates the proliferation and migration of non-small cell lung
cancer through targeting HOXB5. Biochem Biophys Res Commun.
495:1074–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao Y, Song J, Ge J, Song Z, Chen J and Wu
C: MicroRNA-100 suppresses human gastric cancer cell proliferation
by targeting CXCR7. Oncol Lett. 15:453–458. 2018.PubMed/NCBI
|
|
7
|
Kang M, Shi J, Peng N and He S:
MicroRNA-211 promotes non-small-cell lung cancer proliferation and
invasion by targeting MxA. Onco Targets Ther. 10:5667–5675. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang P, Wang L, Rodriguez-Aguayo C, Yuan
Y, Debeb BG, Chen D, Sun Y, You MJ, Liu Y, Dean DC, et al: miR-205
acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat
Commun. 5:56712014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Niu K, Shen W, Zhang Y, Zhao Y and Lu Y:
MiR-205 promotes motility of ovarian cancer cells via targeting
ZEB1. Gene. 574:330–336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhong G and Xiong X: miR-205 promotes
proliferation and invasion of laryngeal squamous cell carcinoma by
suppressing CDK2AP1 expression. Biol Res. 48:602015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Orang AV, Safaralizadeh R, Feizi
Hosseinpour MA and Somi MH: Diagnostic and prognostic value of
miR-205 in colorectal cancer. Asian Pac J Cancer Prev.
15:4033–4037. 2015. View Article : Google Scholar
|
|
13
|
Jin C and Liang R: miR-205 promotes
epithelial-mesenchymal transition by targeting AKT signaling in
endometrial cancer cells. J Obstet Gynaecol Res. 41:1653–1660.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Hao L, Wu J, Zhang J and Su J:
Linarin promotes osteogenic differentiation by activating the
BMP-2/RUNX2 pathway via protein kinase A signaling. Int J Mol Med.
37:901–910. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kayed H, Jiang X, Keleg S, Jesnowski R,
Giese T, Berger MR, Esposito I, Löhr M, Friess H and Kleeff J:
Regulation and functional role of the Runt-related transcription
factor-2 in pancreatic cancer. Br J Cancer. 97:1106–1115. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin QJ, Yang F, Jin C and Fu DL: Current
status and progress of pancreatic cancer in China. World J
Gastroenterol. 21:7988–8003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H
and Zou X: Report of cancer incidence and mortality in China, 2010.
Ann Transl Med. 2:612014.PubMed/NCBI
|
|
19
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chitkara D, Mittal A and Mahato RI: miRNAs
in pancreatic cancer: Therapeutic potential, delivery challenges
and strategies. Adv Drug Deliv Rev. 81:34–52. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi M, Xie D, Gaod Y and Xie K: Targeting
miRNAs for pancreatic cancer therapy. Curr Pharm Des. 20:5279–5286.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bouchie A: First microRNA mimic enters
clinic. Nat Biotechnol. 31:5772013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Orang AV, Safaralizadeh R and Feizi
Hosseinpour MA: Insights into the diverse roles of miR-205 in human
cancers. Asian Pac J Cancer Prev. 15:577–583. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chaudhary AK, Mondal G, Kumar V, Kattel K
and Mahato RI: Chemosensitization and inhibition of pancreatic
cancer stem cell proliferation by overexpression of microRNA-205.
Cancer Lett. 402:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|