Introduction
MicroRNAs (miR/miRNAs) are small endogenous
non-coding RNAs that post-transcriptionally regulate the expression
of target genes by binding to the 3′-untranslated regions of mRNAs,
causing destabilization, degradation, or translation inhibition
(1). Because dysregulation of miRNA
expression has been identified in a number of cancers, some miRNAs
are categorized as oncogenic miRNAs or ‘oncomiRs’, a term used to
describe either tumor suppressors or oncogenes (2–5).
Consequently, miRNAs have been investigated as potential
therapeutic targets for several malignant cancers including
melanoma (6–7). The tumor burden in mice with liver
melanoma metastasis was found to be reduced by anti-miR-182
oligonucleotides that inhibited the upregulated miR-182 in the
tumor cells (6). Inhibition of
miR-383 over-expression suppressed the proliferation, cell cycle
progression and invasion of human epithelial ovarian cancer (EOC)
and immortal EOC cell lines (8).
Over-expression of miR-203 sensitized malignant melanoma cells to
temozolomide drug by targeting glutaminase, which opened new
opportunities for chemotherapy-resistant malignant melanoma
patients (9). Thus, profiling
dysregulated miRNA expression in cancers is an important approach
for detecting potential therapeutic targets.
Simpson et al 2013 (10) suggested significant overlapping may
exist in the clinical and histopathological features of canine and
human mucosal melanomas. miRNA expression has been investigated in
different canine tumors, including B and T-cell lymphoma (11), lymphocytic leukemia (12), transitional cell carcinoma (13), mammary cancer (14), prostate cancer (15) and melanoma (16–18). These
studies indicated that the expression patterns of specific miRNAs
in specific cancers were similar to those in corresponding human
cancers. For example, the upregulation of miR-21 and miR-29b in
canine mammary cancer is consistent with their upregulation in
human breast cancer (14,19,20) and
melanoma (21,22) and miR-145, miR-203, and miR-205 were
found to be downregulated in both canine malignant melanoma (CMM)
and human malignant melanoma (HMM) (16,17). In
the Noguchi et al (17)
studies of HMM, a total of seven downregulated miRNAs were detected
by microarray analysis; three of them were confirmed by
quantitative reverse transcription PCR (qRT-PCR). In almost all HMM
tumors that have been studied, upregulated miRNA expression has
been reported, including the miR-17-92 cluster, miR-222/221, miR-21
and miR-155 (23). Therefore, it is
likely that some miRNAs will be upregulated in oral CMM, similar to
what Starkey et al (18)
reported in canine uveal melanoma. However, until now, no
upregulated miRNAs in oral CMM have been reported. To investigate
this hypothesis, we examined the expression of miRNAs in CMM
tissues obtained from the oral cavity using microarray and qRT-PCR
analyses. Here we report the upregulation of seven miRNAs in CMM
tissues. To understand the biological relevance of miRNAs it is
necessary to identify the target genes with which they interact.
Protein-protein interactions are essential for cells to maintain
systemic biological functions such as replication of DNA,
transcription, translation and signal transduction (24). Dysregulation of proteins may collapse
the homeostasis process leading to complex diseases and miRNAs may
act as master regulators by maintaining the stability of
protein-protein interaction networks (25). So, determining the interactions
between the proteins encoded by targets of dysregulated miRNAs and
other proteins is very important. In this study, we drew a
miRNA-target regulatory interaction network with tumor suppressor
genes, which revealed miR-383 and miR-204 may play roles in the
development of melanoma by avoiding DNA repair and apoptosis.
Materials and methods
Sample collection
The CMM tissues used in this study were obtained
from dogs (n=10) that had undergone biopsy or surgical resection
for diagnosis or treatment at the Veterinary Teaching Hospital,
Kagoshima University (Kagoshima, Japan). All melanoma samples were
obtained from the oral cavity and were histopathologically
diagnosed by two pathologists. Normal oral tissues were obtained
from healthy laboratory beagle dogs (n=12). In addition to the CMM
and normal oral tissues, we obtained a total of 21 canine tumors
and normal tissues to use as microarray reference samples as
follows: Mammary tubulopapillary carcinoma (n=4), mammary benign
mixed tumor (n=4), hepatic cell carcinoma (n=1), squamous cell
carcinoma (n=1), lymphoma (n=1), adenosquamous carcinoma (n=1),
mast cell tumor (n=1), malignant peripheral nerve sheath tumor
(n=1), normal mammary gland tissue (n=4) and normal hepatic tissue
(n=3). The animal experiments were approved by the Kagoshima
University's Laboratory Animal Committee (A10031).
Isolation of total RNA
All the tissues were preserved in RNAlater
(Thermo Fisher Scientific Inc., Waltham, MA, USA) immediately after
biopsy or surgical resection until used for RNA isolation. Total
RNA was isolated from the stored tissues using a
mirVana™ miRNA Isolation kit (Thermo Fisher Scientific
Inc.) according to the manufacturer's instructions. RNA quantity
was measured using either an ND-1000 spectrophotometer (Thermo
Fisher Scientific Inc.) or a NanoPhotometer™ Pearl
(Implen GmbH, München, Germany). RNA quality was verified using an
Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA,
USA) and RNA integrity numbers were determined (26).
Microarray analysis
Three assays were performed (n=3) using the
miRCURY™ LNA microRNA Array, version 11.0 (Exiqon Inc.,
Woburn, MA, USA). In each assay, Hy3 labeled miRNAs from different
CMM tissues but the same references Hy5 labeled miRNAs were used.
The reference miRNAs comprised equal amounts of RNA from 21
reference samples from 10 different tissues (listed in the
Sample collection section), all of which were pooled.
Two-color miRNA-microarrays with 264 identical canine miRNA probes
were used. Signal extraction was performed using Feature Extraction
10.7.3.1 software (Agilent Technologies). To minimize error, each
miRNA was spotted at four different locations on the array and the
average signal intensity value of the four spots was used and
variable coefficients were calculated [standard deviation (SD) of
signal intensity of four spots/average values]. miRNAs with signal
intensity variable coefficients >0.5 or with low signal
intensity (<100) in both the CMM and reference tissues were
excluded from further analysis. The average values of the Hy3/Hy5
(fold change; FC) ratio between the CMM and reference tissues were
compared using the Lowess normalization method (27). miRNAs that had FC ratios >2.0 or
<0.5 were considered to be dysregulated.
qRT-PCR assays
CMM tissues (n=10) and normal oral tissues (n=12)
were used in the qRT-PCRs, which were performed in duplicate using
TaqMan microRNA Assays (Thermo Fisher Scientific Inc.; see Table I for assay details) with 2 ng/µl total
RNA, according to the optimal reagent concentrations and reaction
conditions described in the manufacturer's instructions. The canine
miRNA sequences used for the PCRs were identical to the
corresponding human miRNA sequences (Table I). The qRT-PCRs were carried out using
an Applied Biosystems 7300 Real-Time PCR System (Thermo Fisher
Scientific Inc.). RNU6B, U6 small nuclear RNA, was used as a
quantitative normalization control (13,14).
Relative expression levels were calculated using the comparative
delta Cq method (2−ΔΔCq) (28). Cq values >36.0 were considered as
absence of miRNA expression. The relative expression levels of
miRNAs in the CMM tissues were calculated relative to the average
values in the normal oral tissues, which were assigned a value of
1.0.
 | Table I.miRs used in the reverse
transcription-quantitative polymerase chain reaction assays. |
Table I.
miRs used in the reverse
transcription-quantitative polymerase chain reaction assays.
| A, miRNA
sequences |
|---|
|
|---|
| Assay name | Assay ID | Mature miRNA
sequence | miRBase accession
number |
|---|
| hsa-miR-16 | 000391 |
UAGCAGCACGUAAAUAUUGGCG | MI0000070 |
| hsa-miR-21 | 000397 |
UAGCUUAUCAGACUGAUGUUGA | MI0000077 |
| hsa-miR-29b | 000413 |
UAGCACCAUUUGAAAUCAGUGUU | MI0000105 |
| hsa-miR-92a | 000431 |
UAUUGCACUUGUCCCGGCCUGU | MI0000093 |
| hsa-miR-122 | 002245 |
UGGAGUGUGACAAUGGUGUUUG | MI0000442 |
| hsa-miR-125b | 000449 |
UCCCUGAGACCCUAACUUGUGA | MI0000446 |
| hsa-miR-143 | 002249 |
UGAGAUGAAGCACUGUAGCUC | MI0000459 |
| hsa-miR-204 | 000508 |
UUCCCUUUGUCAUCCUAUGCCU | MI0000284 |
| hsa-miR-205 | 000509 |
UCCUUCAUUCCACCGGAGUCUG | MI0000285 |
| hsa-miR-222 | 002276 |
AGCUACAUCUGGCUACUGGGU | MI0000299 |
| hsa-miR-383 | 000573 |
AGAUCAGAAGGUGAUUGUGGCU | MI0000791 |
|
| B, Control
sequences |
|
| Assay
name | Assay
ID | Control
sequence | NCBI accession
number |
|
| RNU6B | 001093 |
CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT | NR_002752 |
Statistics
In the microarray experiments, P-values and
false discovery rates (FDRs) were analyzed using Welch's test and
the Benjamini-Hochberg correction for multiple hypotheses testing
using R software (29). For the
qRT-PCRs, the miRNA expression levels between CMM and normal oral
tissues were analyzed using the Mann Whitney U-test. Statistical
analyses were performed with JMP 10.0 (SAS Institute, Cary, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Network construction
miRNA targets were predicted using TargetScan 7.1
(30) and 1021 human tumor suppressor
genes (with basic annotations) from the Tumor Suppressor Gene
Database (TSGene; http://bioinfo.uth.edu/TSGene/). A miRNA-target
interaction network was drawn using Cytoscape v3.5 (http://www.cytoscape.org/) (31) and a protein-protein interaction
network of tumor suppressor genes was constructed using STRING
(confidence score, 0.9) (http://string-db.org/) (32). The two networks were merged within
Cytoscape and interconnected nodes were separated to obtain a
co-ordinate network. Analysis of basic network parameters (degree,
betweenness, centroid value and Eigenvector) was done using
Centiscape 2.2 (33). In the network,
a node represents a protein (encoded by a target mRNA) or a miRNA
and a line represents an interaction between a protein and a
miRNA.
Results
Screening of differentially expressed
miRNAs by microarray analysis
The microarray analysis revealed 17 dysregulated
miRNAs in the CMM tissues based on the FC ratios (Table II). Of the 17 miRNAs, 5 were
upregulated (FC ratios >2.0) with no significant FDRs and 12
were downregulated (FC ratios <0.5) and 4 of them had
significant FDRs (P<0.05) (Table
II).
 | Table II.Dysregulated miRNAs identified in
canine malignant melanoma tissues by microarray analysis. |
Table II.
Dysregulated miRNAs identified in
canine malignant melanoma tissues by microarray analysis.
| A, Upregulated
miRNAs |
|---|
|
|---|
| Upregulated
(FC>2.0) |
|---|
|
|---|
| miRNA | FC | FDR |
|---|
| miR-9 | 2.420 | >0.05 |
| miR-149 | 2.022 | >0.05 |
| miR-204 | 2.781 | >0.05 |
| miR-326 | 2.056 | >0.05 |
| miR-383 | 3.581 | >0.05 |
|
| B, Downregulated
miRNAs |
|
|
| Downregulated
(FC<0.5) |
|
| miRNA | FC | FDR |
|
| miR-10 | 0.486 | <0.05 |
| miR-101 | 0.446 | >0.05 |
| miR-122 | 0.060 | <0.05 |
| miR-142 | 0.385 | >0.05 |
| miR-143 | 0.244 | <0.05 |
| miR-195 | 0.391 | >0.05 |
| miR-200c | 0.382 | >0.05 |
| miR-205 | 0.100 | <0.05 |
| miR-328 | 0.299 | >0.05 |
| miR-487b | 0.430 | >0.05 |
| miR-652 | 0.457 | >0.05 |
Confirmation of differentially
expressed miRNAs by qRT-PCR
qRT-PCRs were performed to validate some of the
dysregulated miRNAs from the microarray analysis (Table II). Because none of the upregulated
miRNAs had significant FDRs, we selected the two most highly
upregulated miRNAs, miR-204 and miR-383, for validation. From among
the downregulated miRNAs, we selected three miRNAs (miR-122,
miR-143 and miR-205) that had the most significant FDRs. We also
selected six other miRNAs (miR-16, miR-21, miR-29b, miR-92a,
miR-125b and miR-222) for validation because they were reported to
be dysregulated in cancers other than CMM (13,14,34–36).
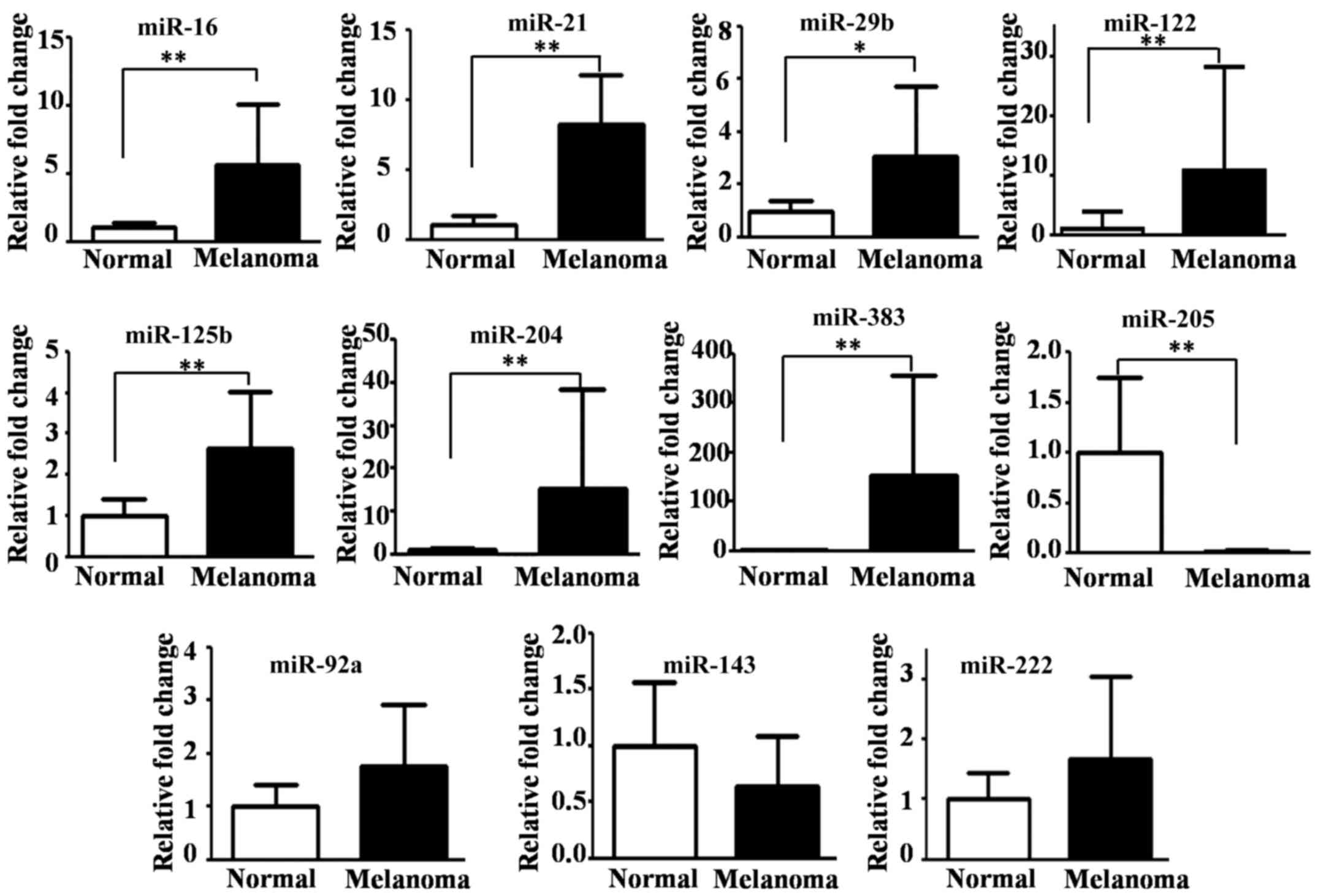
We found that seven miRNAs were significantly
upregulated [P-values from 0.0001 (miR-21) to 0.025 (miR-29b)], but
miR-205 was the only significantly downregulated miRNA
(P<0.0001) in the CMM tissues compared with normal oral tissues
(Fig. 1). No significant differences
were detected in the expression of miR-92a, miR-143 and miR-222
between the CMM and normal oral tissues (Fig. 1).
Of the 17 dysregulated miRNAs identified by
microarray analysis (Table II), only
miR-204, miR-383 and miR-205 were found to be highly differentially
expressed by qRT-PCR. The average FCs for miR-204 and miR-383 were
15.3 and 152.7, respectively, but for miR-205 the average FC was
0.01 (Fig. 1).
The relative expression patterns of miR-204, miR-383
and miR-205 were consistent between the qRT-PCR and microarray
results, but there were discrepancies for some of the other miRNAs.
For example, miR-122 was downregulated (FC<0.5) in the
microarray analysis but significantly upregulated in the qRT-PCR
analysis and miR-143 was downregulated (FC of 0.244) in the
microarray analysis but was not found to be significantly
differentially expressed by qRT-PCR (Fig.
1).
miRNA-target regulatory interaction
network
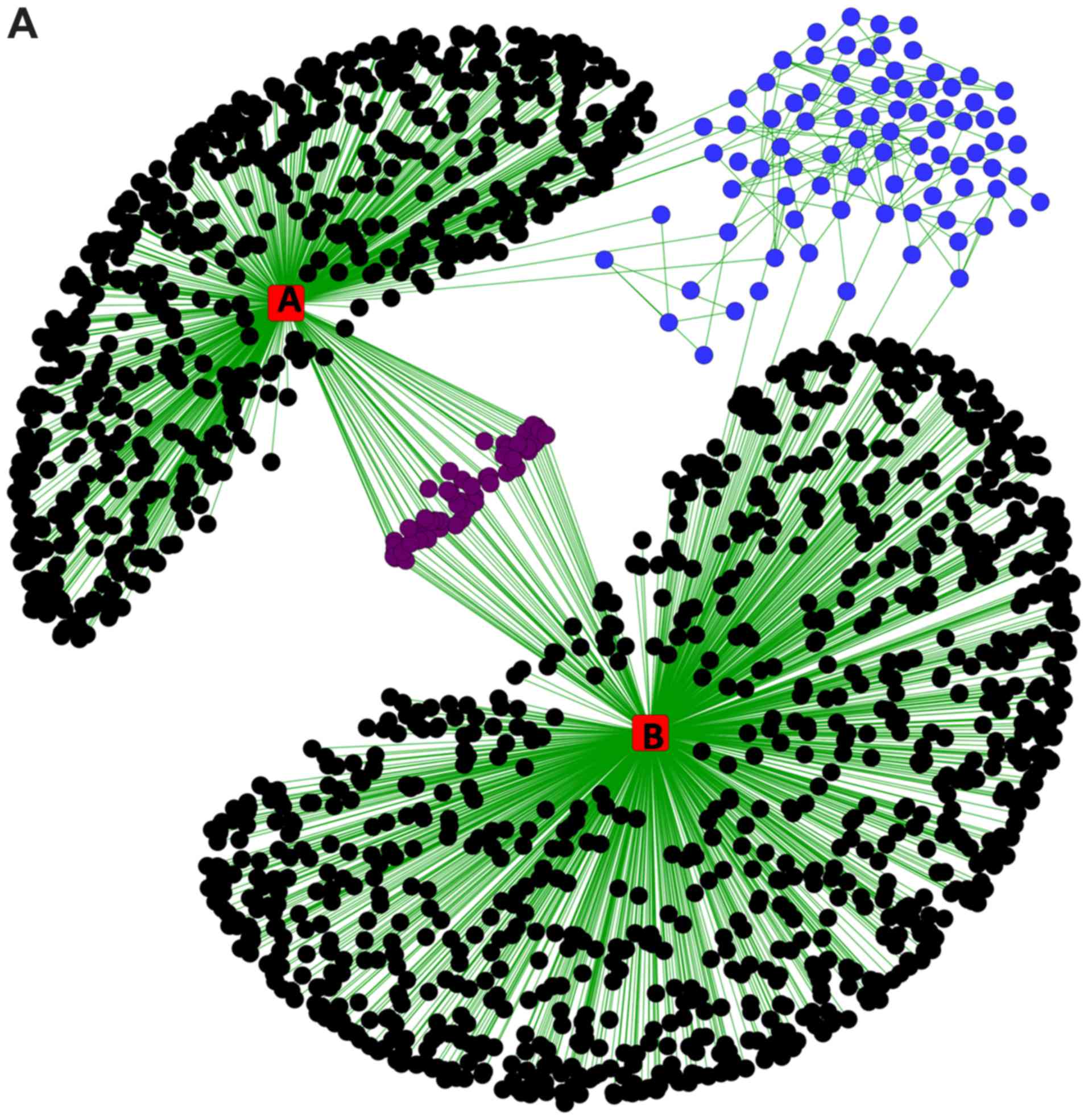
As indicated in Fig.
2A, the STRING protein interaction network revealed that
miR-383 and miR-204 interacted with several common genes
(proteins), as was reported previously (37,38). When
we separated the connected network and calculated the basic
parameters (degree, betweenness, centroid value and eigenvector) by
Centiscape 2.2 through Cytoscape (Fig.
2B), we found all the basic parameters of TP53 (Fig. 3A) had higher value than any of the
others. Further, the basic parameters of miR-383, miR-204, SIRT1,
CDK2 and ATR (Fig. 3B-F) were higher
than the average values, implying these miRNAs and proteins were
the hub nodes of this biological network. In the separated
miRNA-target interaction network we found that ATR and CDK2 were
targets of miR-383 and miR-204 (Fig.
2B). Moreover, miR-204 could regulate the network through TP53
mediated by SIRT1. RBBP7, SMARCB1, and CREBBP were also connected
with several nodes and may be related to the regulation of a small
cluster network.
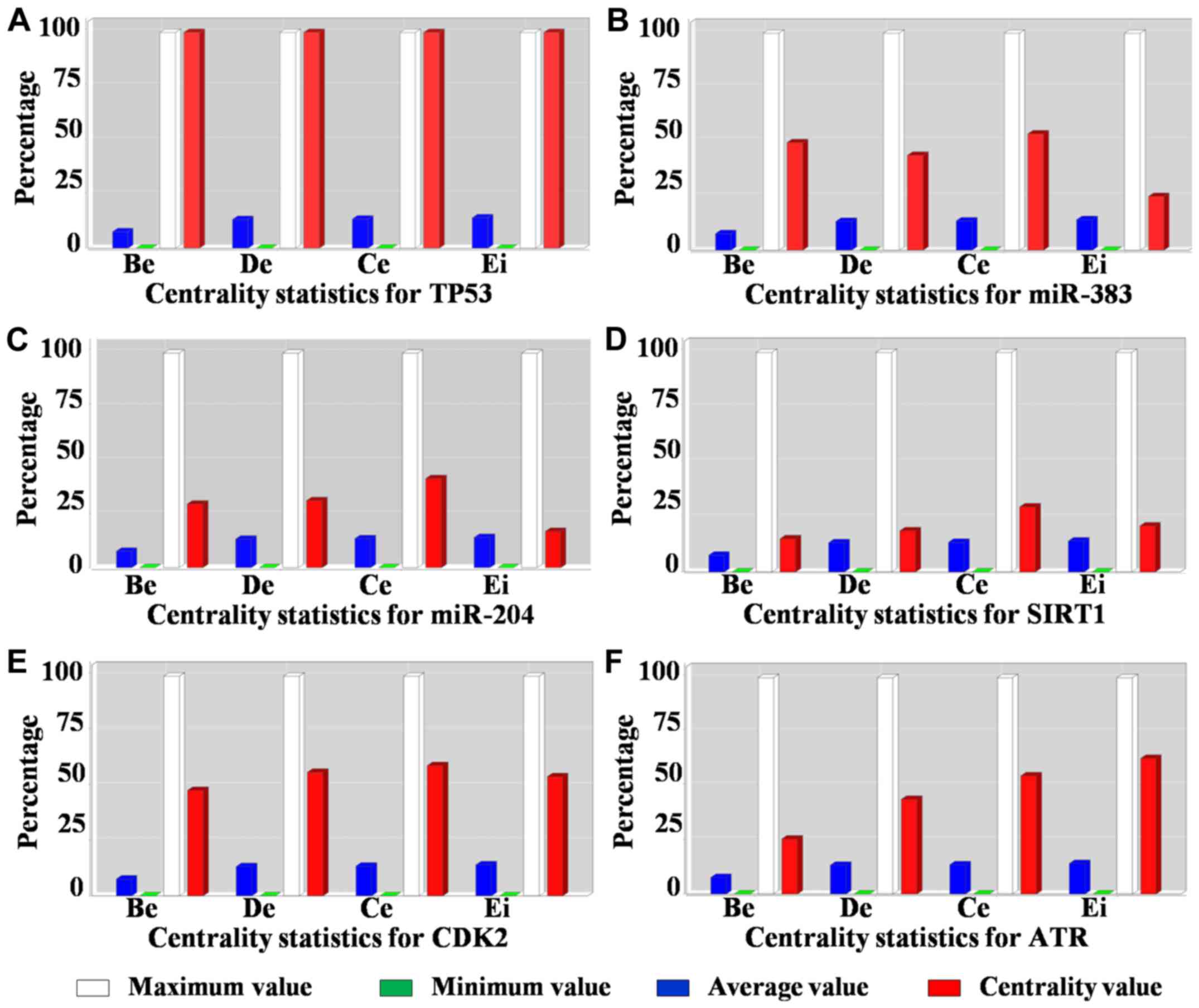 | Figure 3.Centrality measures of the hub nodes
in the miRNA-target regulatory network. Betweenness (Be), degree
(De), centroid value (Ce) and eigenvector (Ei) plots for (A) TP53,
(B) miR-383, (C) miR-204, (D) SIRT1, (E) CDK2 and (F) ATR
respectively. miR/miRNA, microRNA; TP53, tumor protein p53; SIRT1,
sirtuin 1; CDK2, cyclin dependent kinase 2; ATR, ATR
serine/threonine kinase. |
Discussion
Some of the dysregulated miRNAs identified in the
CMM tissues by microarray analysis were validated by qRT-PCR. The
upregulation of seven miRNAs in CMM, namely miR-16, miR-21,
miR-29b, miR-122, miR-125b, miR-204, and miR-383 was demonstrated
here for the first time. In particular, miR-204 and miR-383 showed
extra ordinarily high expression levels in the microarray and
qRT-PCR analyses.
Downregulation of miR-145, miR-205 and miR-203 was
detected in the microarray analysis, which is consistent with
previous studies on CMM (16,17). However, we did not detect
dysregulation of other miRNAs that have been reported previously to
be downregulated (17). These
inconsistencies might be because different microarray platforms
and/or samples were used in the two studies. Noguchi et al
(17) used a CombiMatrix array,
whereas we used a miRCURY™ LNA microRNA Array. Thus,
there were differences in the miRNAs that were spotted on the
arrays. We used CMM tissues from three different dogs and Noguchi
et al (17) used CMM tissue
from only one dog. Finally, in the previous study, miRNA expression
was compared between CMM tissue and normal oral mucosal tissue
(17), whereas we compared CMM
tissues with reference miRNAs from several cancers and normal
tissues. We used mixed miRNA reference samples to avoid biases from
low signal intensities in the microarray data. Using miRNAs from
several different origins means different miRNAs will be included
because miRNA expression is highly dependent on the tissue origin
and status. Our approach should cover a broad range of miRNAs, thus
avoiding misleading FC ratios as a result of weak signals (39). However, because our reference tissues
were mostly tumor samples (70.8%), using this kind of miRNA
reference samples may have caused miRNAs that are commonly
dysregulated in tumors to be overlooked but, importantly, may have
revealed miRNAs that are specifically dysregulated in melanoma.
In this study, the microarray and qRT-PCR results
were consistent for the relative expressions of miR-204, miR-383
and miR-205. However, the discrepant expressions of miR-122 and
miR-143 between the microarray and qRT-PCR results may be explained
by differences in the control samples that were used in the two
experiments; that is, a mixed sample reference in the microarray
analysis and normal oral tissues in the qRT-PCRs. For the same
reason, differential expression of miR-16, miR-21, miR-29b and
miR-125b was not detected in the microarray analysis but was
detected by qRT-PCR. miR-21 and miR-29b are known to be upregulated
in several tumors; for example, miR-21 in mouse BL/6 melanoma cells
(40), miR-29b in human breast cancer
(20) and both miRNAs in canine
mammary cancer (14). These findings
indicate that miR-21 and miR-29b are common oncomiRs in several
species. Thus, the microarray screening method that we used may
have masked the differential expression of these miRNAs because
they are not specific to melanoma but commonly shared among several
kinds of tumors.
While the significant downregulation of miR-205 can
be explained, upregulation of miR-204 and miR-383 expression has
not been reported in CMM until now. Indeed, miR-204 was reported to
be upregulated in old HMM patients compared with young HMM patients
(41); however, no comparison between
melanoma and normal tissue was performed and the target mRNA was
not defined. In another study, miR-204 was found to be
downregulated in malignant melanoma compared with benign nevi
(42), but the age of the patients
was not considered and the comparisons were between malignant
melanoma and benign nevi tissues. In prostate cancer and breast
cancer studies, miR-204 was reported to be both up- and
downregulated (43–47), maybe because of different experimental
designs and individual identity.
TP53 is a well-known tumor suppressor gene located
in the center of the network with a high centroid value (Fig. 3A). SIRT1, an indirect regulator of
TP53, is a direct target of miR-204 in the network and has been
reported to be downregulated in canine melanoma (48). SIRT1 acts as a tumor suppressor via
β-catenin and has reminiscent effects on TP53 in colon cancer
(49). Abnormal expression of
β-catenin was reported in melanoma (50,51), so
the miR-204-mediated downregulation of SIRT1 revealed in the
network may cause β-catenin-mediated cell survival by evading TP53
in melanoma.
Up-regulation of miR-383 expression has been
observed in primary HMM tumor cell lines compared with normal human
epidermal melanocytes (52). In their
study, Mueller et al (52)
found that miR-383 was downregulated in snail stable knockdown
melanoma cells by transfection of an antisense snail plasmid
construct, named as-snail, compared with the parental melanoma cell
line. Snail belongs to the snail superfamily of zinc finger
transcription factors and is involved in the development of
malignant melanoma through direct repression of E-cadherin
expression (53). Indeed, the
transcriptional profile of the as-snail cells was reported to be
more similar to normal melanocytes than malignant melanoma cells
(52). However, the detailed
biological functions of miR-383 have not been reported so far. In
our study, miR-383 was upregulated in CMM tissues. Liao et
al (54) showed that ATR was the
direct target of miR-383 and ATR was found to play a central role
in the ATM/ATR pathway involved in DNA damage recognition and
initial phosphorylation (55). Liao
et al (54) also showed that
GADD45γ, MDC1, and H2AX were all negatively correlated with miR-383
expression. Moreover, a recent study showed that loss of function
or mutations of ATR lead to the development of melanoma (56). In testicular embryonal carcinoma
miR-383 overexpression was found to reduce CDK2 expression at the
protein level, which was also found to be necessary for proper DNA
repair (57). Furthermore, CREB
binding protein, a known co-activator of TP53, was found to be a
direct target of miR-383 (58). There
is also a possibility that miR-383 has indirect control over
apoptosis via TP53 inhibition through CDK2. So, our network
analysis and the above discussion suggest that miR-383 may be
involved in DNA damage repair and apoptosis phenomena in melanoma.
In this study, we demonstrated the dysregulation of 17 miRNAs in
CMM and investigated the probable biological functions of these
miRNAs based on their target genes. Our study is valid not only for
dog but also for human because dog has been considered as a good
preclinical model for human melanoma (10). Further studies are required to clarify
the functions of the dysregulated miRNAs by for example, detecting
the actual target genes and their pathways and analyzing their
differential expression patterns in established canine melanoma
cell lines (59,60) to determine the roles of the
miRNA-target interactions in CMM tumor genesis and therapy.
We have demonstrated the upregulation of potential
oncomiRs, miR-16, miR-21, miR-29b, miR-122, miR-125b, miR-204 and
miR-383 in CMM tissues. In particular, the strong upregulation of
miR-383 in CMM tissues compared with normal oral tissues identified
by microarray screening was confirmed by qRT-PCR. We conclude that
miR-383 and miR-204 may promote melanoma development by regulating
both the DNA repair/checkpoint and apoptosis. To identify
therapeutic targets in melanoma, further studies are required to
verify the biological significance of the miRNA target genes.
Acknowledgements
The authors would like to thank Dr Margaret Biswas
for editing a draft of this manuscript.
Funding
The present study was supported by the Japan Society
for the Promotion of Science KAKENHI (grant nos. 17H03926,
15H14872, 25292180 and 22780283).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
NU, MR, TM and NaM were involved in designing the
study. NU, MR, TM, TI, NoM and HK collaborated in the data
analysis. NaM acquired the funding. NU, MR, YCL, TM, TI, HK, YM and
NaM performed the experiments. NaM was involved in project
administration. NU, TI, NoM and HK acquired the resources. NoM, YM
and NaM supervised the study. NaM was involved in data validation.
TM wrote the original draft of the manuscript. NU, MR, YM and NaM
revised and edited the original draft. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Informed consent to use the specimens in this study
was obtained from the dog patient's owners. The animal experiments
were approved by the Kagoshima University's Laboratory Animal
Committee (approval no. A10031).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Manikandan J, Aarthi JJ, Kumar SD and
Pushparaj PN: OncomiRs: The potential role of non-coding microRNAs
in understanding cancer. Bioinformation. 2:330–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huynh C, Segura MF, Gaziel-Sovran A,
Menendez S, Darvishian F, Chiriboga L, Levin B, Meruelo D, Osman I,
Zavadil J, et al: Efficient in vivo microRNA targeting of liver
metastasis. Oncogene. 30:1481–1488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun V, Zhou WB, Majid S, Kashani-Sabet M
and Dar AA: MicroRNA-mediated regulation of melanoma. Br J
Dermatol. 171:234–241. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Dou Y and Sheng M: Inhibition of
microRNA-383 has tumor suppressive effect in human epithelial
ovarian cancer through the action on caspase-2 gene. Biomed
Pharmacother. 83:1286–1294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang X, Zhu W, Zhang H and Lian S:
Sensitization of melanoma cells to temozolomide by overexpression
of microRNA 203 through direct targeting of glutaminase-mediated
glutamine metabolism. Clin Exp Dermatol. 42:614–621. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Simpson RM, Bastian BC, Michael HT,
Webster JD, Prasad ML, Conway CM, Prieto VM, Gary JM, Goldschmidt
MH, Esplin DG, Smedley RC, et al: Sporadic naturally occurring
melanoma in dogs as a preclinical model for human melanoma. Pigment
Cell Melanoma Res. 27:37–47. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Uhl E, Krimer P, Schliekelman P, Tompkins
SM and Suter S: Identification of altered MicroRNA expression in
canine lymphoid cell lines and cases of B- and T-cell lymphomas.
Genes Chromosomes Cancer. 50:950–967. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gioia G, Mortarino M, Gelain ME, Albonico
F, Ciusani E, Forno I, Marconato L, Martini V and Comazzi S:
Immunophenotype-related microRNA expression in canine chronic
lymphocytic leukemia. Vet Immunol Immunopathol. 142:228–235. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vinall RL, Kent MS and deVere White RW:
Expression of microRNAs in urinary bladder samples obtained from
dogs with grossly normal bladders, inflammatory bladder disease, or
transitional cell carcinoma. Am J Vet Res. 73:1626–1633. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boggs RM, Wright ZM, Stickney MJ, Porter
WW and Murphy KE: MicroRNA expression in canine mammary cancer.
Mamm Genome. 19:561–569. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kobayashi M, Saito A, Tanaka Y, Michishita
M, Kobayashi M, Irimajiri M, Kaneda T, Ochiai K, Bonkobara M,
Takahashi K, et al: MicroRNA expression profiling in canine
prostate cancer. J Vet Med Sci. 79:719–725. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Noguchi S, Mori T, Hoshino Y, Yamada N,
Nakagawa T, Sasaki N, Akao Y and Maruo K: Comparative study of
anti-oncogenic microRNA-145 in canine and human malignant melanoma.
J Vet Med Sci. 74:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Noguchi S, Mori T, Hoshino Y, Yamada N,
Maruo K and Akao Y: MicroRNAs as tumour suppressors in canine and
human melanoma cells and as a prognostic factor in canine
melanomas. Vet Comp Oncol. 11:113–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Starkey MP, Compston-Garnett L, Malho P,
Dunn K and Dubielzig R: Metastasis-associated microRNA expression
in canine uveal melanoma. Vet Comp Oncol. 16:81–89. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Bian Z, Wei D and Zhang JG:
miR-29b regulates migration of human breast cancer cells. Mol Cell
Biochem. 352:197–207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Latchana N, Ganju A, Howard JH and Carson
WE III: MicroRNA dysregulation in melanoma. Surg Oncol. 25:184–189.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmitt MJ, Philippidou D, Reinsbach SE,
Margue C, Wienecke-Baldacchino A, Nashan D, Behrmann I and Kreis S:
Interferon-γ-induced activation of signal transducer and activator
of transcription 1 (STAT1) up-regulates the tumor suppressing
microRNA-29 family in melanoma cells. Cell Commun Signal.
10:412012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hartwell LH, Hopfield JJ, Leibler S and
Murray AW: From molecular to modular cell biology. Nature.
402:(6761 Suppl)C47–C52. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu W, Yang L and Du Z: MicroRNA
regulation and tissue-specific protein interaction network. PLoS
One. 6:e253942011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schroeder A, Mueller O, Stocker S,
Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M
and Ragg T: The RIN: An RNA integrity number for assigning
integrity values to RNA measurements. BMC Mol Biol. 7:32006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao Y, Wang E, Liu H, Rotunno M, Koshiol
J, Marincola FM, Landi MT and McShane LM: Evaluation of
normalization methods for two-channel microRNA microarrays. J
Transl Med. 8:692010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reiner A, Yekutieli D and Benjamini Y:
Identifying differentially expressed genes using false discovery
rate controlling procedures. Bioinformatics. 19:368–375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lopes CT, Franz M, Kazi F, Donaldson SL,
Morris Q and Bader GD: Cytoscape Web: An interactive web-based
network browser. Bioinformatics. 26:2347–2348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scardoni G, Petterlini M and Laudanna C:
Analyzing biological network parameters with CentiScaPe.
Bioinformatics. 25:2857–2859. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Felicetti F, Errico MC, Bottero L,
Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini
M, Colombo MP, et al: The promyelocytic leukemia zinc
finger-microRNA-221/-222 pathway controls melanoma progression
through multiple oncogenic mechanisms. Cancer Res. 68:2745–2754.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kappelmann M, Kuphal S, Meister G,
Vardimon L and Bosserhoff AK: MicroRNA miR-125b controls melanoma
progression by direct regulation of c-Jun protein expression.
Oncogene. 32:2984–2991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H
and Hu C: Circulating microRNA-92a and microRNA-21 as novel
minimally invasive biomarkers for primary breast cancer. J Cancer
Res Clin Oncol. 139:223–229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Creighton CJ, Hernandez-Herrera A,
Jacobsen A, Levine DA, Mankoo P, Schultz N, Du Y, Zhang Y, Larsson
E, Sheridan R, et al: Integrated analyses of microRNAs demonstrate
their widespread influence on gene expression in high-grade serous
ovarian carcinoma. PLoS One. 7:e345462012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: Suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Parker BJ, Günter S and Bedo J:
Stratification bias in low signal microarray studies. BMC
Bioinformatics. 8:3262007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang CH, Yue J, Pfeffer SR, Handorf CR and
Pfeffer LM: MicroRNA miR-21 regulates the metastatic behavior of
B16 melanoma cells. J Biol Chem. 286:39172–39178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jukic DM, Rao UN, Kelly L, Skaf JS,
Drogowski LM, Kirkwood JM and Panelli MC: Microrna profiling
analysis of differences between the melanoma of young adults and
older adults. J Transl Med. 8:272010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J,
Ruan H, Ma S and Xu B: miR-204-5p acts as a tumor suppressor by
targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in
malignant melanoma. Onco Targets Ther. 10:1237–1246. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ding M, Lin B, Li T, Liu Y, Li Y, Zhou X,
Miao M, Gu J, Pan H, Yang F, et al: A dual yet opposite
growth-regulating function of miR-204 and its target XRN1 in
prostate adenocarcinoma cells and neuroendocrine-like prostate
cancer cells. Oncotarget. 6:7686–7700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Findlay VJ, Turner DP, Moussa O and Watson
DK: MicroRNA-mediated inhibition of prostate-derived Ets factor
messenger RNA translation affects prostate-derived Ets factor
regulatory networks in human breast cancer. Cancer Res.
68:8499–8506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Imam JS, Plyler JR, Bansal H, Prajapati S,
Bansal S, Rebeles J, Chen HI, Chang YF, Panneerdoss S, Zoghi B, et
al: Genomic loss of tumor suppressor miRNA-204 promotes cancer cell
migration and invasion by activating AKT/mTOR/Rac1 signaling and
actin reorganization. PLoS One. 7:e523972012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li W, Jin X, Zhang Q, Zhang G, Deng X and
Ma L: Decreased expression of miR-204 is associated with poor
prognosis in patients with breast cancer. Int J Clin Exp Pathol.
7:3287–3292. 2014.PubMed/NCBI
|
|
47
|
Turner DP, Findlay VJ, Moussa O,
Semenchenko VI, Watson PM, LaRue AC, Desouki MM, Fraig M and Watson
DK: Mechanisms and functional consequences of PDEF protein
expression loss during prostate cancer progression. Prostate.
71:1723–1735. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Marfe G, De Martino L, Tafani M,
Irno-Consalvo M, Pasolini MP, Navas L, Papparella S, Gambacurta A
and Paciello O: A multicancer-like syndrome in a dog characterized
by p53 and cell cycle-checkpoint kinase 2 (CHK2) mutations and
sirtuin gene (SIRT1) down-regulation. Res Vet Sci. 93:240–245.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Firestein R, Blander G, Michan S,
Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S,
de Cabo R, Fuchs C, et al: The SIRT1 deacetylase suppresses
intestinal tumorigenesis and colon cancer growth. PLoS One.
3:e20202008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pećina-Slaus N, Zigmund M, Kusec V, Martić
TN, Cacić M and Slaus M: E-cadherin and beta-catenin expression
patterns in malignant melanoma assessed by image analysis. J Cutan
Pathol. 34:239–246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rubinfeld B, Robbins P, El-Gamil M, Albert
I, Porfiri E and Polakis P: Stabilization of beta-catenin by
genetic defects in melanoma cell lines. Science. 275:1790–1792.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mueller DW, Rehli M and Bosserhoff AK:
miRNA expression profiling in melanocytes and melanoma cell lines
reveals miRNAs associated with formation and progression of
malignant melanoma. J Invest Dermatol. 129:1740–1751. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kuphal S, Palm HG, Poser I and Bosserhoff
AK: Snail-regulated genes in malignant melanoma. Melanoma Res.
15:305–313. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liao XH, Zheng L, He HP, Zheng DL, Wei ZQ,
Wang N, Dong J, Ma WJ and Zhang TC: STAT3 regulated ATR via
microRNA-383 to control DNA damage to affect apoptosis in A431
cells. Cell Signal. 27:2285–2295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Rouse J and Jackson SP: Interfaces between
the detection, signaling, and repair of DNA damage. Science.
297:547–551. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen CF, Ruiz-Vega R, Vasudeva P, Espitia
F, Krasieva TB, de Feraudy S, Tromberg BJ, Huang S, Garner CP, Wu
J, et al: ATR mutations promote the growth of melanoma tumors by
modulating the immune microenvironment. Cell Rep. 18:2331–2342.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Satyanarayana A and Kaldis P: A dual role
of Cdk2 in DNA damage response. Cell Div. 4:92009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Grossman SR: p300/CBP/p53 interaction and
regulation of the p53 response. Eur J Biochem. 268:2773–2778. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ohashi E, Hong SH, Takahashi T, Nakagawa
T, Mochizuki M, Nishimura R and Sasak N: Effect of retinoids on
growth inhibition of two canine melanoma cell lines. J Vet Med Sci.
63:83–86. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Inoue K, Ohashi E, Kadosawa T, Hong SH,
Matsunaga S, Mochizuki M, Nishimura R and Sasaki N: Establishment
and characterization of four canine melanoma cell lines. J Vet Med
Sci. 66:1437–1440. 2004. View Article : Google Scholar : PubMed/NCBI
|