Introduction
Osteosarcoma is one of the most common clinical
malignant tumors in orthopedics. It is a kind of primary bone
tissue tumor with a high degree of malignancy, which often occurs
in children and adolescents and has the characteristics of strong
invasion, rapid metastasis and high mortality (1,2). At
present, there is no ideal treatment for osteosarcoma. The commonly
used methods include surgery, radiotherapy and chemotherapy, but
the curative effect is not good. According to statistics, the
5-year survival rate of osteosarcoma is only 50–60% (3). Fragile histidine triad (FHIT) gene is an
important member of histidine triad gene family. It is considered
as a tumor suppressor gene, whose abnormal expression is associated
with a variety of malignant tumors, such as lung, cancer and breast
cancer. Studies have shown that (4,5) FHIT gene
plays an important regulatory role in the process of cell
proliferation and apoptosis. However, the role of FHIT in
osteosarcoma remains unclear. The purpose of the present study was
to investigate the role of FHIT gene in the regulation of
proliferation and apoptosis of Saos2 osteosarcoma cells, clarify
its relationship with proliferation and apoptosis of osteosarcoma
cells, and demonstrate that FHIT gene is as an effective target for
the treatment of osteosarcoma.
Materials and methods
Materials and reagents
hFOB1.19 normal human osteoblastic cell line, Saos2
osteosarcoma cell line (both from American Type Culture Collection,
Manassas, VA, USA), rabbit anti-epidermal growth factor receptor
(EGFR), human estrogen receptor-2 (HER-2) monoclonal antibodies and
secondary goat anti-rabbit polyclonal antibody (cat. nos. ab52894,
ab134182, ab6721; Abcam, Cambridge, MA, USA), Roswell Park Memorial
Institute (RPMI)-1640 medium, Dulbecco's modified Eagles medium
(DMEM-F12) (both from HyClone; GE Healthcare Life Sciences, Logan,
UT, USA), fetal bovine serum (FBS), trypsin (both from Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), penicillin,
streptomycin (both from Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), Cell Counting Kit 8 (CCK8) (Beyotime Institute of
Biotechnology, Hangzhou, China), AceQ reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Synergy Brands (SYBR) Green Master Mix kit (Vazyme, Nanjing,
China), HiScript II Q reverse transcription (RT) SuperMix for
RT-qPCR [+genomic deoxyribonucleic acid (gDNA) wiper] kit (Vazyme),
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI)
apoptosis detection kit (Beyotime Institute of Biotechnology),
restriction endonuclease EcoRI and restriction endonuclease
BamHI (both from Promega Corp., Madison, WI, USA).
The study was approved by the Ethics Committee of
Zhoupu Hospital affiliated to Shanghai University of Medicine and
Health Science (Shanghai, China).
Instruments
CO2 cell incubator (Thermo Fisher
Scientific, Inc.), fluorescence microscope (Leica DMI 4000B/DFC425;
Leica Microsystems GmbH, Wetzlar, Germany), NanoDrop ND-1000
spectrophotometer (NanoDrop Technology; Thermo Fisher Scientific,
Inc., Wilmington, DE, USA), fluorescence RT-qPCR instrument (ABI
7500; Applied Biosystems, Foster City, CA, USA), microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA), FACS flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA), Image Lab and
Image-Pro image analysis systems (Bio-Rad Laboratories, Inc.).
Cell culture
hFOB1.19 cells were cultured in DMEM-F12 complete
medium containing 10% FBS and 1% penicillin and streptomycin. Saos2
cells were cultured in RPMI complete medium containing 10% FBS and
1% penicillin and streptomycin. All cells were cultured in an
incubator with 5% CO2 at 37°C, and the medium was
changed once every 3 days, followed by passage when 80% of them
were fused.
Cell passage
When 80% of the cultured cells were fused, the cell
culture medium was removed, followed by digestion with 0.125%
trypsin for 1 min and then gentle pipetting to digest the adherent
cells. RIPM-1640 medium containing 10% FBS was added to stop
digestion, followed by centrifugation at 1,200 × g for 5 min at
4°C. Then, the supernatant was removed, the medium was added for
resuspension, and the cells continued to be cultured. Finally, the
cells were collected for experiments when they were passaged to the
third generation.
Cell treatment and transfection
Cells were divided into hFOB, Saos2, transfection
and no-load transfection groups. hFOB cells were routinely cultured
in hFOB group without any treatment. Saos2 cells were routinely
cultured in Saos2 group without any treatment. pcDNA3.1-FHIT
overexpression vectors containing FHIT gene were transfected into
the cultured cells in the transfection group. pcDNA3.1 vectors were
transfected into the cultured cells in the no-load transfection
group.
Transfection methods: pcDNA3.1-FHIT overexpression
vectors were constructed, followed by amplification of FHIT gene in
hFOB1.9 cell line and identification of pMD10-T vector sequencing.
pMD10-T recombinant vectors containing correct FHIT gene fragments
were dually digested with EcoRI and BamHI, and cloned
into pcDNA3.1 eukaryotic expression vectors. The overexpression
plasmids containing correct sequences were mixed with liposomes,
diluted with RPMI medium and allowed to rest at room temperature
for 20 min. Finally, the cells were implanted and cultured with
cell culture medium for 48 h.
Western blot analysis
After transfection, the cells in each group were
collected with ProteoPrep® Total Extraction Sample kit
(Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) and added with
lysis buffer, followed by ice bath for 60 min and centrifugation at
14,000 × g for 10 min at 4°C, and the protein was quantified by
bicinchoninic acid (BCA) method. The standard curve and absorbance
were measured by the microplate reader and the protein
concentration was calculated. After denaturation of the protein,
the samples were loaded (10 µl per lane) and separated by 15%
sodium dodecyl sulfate-polyacrylamide gel electropheresis
(SDS-PAGE) with corresponding concentration. When the marker
protein ran to the bottom of the glass plate and the sample protein
was basically in a straight line sinking to the bottom, the gel
running was stopped. The samples were transferred onto a
polyvinylidene fluoride (PVDF) membrane, sealed, rinsed with
phosphate-buffered saline with Tween-20 (PBST) 3 times, and then
sealed with sealing solution for 1.5 h. Primary anti-human EGFR,
HER-2 monoclonal antibodies (1:1,000) and secondary antibody
(1:1,000) were added successively. The membrane was rinsed with
Tris-buffered saline with Tween-20 (TBST) at each step interval.
After being rinsed with TBST to remove the secondary antibody,
color development began. The membrane was placed in the
chemiluminescence reagent (Beyotime Institute of Biotechnology,
Shanghai, China) for reaction for 1 min and then developed in dark
conditions. Finally, gel scanning imaging system (Bio-Rad
Laboratories, Inc.) was used for analysis.
RT-qPCR detection
Total RNA was extracted using TRIzol (Thermo Fisher
Scientific, Inc.) from spare bone blocks stored at −20°C. The
extracted total RNA was reversely transcribed into complementary
DNA (cDNA) using RT kit (ABclonal Biotech Co., Ltd., Wuhan, China),
and the reaction system was 20 µl. Fast SYBR Green Master Mix was
used. The reaction conditions were as follows: reaction at 51°C for
2 min, predenaturation at 96°C for 10 min, denaturation at 96°C for
10 sec, annealing at 60°C for 30 sec, 40 cycles. The relative
expression of FHIT messenger ribonucleic acid (mRNA) was
calculated. The results were analyzed by using the
2−ΔΔCq method (6). Primer
sequences are shown in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Name | Primer sequences |
|---|
| FHIT | F:
5′-TTGGGGCGCGGGTTTGGGTTTTTAC-3′ |
|
| R:
5′-CGTAAACGACGCCGACCCCACTA-3′ |
| GAPDH | F:
5′-ACGGCAAGTTCAACGGCACAG-3′ |
|
| R:
5′-GAAGACGCCAGTAGACTCCACGAC-3′ |
Detection of cell proliferation by
CCK8
After cell transfection, the cells in each group
were inoculated into a 96-well plate at a density of
3×103/ml. Three replicate wells were set and each well
was added with 100 µl of complete medium. The cells were cultured
in an incubator with 5% CO2 at 37°C for 24 h. Each well
was added with 10 µl of CCK8 solution, followed by incubation under
the above conditions for 1 h. The optical density (OD) value was
measured at a wavelength of 450 nm to calculate the cell
proliferation rate.
Detection of apoptosis by flow
cytometry
After cell transfection, the cells in each group
were collected and rinsed with PBS, and then the supernatant was
discarded. The cells were resuspended with binding buffer and the
concentration was adjusted to 1×105/ml. Annexin V-FITC
solution (10 µl) and 5 µl PI solution were added and mixed well.
After reaction in the dark at room temperature for 15 min, the
mixture was detected by flow cytometry. Data were analyzed by
cytomics fc 500 (Beckman Coulter, Inc., Atlanta, GA, USA).
Statistical analysis
In this study, Statistical Product and Service
Solutions (SPSS) 20.0 software (IBM Corp., Armonk, NY, USA) was
used for statistical analysis. Enumeration data are presented as
mean ± standard deviation. t-test was used for data in line with
the normal distribution and homogeneity of variance, corrected
t-test was used for data in line with the normal distribution and
heterogeneity of variance, and non-parametric test was used for
data in line with the abnormal distribution and heterogeneity of
variance. Chi-square test was used for enumeration data.
Results
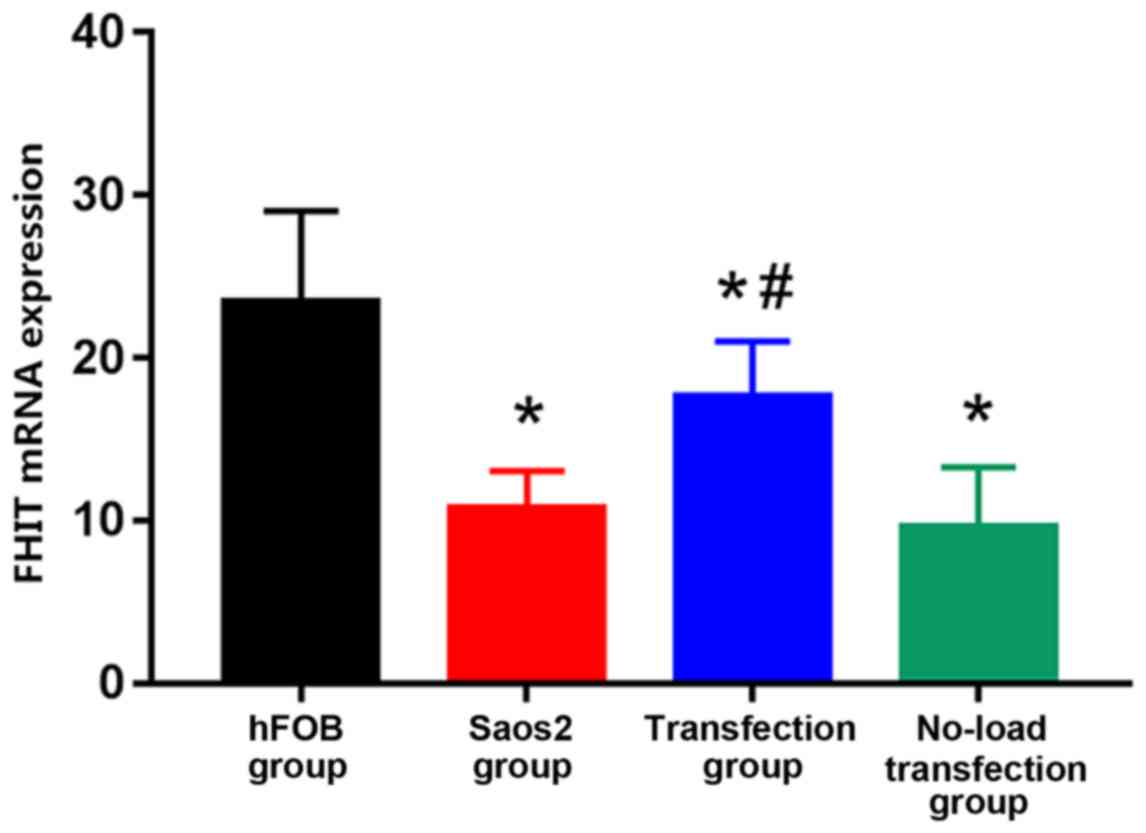
Detection of FHIT mRNA expression via
RT-qPCR
The expression of FHIT mRNA was the highest in hFOB
group and the lowest in Saos2 group. Compared with that in hFOB
group, the expression levels of FHIT mRNA in the other groups were
significantly decreased, and the differences were statistically
significant (P<0.05). Compared with that in Saos2 group, the
expression of FHIT mRNA in transfection group was significantly
increased, and the difference was statistically significant
(P<0.05) (Fig. 1). The results
suggested that FHIT mRNA is highly expressed in normal osteoblast
cell line, but lowly expressed in Saos2 osteosarcoma cell line. The
transfection of pcDNA3.1-FHIT overexpression vectors containing
FHIT gene into Saos2 cells can promote the expression of FHIT mRNA,
indicating that the transfection method is effective.
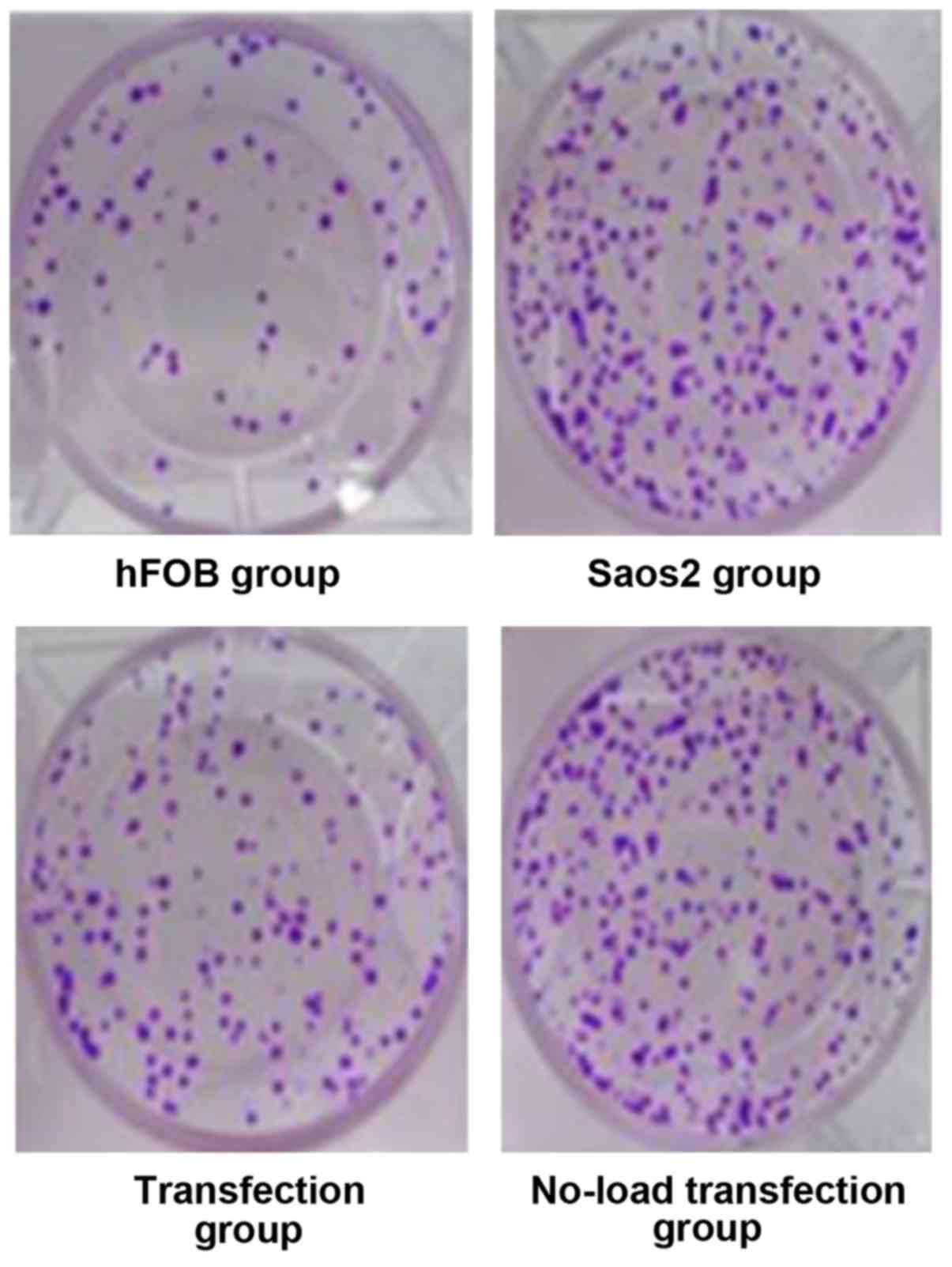
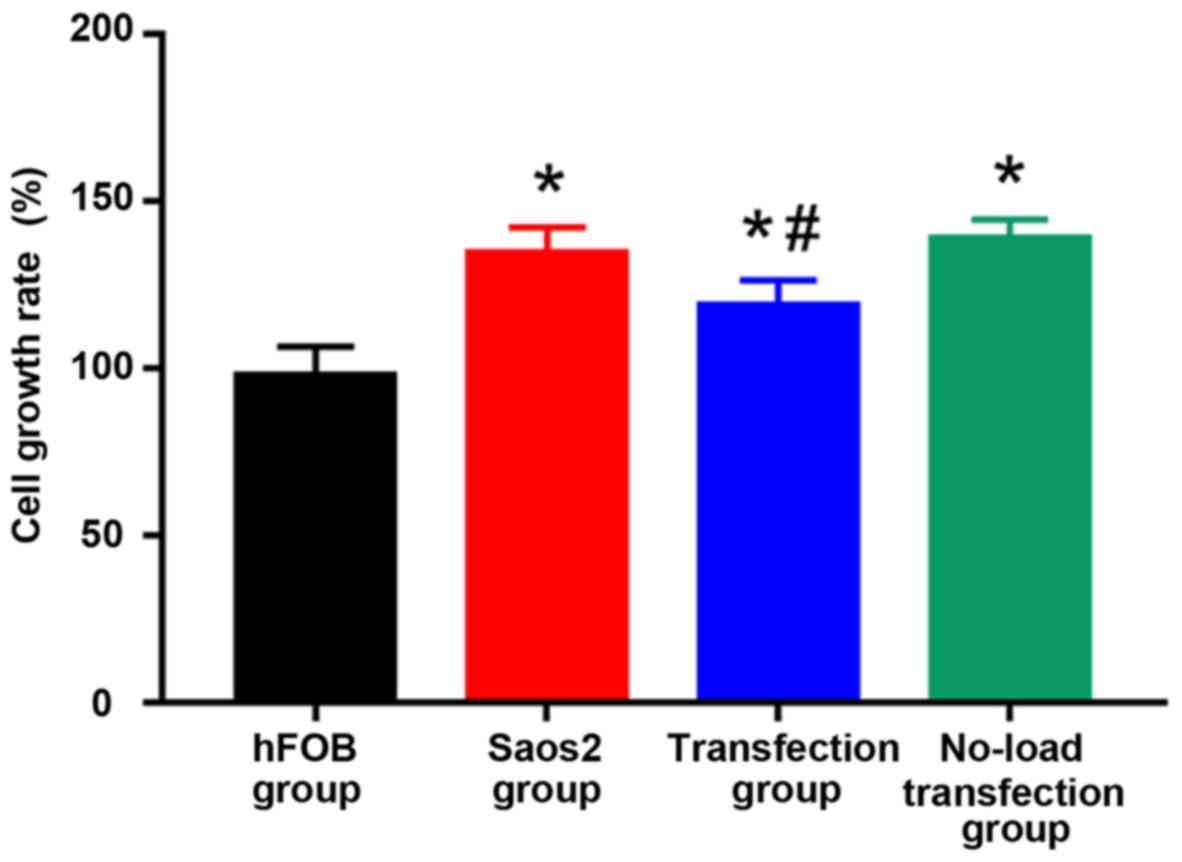
Detection of cell proliferation by
CCK8 assay
Cell growth is shown in Fig. 2 hFOB group had moderate cell density,
Saos2 group and no-load transfection group had high cell density
with concentrated cells. The cell density in the transfection group
was lower than that in Saos2 and no-load transfection groups. CCK8
results are shown in Fig. 3: Compared
with that in hFOB group, cell proliferation was significantly
increased in the other groups, and the differences were
statistically significant (P<0.05). Compared with that in Saos2
group, cell proliferation was significantly decreased in
transfection group, and the difference was statistically
significant (P<0.05). The results suggested that the
proliferation of normal osteoblast cell line is lower, and that of
Saos2 osteosarcoma cell line is higher. The transfection of
pcDNA3.1-FHIT overexpression vectors containing FHIT gene into
Saos2 cells can inhibit the proliferation of Saos2 osteosarcoma
cells.
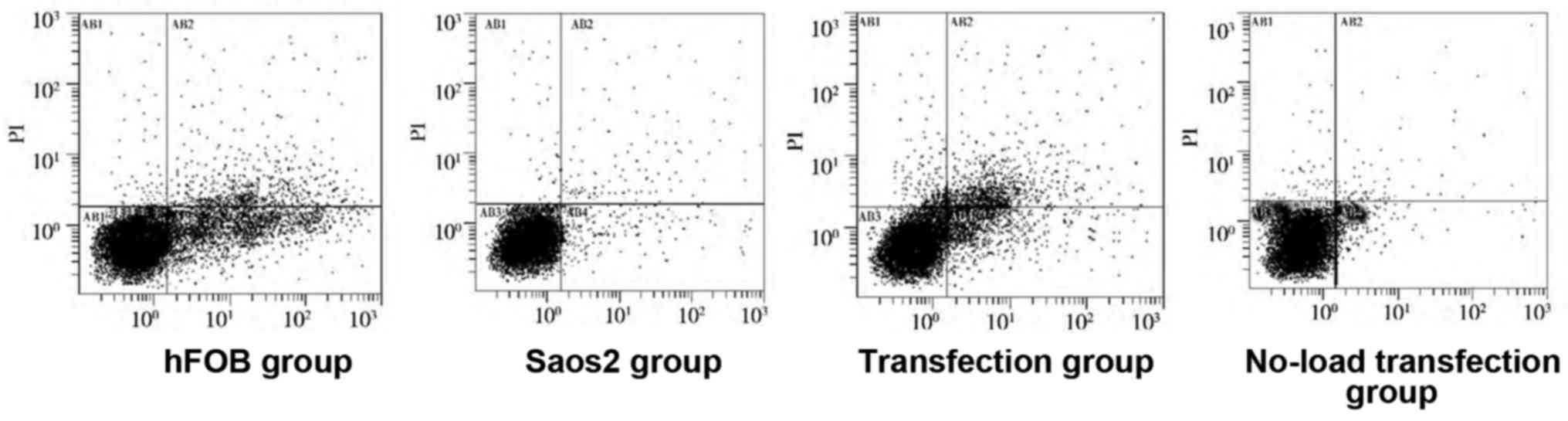
Detection of apoptosis by flow
cytometry
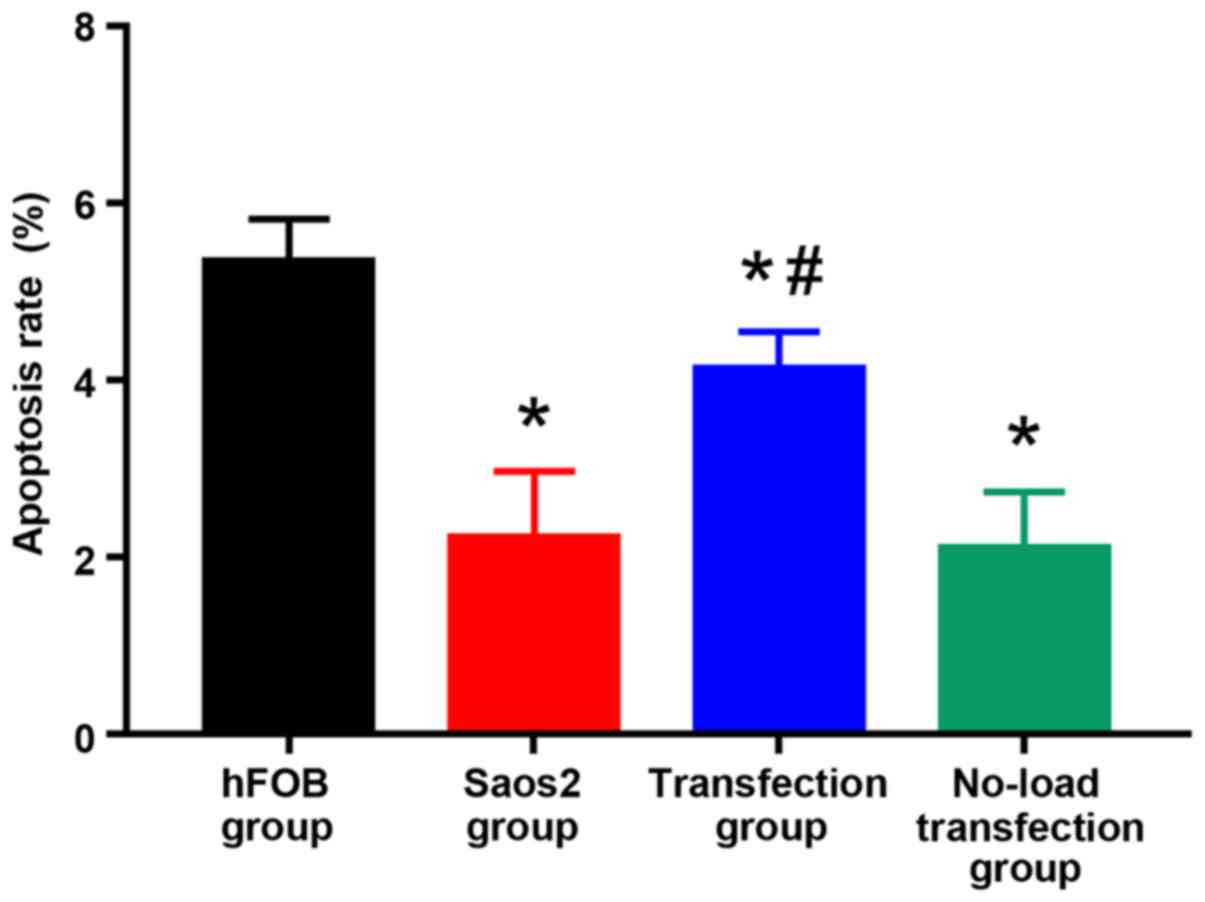
As shown in Figs. 4
and 5, compared with that in hFOB
group, the apoptosis rates in the other groups were significantly
decreased, and there were statistically significant differences
(P<0.05). Compared with that in Saos2 group, the apoptosis rate
in transfection group was significantly increased, and the
difference was statistically significant (P<0.05). These results
indicated that the apoptosis rate of normal osteoblast cell line is
higher, and that of Saos2 osteosarcoma cell line is lower. The
transfection of pcDNA3.1-FHIT overexpression vectors containing
FHIT gene into Saos2 cells can promote apoptosis of Saos2
osteosarcoma cells.
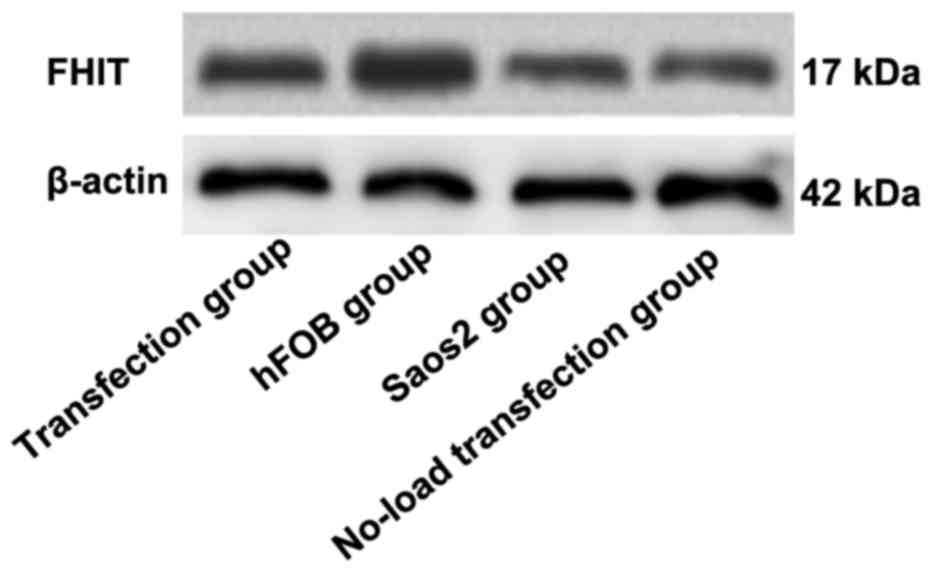
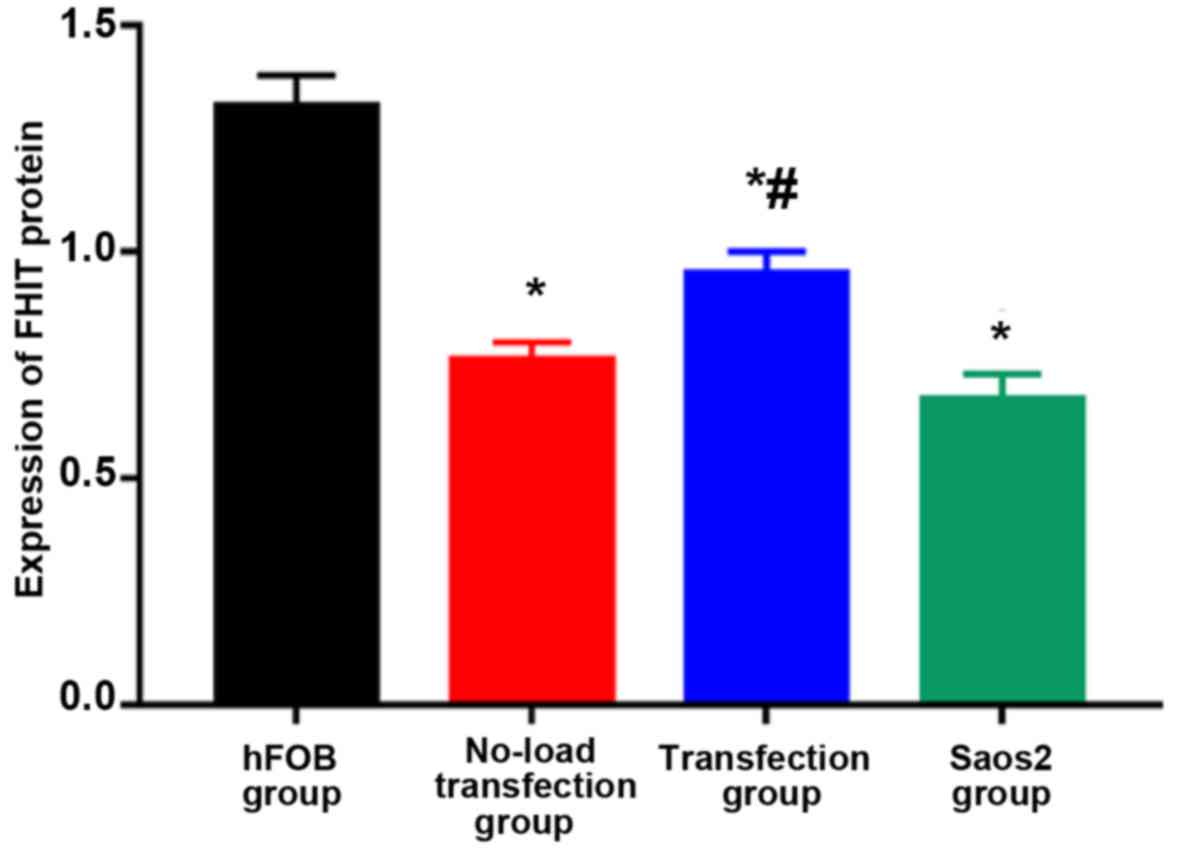
Detection of FHIT protein expression
via western blot analysis
As shown in Figs. 6
and 7, the highest expression of FHIT
protein was found in hFOB group and the lowest expression was in
Saos2 group. Compared with that in hFOB group, the expressions of
FHIT protein in the other groups were significantly decreased, and
the differences were statistically significant (P<0.05).
Compared with that in Saos2 group, the expression of FHIT protein
in transfection group was significantly increased, and there was a
statistically significant difference (P<0.05). The results
indicated that FHIT protein is highly expressed in normal
osteoblast cell line, but lowly expressed in Saos2 osteosarcoma
cell line. The transfection of pcDNA3.1-FHIT overexpression vectors
containing FHIT gene into Saos2 cells can promote FHIT protein
expression.
Discussion
Osteosarcoma is a common tumor with a high degree of
malignancy, mainly derived from mesenchymal tissues (6). Osteosarcoma mainly occurs in adolescents
and the growth is rapid, the metastasis and mortality rates are
high, and the prognosis is poor (7,8). With the
progress of society and the development of science and technology,
some achievements have been made in the treatment of osteosarcoma,
and the 5-year survival rate is about 50–60% (9). However, there are still no effective and
ideal methods for the treatment of osteosarcoma. At present, it is
believed that abnormal expression and functional changes of
multiple genes are important reasons for the rapid development of
osteosarcoma (10). The FHIT gene is
located on human chromosome 3p14.2 and encodes a protein with 16.8
kDa in size. The protein plays an important regulatory role in the
process of DNA repair and cell cycle. It is currently believed that
the FHIT gene is closely related to the occurrence of many tumors.
A study found that retransfection of FHIT gene in esophageal, lung
and breast cancer cell lines with FHIT gene knockout can
effectively inhibit the growth of cancer cells (11). As an expression product of FHIT gene,
FHIT protein can significantly inhibit the growth of human colon
cancer cell line and promote its apoptosis (12). Therefore, FHIT gene is considered to
be an effective tumor suppressor gene. At the same time, there is a
close correlation between FHIT gene and apoptosis. Transfection of
FHIT gene into lung cancer cells lacking FHIT gene can promote
apoptosis of lung cancer cells and block the cell cycle at G0/G1
phase (13). In addition, adenovirus
vector-mediated FHIT gene plays an important role in regulating the
growth, proliferation, apoptosis and long cell cycle of human tumor
cells and nude mouse tumor cells. Overexpression of adenovirus
vector-mediated FHIT gene in tumor cells can effectively inhibit
the growth of tumor cells, but has no obvious effects on normal
cells (14). At the same time,
overexpression of adenovirus vector-mediated FHIT gene in tumor
cells can effectively promote apoptosis of tumor cells, change the
process of cell cycle and increase the number of apoptotic tumor
cells (15,16). Overexpressed FHIT gene and its
expression product FHIT protein can be obtained by injecting
adenovirus vector-mediated FHIT gene into subcutaneous tumor of
nude mice, and tumor growth can be inhibited at the same time,
indicating that adenovirus vector-mediated FHIT gene can be highly
expressed in tumor tissues and inhibit tumor growth (17,18).
Therefore, FHIT gene is involved in the regulation of cell
apoptosis and cell cycle process. The inhibitory effect of FHIT
gene on tumor growth may be achieved by promoting apoptosis of
tumor cells and inhibiting the growth of tumor cells (19,20). In
this study, it was found that FHIT gene was abnormally expressed in
Saos2 osteosarcoma cell line, and the FHIT gene expression was
suppressed in Saos2 osteosarcoma cell line compared with that in
normal osteoblast cell line, including transcription and
translation. Plasmid vectors carrying target gene fragments are
transfected into cells to realize the stable expression of products
of target genes in cells. This cell transfection method is reliable
and stable, and is widely used in the cell transfection technology.
In this study, plasmid vectors carrying FHIT gene were transfected
into Saos2 cells. It was found that the transfected FHIT gene could
be stably expressed in Saos2 cells, which increased the expression
of FHIT gene mRNA and protein, indicating that the cell
transfection was successful. At the same time, this study
manifested that Saos2 osteosarcoma cell line had a high
proliferation rate and a low apoptosis rate compared with normal
osteoblast cell line. However, after transfection with FHIT gene,
the proliferation rate of the transfected Saos2 osteosarcoma cells
was decreased and the apoptosis rate was increased. These results
suggest that FHIT gene regulates the proliferation and apoptosis of
Saos2 osteosarcoma cells, inhibits the proliferation and promotes
apoptosis of osteosarcoma cells.
Acknowledgements
Not applicable.
Funding
The study was supported by the Shanghai Medical Key
Specialty Construction Fund (ZK2015A14), the Seed Fund Program of
Shanghai University of Medicine & Health Sciences
(HMSF-17-22-040), the Construction of Key Discipline Group of
Sanitary System of Shanghai Pudong New District (PWZxq2017-12), and
the Construction of ‘the most important’ Discipine of Zhoupu
Hospital of Shanghai Pudong New District (ZP-XK-2015a-2).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZX wrote the manuscript and was responsible for the
cell culture. JW assisted with cell treatment and transfection. PC
and XZ performed western blot analysis and RT-qPCR. CY was in
charge of CCK8. BW contributed to flow cytometry. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Zhoupu Hospital affiliated to Shanghai University of Medicine and
Health Science (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Novello C, Pazzaglia L, Cingolani C, Conti
A, Quattrini I, Manara MC, Tognon M, Picci P and Benassi MS: miRNA
expression profile in human osteosarcoma: Role of miR-1 and
miR-133b in proliferation and cell cycle control. Int J Oncol.
42:667–675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tang J, Shen L, Yang Q and Zhang C:
Overexpression of metadherin mediates metastasis of osteosarcoma by
regulating epithelial-mesenchymal transition. Cell Prolif.
47:427–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Díaz-Rodríguez L, García-Martínez O,
Morales MA, Rodríguez-Pérez L, Rubio-Ruiz B and Ruiz C: Effects of
indomethacin, nimesulide, and diclofenac on human MG-63
osteosarcoma cell line. Biol Res Nurs. 14:98–107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sard L, Accornero P, Tornielli S, Delia D,
Bunone G, Campiglio M, Colombo MP, Gramegna M, Croce CM, Pierotti
MA, et al: The tumor-suppressor gene FHIT is involved in the
regulation of apoptosis and in cell cycle control. Proc Natl Acad
Sci USA. 96:8489–8492. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ji L, Fang B, Yen N, Fong K, Minna JD and
Roth JA: Induction of apoptosis and inhibition of tumorigenicity
and tumor growth by adenovirus vector-mediated fragile histidine
triad (FHIT) gene overexpression. Cancer Res. 59:3333–3339.
1999.PubMed/NCBI
|
|
6
|
Ta HT, Dass CR, Choong PF and Dunstan DE:
Osteosarcoma treatment: State of the art. Cancer Metastasis Rev.
28:247–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sampo M, Koivikko M, Taskinen M, Kallio P,
Kivioja A, Tarkkanen M and Böhling T: Incidence, epidemiology and
treatment results of osteosarcoma in Finland - a nationwide
population-based study. Acta Oncol. 50:1206–1214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jaffe N: Osteosarcoma: Review of the past,
impact on the future. The American experience. Cancer Treat Res.
152:239–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Strauss SJ, Ng T, Mendoza-Naranjo A,
Whelan J and Sorensen PH: Understanding micrometastatic disease and
Anoikis resistance in Ewing family of tumors and osteosarcoma.
Oncologist. 15:627–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zuo H, Chan GP, Zhu J, Yeung WW, Chan AS,
Ammer H and Wong YH: Activation state-dependent interaction between
Gαq subunits and the Fhit tumor suppressor. Cell Commun Signal.
11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morikawa H, Nakagawa Y, Hashimoto K, Niki
M, Egashira Y, Hirata I, Katsu K and Akao Y: Frequent altered
expression of fragile histidine triad protein in human colorectal
adenomas. Biochem Biophys Res Commun. 278:205–210. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu B, Ying X, Wang J, Piriyapongsa J,
Jordan IK, Sheng J, Yu F, Zhao P, Li Y, Wang H, et al:
Identification of a tumor-suppressive human-specific microRNA
within the FHIT tumor-suppressor gene. Cancer Res. 74:2283–2294.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Siprashvili Z, Sozzi G, Barnes LD, McCue
P, Robinson AK, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L,
et al: Replacement of Fhit in cancer cells suppresses
tumorigenicity. Proc Natl Acad Sci USA. 94:13771–13776. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garinis GA, Gorgoulis VG, Mariatos G,
Zacharatos P, Kotsinas A, Liloglou T, Foukas P, Kanavaros P,
Kastrinakis NG, Vassilakopoulos T, et al: Association of allelic
loss at the FHIT locus and p53 alterations with tumour kinetics and
chromosomal instability in non-small cell lung carcinomas (NSCLCs).
J Pathol. 193:55–65. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin HY, Hung SK, Lee MS, Chiou WY, Huang
TT, Tseng CE, Shih LY, Lin RI, Lin JM, Lai YH, et al: DNA methylome
analysis identifies epigenetic silencing of FHIT as a determining
factor for radiosensitivity in oral cancer: An outcome-predicting
and treatment-implicating study. Oncotarget. 6:915–934. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gemma A, Hagiwara K, Ke Y, Burke LM, Khan
MA, Nagashima M, Bennett WP and Harris CC: FHIT mutations in human
primary gastric cancer. Cancer Res. 57:1435–1437. 1997.PubMed/NCBI
|
|
18
|
Sevignani C, Calin GA, Cesari R, Sarti M,
Ishii H, Yendamuri S, Vecchione A, Trapasso F and Croce CM:
Restoration of fragile histidine triad (FHIT) expression induces
apoptosis and suppresses tumorigenicity in breast cancer cell
lines. Cancer Res. 63:1183–1187. 2003.PubMed/NCBI
|
|
19
|
Fang JM, Arlt MF, Burgess AC, Dagenais SL,
Beer DG and Glover TW: Translocation breakpoints in FHIT and FRA3B
in both homologs of chromosome 3 in an esophageal adenocarcinoma.
Genes Chromosomes Cancer. 30:292–298. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huiping C, Jonasson JG, Agnarsson BA,
Sigbjornsdottir BI, Huebner K and Ingvarsson S: Analysis of the
fragile histidine triad (FHIT) gene in lobular breast cancer. Eur J
Cancer. 36:1552–1557. 2000. View Article : Google Scholar : PubMed/NCBI
|