|
1
|
Lobo NA, Shimono Y, Qian D and Clarke MF:
The biology of cancer stem cells. Annu Rev Cell Dev Biol.
23:675–699. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
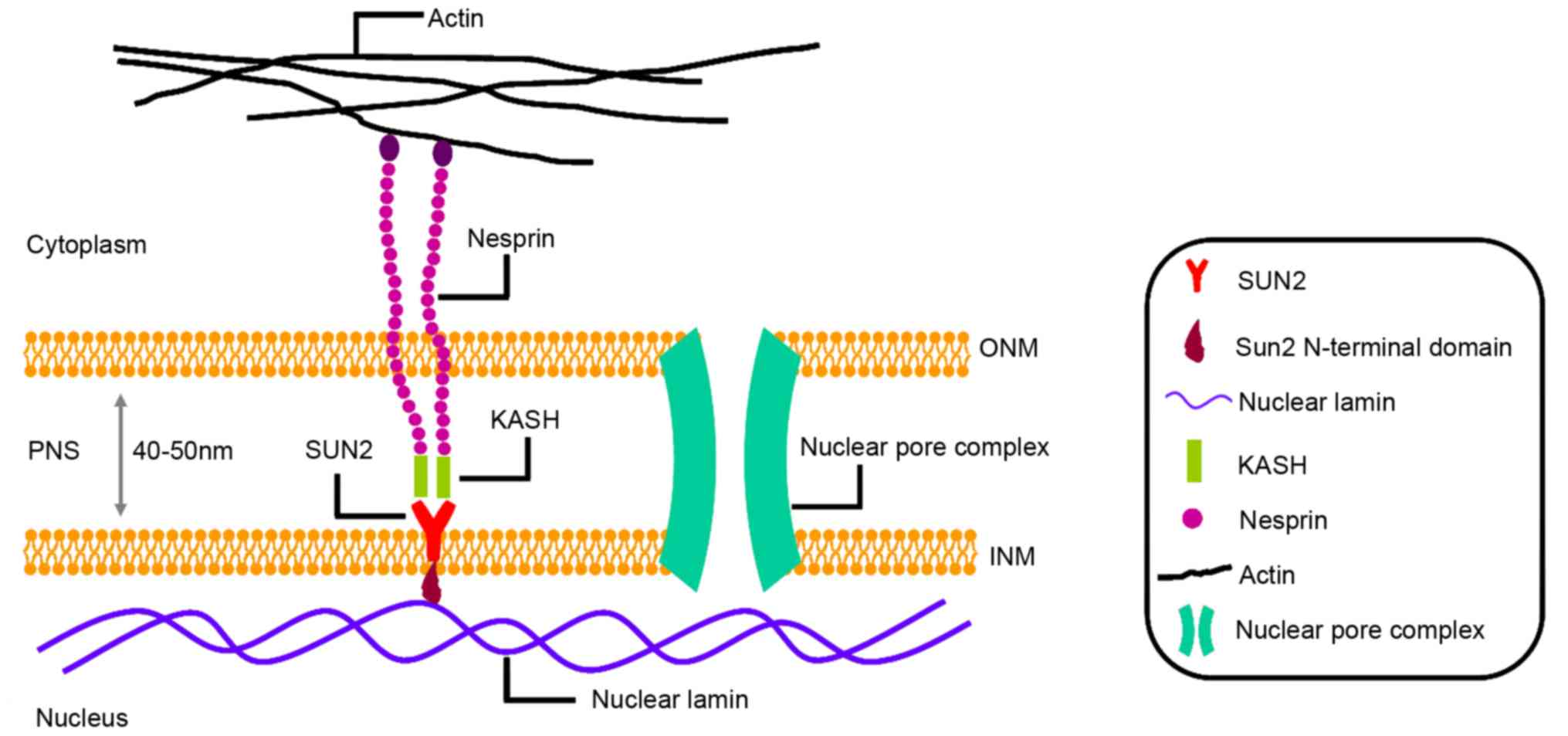
|
|
3
|
Liu Y, Li Z, Wu L, Wang Z, Wang X, Yu Y,
Zhao Q and Luo F: MiRNA-125a-5p: A regulator and predictor of
gefitinib's effect on nasopharyngeal carcinoma. Cancer Cell Int.
14:242014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qi X, Li J, Zhou C, Lv C and Tian M:
MicroRNA-320a inhibits cell proliferation, migration and invasion
by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett.
588:3732–3738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheung CC, Chung GT, Lun SW, To KF, Choy
KW, Lau KM, Siu SP, Guan XY, Ngan RK, Yip TT, et al: miR-31 is
consistently inactivated in EBV-associated nasopharyngeal carcinoma
and contributes to its tumorigenesis. Mol Cancer. 13:1842014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu X, Yu X, Xie J, Zhan M, Yu Z, Xie L,
Zeng H, Zhang F, Chen G, Yi X and Zheng J: ANGPTL2/LILRB2 signaling
promotes the propagation of lung cancer cells. Oncotarget.
6:21004–21015. 2015.PubMed/NCBI
|
|
8
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hecht I, Natan S, Zaritsky A, Levine H,
Tsarfaty I and Ben-Jacob E: The motility-proliferation-metabolism
interplay during metastatic invasion. Sci Rep. 5:135382015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsieh TH, Chien CL, Lee YH, Lin CI, Hsieh
JY, Chao ME, Liu DJ, Chu SS, Chen W, Lin SC, et al: Downregulation
of SUN2, a novel tumor suppressor, mediates miR-221/222-induced
malignancy in central nervous system embryonal tumors.
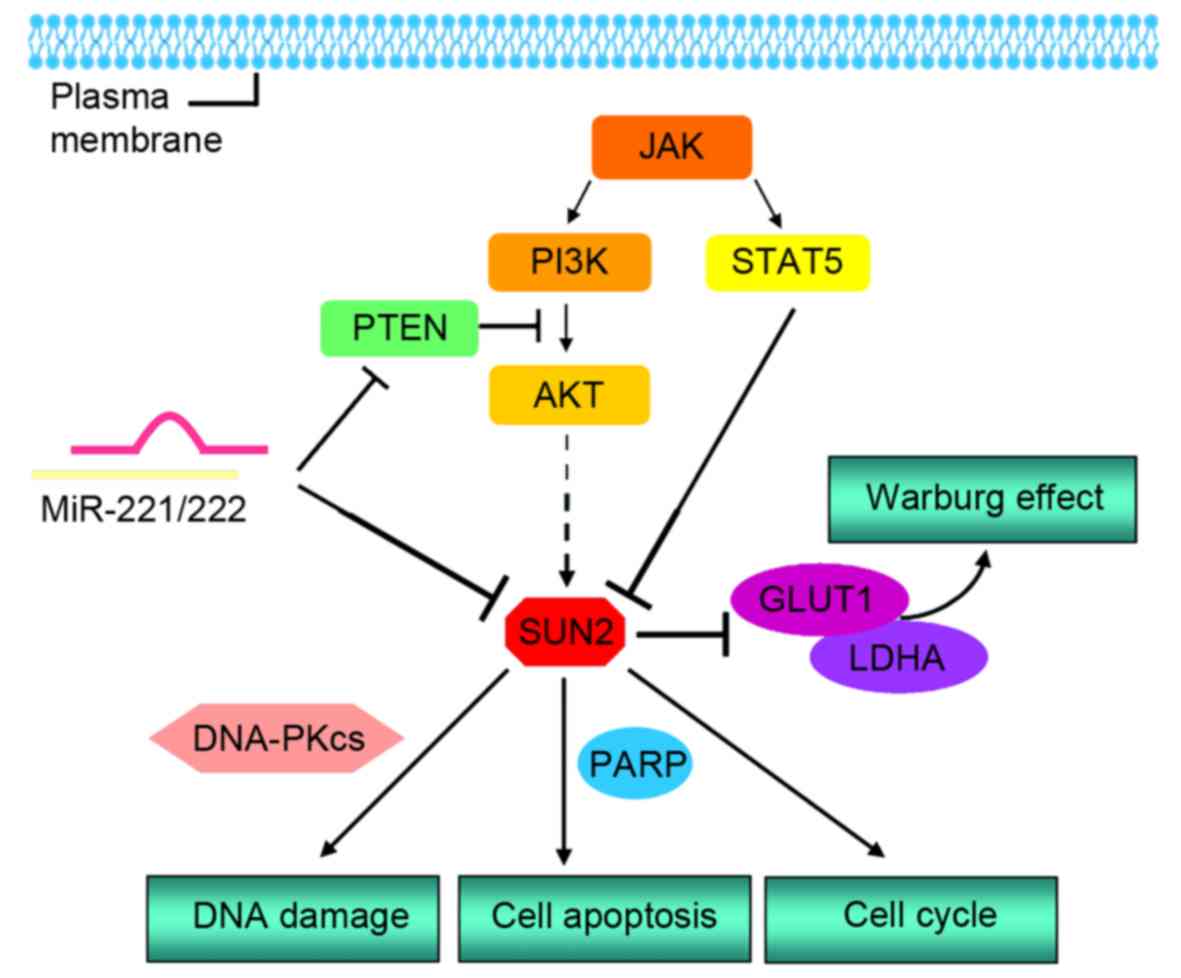
Carcinogenesis. 35:2164–2174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsumoto A, Hieda M, Yokoyama Y, Nishioka
Y, Yoshidome K, Tsujimoto M and Matsuura N: Global loss of a
nuclear lamina component, lamin A/C, and LINC complex components
SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med.
4:1547–1557. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lv XB, Liu L, Cheng C, Yu B, Xiong L, Hu
K, Tang J, Zeng L and Sang Y: SUN2 exerts tumor suppressor
functions by suppressing the Warburg effect in lung cancer. Sci
Rep. 5:179402015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meinke P, Nguyen TD and Wehnert MS: The
LINC complex and human disease. Biochem Soc Trans. 39:1693–1697.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khatau SB, Hale CM, Stewart-Hutchinson PJ,
Patel MS, Stewart CL, Searson PC, Hodzic D and Wirtz D: A
perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci
USA. 106:19017–19022. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Zhu WG and Xu X: Ubiquitin-like
modifications in the DNA damage response. Mutat Res. 803–805.
56–75. 2017.
|
|
16
|
Lei K, Zhu X, Xu R, Shao C, Xu T, Zhuang Y
and Han M: Inner nuclear envelope proteins SUN1 and SUN2 play a
prominent role in the DNA damage response. Curr Biol. 22:1609–1615.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Du X, Cai Z and Greene MI:
Characterization of the structures involved in localization of the
SUN proteins to the nuclear envelope and the centrosome. DNA Cell
Biol. 25:554–562. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hodzic DM, Yeater DB, Bengtsson L, Otto H
and Stahl PD: Sun2 is a novel mammalian inner nuclear membrane
protein. J Biol Chem. 279:25805–25812. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Padmakumar VC, Libotte T, Lu W, Zaim H,
Abraham S, Noegel AA, Gotzmann J, Foisner R and Karakesisoglou I:
The inner nuclear membrane protein Sun1 mediates the anchorage of
Nesprin-2 to the nuclear envelope. J Cell Scie. 118:3419–3430.
2005. View Article : Google Scholar
|
|
20
|
Dreger M, Bengtsson L, Schöneberg T, Otto
H and Hucho F: Nuclear envelope proteomics: Novel integral membrane
proteins of the inner nuclear membrane. Proc Natl Acad Sci USA.
98:11943–11948. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kennedy C, Sebire K, de Kretser DM and
O'Bryan MK: Human sperm associated antigen 4 (SPAG4) is a potential
cancer marker. Cell Tissue Res. 315:279–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tzur YB, Wilson KL and Gruenbaum Y:
SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the
cytoskeleton. Nat Rev Mol Cell Biol. 7:782–788. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Starr DA and Fischer JA: KASH'n Karry: The
KASH domain family of cargo-specific cytoskeletal adaptor proteins.
Bioessays. 27:1136–1146. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hagan I and Yanagida M: The product of the
spindle formation gene sad1+ associates with the fission yeast
spindle pole body and is essential for viability. J Cell Biol.
129:1033–1047. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malone CJ, Fixsen WD, Horvitz HR and Han
M: UNC-84 localizes to the nuclear envelope and is required for
nuclear migration and anchoring during C. elegans development.
Development. 126:3171–3181. 1999.PubMed/NCBI
|
|
26
|
Zhou Z, Du X, Cai Z, Song X, Zhang H,
Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, et al: Structure
of Sad1-UNC84 homology (SUN) domain defines features of molecular
bridge in nuclear envelope. J Biol Chem. 287:5317–5326. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Starr DA and Fridolfsson HN: Interactions
between nuclei and the cytoskeleton are mediated by SUN-KASH
nuclear-envelope bridges. Annu Rev Cell Dev Biol. 26:421–444. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stewart-Hutchinson PJ, Hale CM, Wirtz D
and Hodzic D: Structural requirements for the assembly of LINC
complexes and their function in cellular mechanical stiffness. Exp
Cell Res. 314:1892–1905. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Crisp M, Liu Q, Roux K, Rattner JB,
Shanahan C, Burke B, Stahl PD and Hodzic D: Coupling of the nucleus
and cytoplasm: Role of the LINC complex. J Cell Biol. 172:41–53.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Haque F, Lloyd DJ, Smallwood DT, Dent CL,
Shanahan CM, Fry AM, Trembath RC and Shackleton S: SUN1 interacts
with nuclear lamin A and cytoplasmic nesprins to provide a physical
connection between the nuclear lamina and the cytoskeleton. Mol
Cell Biol. 26:3738–3751. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schmitt J, Benavente R, Hodzic D, Höög C,
Stewart CL and Alsheimer M: Transmembrane protein Sun2 is involved
in tethering mammalian meiotic telomeres to the nuclear envelope.
Proc Natl Acad Sci USA. 104:7426–7431. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee KK, Starr D, Cohen M, Liu J, Han M,
Wilson KL and Gruenbaum Y: Lamin-dependent localization of UNC-84,
a protein required for nuclear migration in Caenorhabditis
elegans. Mol Biol Cell. 13:892–901. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Starr DA and Han M: ANChors away: An actin
based mechanism of nuclear positioning. J Cell Scie. 116:211–216.
2003. View Article : Google Scholar
|
|
34
|
Ostlund C, Folker ES, Choi JC, Gomes ER,
Gundersen GG and Worman HJ: Dynamics and molecular interactions of
linker of nucleoskeleton and cytoskeleton (LINC) complex proteins.
J Cell Sci. 122:4099–4108. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chistiakov DA, Sobenin IA, Orekhov AN and
Bobryshev YV: Human miR-221/222 in physiological and
atherosclerotic vascular remodeling. Biomed Res Int.
2015:3545172015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song J, Ouyang Y, Che J, Li X, Zhao Y,
Yang K, Zhao X, Chen Y, Fan C and Yuan W: Potential value of
miR-221/222 as diagnostic, prognostic and therapeutic biomarkers
for diseases. Front Immunol. 8:562017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu Z, Wu L, Weng D, Xu D, Geng J and Zhao
F: Reduced expression of lamin A/C correlates with poor
histological differentiation and prognosis in primary gastric
carcinoma. J Exp Clin Cancer Res. 28:82009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Broers JL, Raymond Y, Rot MK, Kuijpers H,
Wagenaar SS and Ramaekers FC: Nuclear A-type lamins are
differentially expressed in human lung cancer subtypes. Am J
Pathol. 143:211–220. 1993.PubMed/NCBI
|
|
39
|
Stadelmann B, Khandjian E, Hirt A, Lüthy
A, Weil R and Wagner HP: Repression of nuclear lamin A and C gene
expression in human acute lymphoblastic leukemia and non-Hodgkin's
lymphoma cells. Leuk Res. 14:815–821. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agrelo R, Setien F, Espada J, Artiga MJ,
Rodriguez M, Pérez-Rosado A, Sanchez-Aguilera A, Fraga MF, Piris MA
and Esteller M: Inactivation of the lamin A/C gene by CpG island
promoter hypermethylation in hematologic malignancies, and its
association with poor survival in nodal diffuse large B-cell
lymphoma. J Clin Oncol. 23:3940–3947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Willis ND, Cox TR, Rahman-Casañs SF, Smits
K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M,
Wilson RG, de Bruïne A and Hutchison CJ: Lamin A/C is a risk
biomarker in colorectal cancer. PLoS One. 3:e29882008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kong L, Schäfer G, Bu H, Zhang Y and
Klocker H: Lamin A/C protein is overexpressed in tissue-invading
prostate cancer and promotes prostate cancer cell growth, migration
and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis.
33:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tilli CM, Ramaekers FC, Broers JL,
Hutchison CJ and Neumann HA: Lamin expression in normal human skin,
actinic keratosis, squamous cell carcinoma and basal cell
carcinoma. Br J Dermatol. 148:102–109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Venables RS, McLean S, Luny D, Moteleb E,
Morley S, Quinlan RA, Lane EB and Hutchison CJ: Expression of
individual lamins in basal cell carcinomas of the skin. Br J
Cancer. 84:512–519. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ellenbroek SI and van Rheenen J: Imaging
hallmarks of cancer in living mice. Nat Rev Cancer. 14:406–418.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ciccia A and Elledge SJ: The DNA damage
response: Making it safe to play with knives. Mol Cell. 40:179–204.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Paull TT, Rogakou EP, Yamazaki V,
Kirchgessner CU, Gellert M and Bonner WM: A critical role for
histone H2AX in recruitment of repair factors to nuclear foci after
DNA damage. Curr Biol. 10:886–895. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Harper JW and Elledge SJ: The DNA damage
response: Ten years after. Mol Cell. 28:739–745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sobol RW, Horton JK, Kühn R, Gu H, Singhal
RK, Prasad R, Rajewsky K and Wilson SH: Requirement of mammalian
DNA polymerase-beta in base-excision repair. Nature. 379:183–186.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Majidinia M and Yousefi B: DNA repair and
damage pathways in breast cancer development and therapy. DNA
Repair (Amst). 54:22–29. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu
T, Xu R and Han M: SUN1/2 and Syne/Nesprin-1/2 complexes connect
centrosome to the nucleus during neurogenesis and neuronal
migration in mice. Neuron. 64:173–187. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Davidson D, Amrein L, Panasci L and Aloyz
R: Small molecules, inhibitors of DNA-PK, targeting DNA repair, and
beyond. Front Pharmacol. 4:52013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Stinson S, Lackner MR, Adai AT, Yu N, Kim
HJ, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, et
al: miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1)
promotes epithelial-to-mesenchymal transition in breast cancer. Sci
Signal. 4:pt52011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hwang MS, Yu N, Stinson SY, Yue P, Newman
RJ, Allan BB and Dornan D: miR-221/222 targets adiponectin receptor
1 to promote the epithelial-to-mesenchymal transition in breast
cancer. PLoS One. 8:e665022013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang
Z, Li L, Qin J, Chen Y, Cho WC, et al: miR-221/222 promotes S-phase
entry and cellular migration in control of basal-like breast cance.
Molecules. 19:7122–7137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gan R, Yang Y, Yang X, Zhao L, Lu J and
Meng QH: Downregulation of miR-221/222 enhances sensitivity of
breast cancer cells to tamoxifen through upregulation of TIMP3.
Cancer Gene Ther. 21:290–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pichiorri F, Palmieri D, De Luca L,
Consiglio J, You J, Rocci A, Talabere T, Piovan C, Lagana A,
Cascione L, et al: In vivo NCL targeting affects breast cancer
aggressiveness through miRNA regulation. J Exp Med. 210:951–968.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Falkenberg N, Anastasov N, Rappl K,
Braselmann H, Auer G, Walch A, Huber M, Höfig I, Schmitt M, Höfler
H, et al: MiR-221/-222 differentiate prognostic groups in advanced
breast cancers and influence cell invasion. Br J Cancer.
109:2714–2723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gramantieri L, Fornari F, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L and
Negrini M: MicroRNA-221 targets Bmf in hepatocellular carcinoma and
correlates with tumor multifocality. Clin Cancer Res. 15:5073–5081.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bae HJ, Jung KH, Eun JW, Shen Q, Kim HS,
Park SJ, Shin WC, Yang HD, Park WS, Lee JY and Nam SW: MicroRNA-221
governs tumor suppressor HDAC6 to potentiate malignant progression
of liver cancer. J Hepatol. 63:408–419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Callegari E, Elamin BK, Giannone F,
Milazzo M, Altavilla G, Fornari F, Giacomelli L, D'Abundo L,
Ferracin M, Bassi C, et al: Liver tumorigenicity promoted by
microRNA-221 in a mouse transgenic model. Hepatology. 56:1025–1033.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li J, Wang Y, Yu W, Chen J and Luo J:
Expression of serum miR-221 in human hepatocellular carcinoma and
its prognostic significance. Biochem Biophys Res Commun. 406:70–73.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Duan M, Yao H, Hu G, Chen X, Lund AK and
Buch S: HIV Tat induces expression of ICAM-1 in HUVECs:
implications for miR-221/-222 in HIV-associated cardiomyopathy.
PLoS One. 8:e601702013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer
Res. 3:465–477. 2013.PubMed/NCBI
|
|
66
|
Passadouro M, Pedroso de Lima MC and
Faneca H: MicroRNA modulation combined with sunitinib as a novel
therapeutic strategy for pancreatic cancer. Int J Nanomedicine.
9:3203–3217. 2014.PubMed/NCBI
|
|
67
|
Tanaka R, Tomosugi M, Horinaka M, Sowa Y
and Sakai T: Metformin causes G1-phase arrest via down-regulation
of MiR-221 and enhances TRAIL sensitivity through DR5 Up-regulation
in pancreatic cancer cells. PLoS One. 10:e01257792015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lee C, He H, Jiang Y, Di Y, Yang F, Li J,
Jin C and Fu D: Elevated expression of tumor miR-222 in pancreatic
cancer is associated with Ki67 and poor prognosis. Med Oncol.
30:7002013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu W, Song N, Yao H, Zhao L, Liu H and Li
G: miR-221 and miR-222 simultaneously target RECK and regulate
growth and invasion of gastric cancer cells. Med Sci Monit.
21:2718–2725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F,
Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z and Chun-Sheng
K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li
JY, Yuasa Y, Kang D, Kim YS and You WC: Identification of serum
microRNAs as novel non-invasive biomarkers for early detection of
gastric cancer. PLoS One. 7:e336082012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fu Z, Qian F, Yang X, Jiang H, Chen Y and
Liu S: Circulating miR-222 in plasma and its potential diagnostic
and prognostic value in gastric cancer. Med Oncol. 31:1642014.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sun K, Wang W, Zeng JJ, Wu CT, Lei ST and
Li GX: MicroRNA-221 inhibits CDKN1C/p57 expression in human
colorectal carcinoma. Acta Pharmacol Sin. 32:375–384. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Qin J and Luo M: MicroRNA-221 promotes
colorectal cancer cell invasion and metastasis by targeting RECK.
FEBS Lett. 588:99–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang
Y, Liu Z, Cao X, Chen P, Liu Z, et al: A microRNA 221- and
222-mediated feedback loop maintains constitutive activation of
NFκB and STAT3 in colorectal cancer cells. Gastroenterology.
147:847–859, e811. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ and
Li GX: Anti-miRNA-221 sensitizes human colorectal carcinoma cells
to radiation by upregulating PTEN. World J Gastroenterol.
19:9307–9317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye
S, Ling S, Jiang L, Tian Y and Lin TY: Circulating miR-221 directly
amplified from plasma is a potential diagnostic and prognostic
marker of colorectal cancer and is correlated with p53 expression.
J Gastroenterol Hepatol. 25:1674–1680. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang C, Zhang J, Hao J, Shi Z, Wang Y,
Han L, Yu S, You Y, Jiang T, Wang J, et al: High level of
miR-221/222 confers increased cell invasion and poor prognosis in
glioma. J Transl Med. 10:1192012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Medina R, Zaidi SK, Liu CG, Stein JL, van
Wijnen AJ, Croce CM and Stein GS: MicroRNAs 221 and 22χ2 bypass
quiescence and compromise cell survival. Cancer Res. 68:2773–2780.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang C, Kang C, You Y, Pu P, Yang W, Zhao
P, Wang G, Zhang A, Jia Z, Han L and Jiang H: Co-suppression of
miR-221/222 cluster suppresses human glioma cell growth by
targeting p27kip1 in vitro and in vivo. Int J Oncol. 34:1653–1660.
2009.PubMed/NCBI
|
|
82
|
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han
L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, et al: MiR-221 and
miR-222 target PUMA to induce cell survival in glioblastoma. Mol
Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Quintavalle C, Garofalo M, Zanca C, Romano
G, Iaboni M, del Basso De Caro M, Martinez-Montero JC, Incoronato
M, Nuovo G, Croce CM and Condorelli G: miR-221/222 overexpession in
human glioblastoma increases invasiveness by targeting the protein
phosphate PTPµ. Oncogene. 31:858–868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chen L, Zhang J, Han L, Zhang A, Zhang C,
Zheng Y, Jiang T, Pu P, Jiang C and Kang C: Downregulation of
miR-221/222 sensitizes glioma cells to temozolomide by regulating
apoptosis independently of p53 status. Oncol Rep. 27:854–860.
2012.PubMed/NCBI
|
|
85
|
Li W, Guo F, Wang P, Hong S and Zhang C:
miR-221/222 confers radioresistance in glioblastoma cells through
activating Akt independent of PTEN status. Curr Mol Med.
14:185–195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Di Martino MT, Gullà A, Cantafio ME,
Lionetti M, Leone E, Amodio N, Guzzi PH, Foresta U, Conforti F,
Cannataro M, et al: In vitro and in vivo anti-tumor activity of
miR-221/222 inhibitors in multiple myeloma. Oncotarget. 4:242–255.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Di Martino MT, Gullà A, Gallo Cantafio ME,
Altomare E, Amodio N, Leone E, Morelli E, Lio SG, Caracciolo D,
Rossi M, et al: In vitro and in vivo activity of a novel locked
nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma
cells. PLoS One. 9:e896592014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gullà A, Di Martino MT, Gallo Cantafio ME,
Morelli E, Amodio N, Botta C, Pitari MR, Lio SG, Britti D, Stamato
MA, et al: A 13 mer LNA-i-miR-221 inhibitor restores drug
sensitivity in melphalan-refractory multiple myeloma cells. Clin
Cancer Res. 22:1222–1233. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Huang JJ, Yu J, Li JY, Liu YT and Zhong
RQ: Circulating microRNA expression is associated with genetic
subtype and survival of multiple myeloma. Med Oncol. 29:2402–2408.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kanemaru H, Fukushima S, Yamashita J,
Honda N, Oyama R, Kakimoto A, Masuguchi S, Ishihara T, Inoue Y,
Jinnin M and Ihn H: The circulating microRNA-221 level in patients
with malignant melanoma as a new tumor marker. J Dermatol Sci.
61:187–193. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Felicetti F, De Feo A, Coscia C, Puglisi
R, Pedini F, Pasquini L, Bellenghi M, Errico MC, Pagani E and Carè
A: Exosome-mediated transfer of miR-222 is sufficient to increase
tumor malignancy in melanoma. J Transl Med. 14:562016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Alamolhodaei NS, Behravan J, Mosaffa F and
Karimi G: MiR 221/222 as new players in tamoxifen resistance. Curr
Pharm Des. 22:6946–6955. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Linher-Melville K and Singh G: The complex
roles of STAT3 and STAT5 in maintaining redox balance: Lessons from
STAT-mediated xCT expression in cancer cells. Mol Cell Endocrinol.
451:40–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li F, He X, Ye D, Lin Y, Yu H, Yao C,
Huang L, Zhang J, Wang F, Xu S, et al: NADP(+)-IDH mutations
promote hypersuccinylation that impairs mitochondria respiration
and induces apoptosis resistance. Mol Cell. 60:661–675. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu
ML, Hsu CM and Yang MY: Altered expression of SIRT gene family in
head and neck squamous cell carcinoma. Tumour Biol. 34:1847–1854.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kim DH, Kwak Y, Kim ND and Sim T:
Antitumor effects and molecular mechanisms of ponatinib on
endometrial cancer cells harboring activating FGFR2 mutations.
Cancer Biol Ther. 17:65–78. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Xiangyun Y, Xiaomin N, Linping G, Yunhua
X, Ziming L, Yongfeng Y, Zhiwei C and Shun L: Desuccinylation of
pyruvate kinase M2 by SIRT5 contributes to antioxidant response and
tumor growth. Oncotarget. 8:6984–6993. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Osborne B, Bentley NL, Montgomery MK and
Turner N: The role of mitochondrial sirtuins in health and disease.
Free Radic Biol Med. 100:164–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lu W, Zuo Y, Feng Y and Zhang M: SIRT5
facilitates cancer cell growth and drug resistance in non-small
cell lung cancer. Tumour Biol. 35:10699–10705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Kyrylenko S, Kyrylenko O, Suuronen T and
Salminen A: Differential regulation of the Sir2 histone deacetylase
gene family by inhibitors of class I and II histone deacetylases.
Cell Mol Life Sci. 60:1990–1997. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ding S, Khoury-Hanold W, Iwasaki A and
Robek MD: Epigenetic reprogramming of the type III interferon
response potentiates antiviral activity and suppresses tumor
growth. PLoS Biol. 12:e10017582014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Vanhaecke T, Papeleu P, Elaut G and
Rogiers V: Trichostatin A-like hydroxamate histone deacetylase
inhibitors as therapeutic agents: toxicological point of view. Curr
Med Chem. 11:1629–1643. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhou W, Liotta LA and Petricoin EF: The
Warburg effect and mass spectrometry-based proteomic analysis.
Cancer Genomics Proteomics. 14:211–218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Sica A, Strauss L, Consonni FM, Travelli
C, Genazzani A and Porta C: Metabolic regulation of suppressive
myeloid cells in cancer. Cytokine Growth Factor Rev. 35:27–35.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Cairns RA: Drivers of the Warburg
phenotype. Cancer J. 21:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
He X, Li C, Ke R, Luo L and Huang D:
Down-regulation of adenosine monophosphate-activated protein kinase
activity: A driver of cancer. Tumour Biol. 39:10104283176975762017.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo
F, Lyssiotis CA, Aldape K, Cantley LC and Lu Z: ERK1/2-dependent
phosphorylation and nuclear translocation of PKM2 promotes the
Warburg effect. Nat Cell Biol. 14:1295–1304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Liu J, Zhang C, Wu R, Lin M, Liang Y, Liu
J, Wang X, Yang B and Feng Z: RRAD inhibits the Warburg effect
through negative regulation of the NF-κB signaling. Oncotarget.
6:14982–14992. 2015.PubMed/NCBI
|
|
109
|
Dueregger A, Schöpf B, Eder T, Höfer J,
Gnaiger E, Aufinger A, Kenner L, Perktold B, Ramoner R, Klocker H
and Eder IE: Differential utilization of dietary fatty acids in
benign and malignant cells of the prostate. PLoS One.
10:e01357042015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
He JX, Yang CH and Miao ZH:
Poly(ADP-ribose) polymerase inhibitors as promising cancer
therapeutics. Acta Pharmacol Sin. 31:1172–1180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Livraghi L and Garber JE: PARP inhibitors
in the management of breast cancer: current data and future
prospects. BMC Med. 13:1882015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Evans T and Matulonis U: PARP inhibitors
in ovarian cancer: Evidence, experience and clinical potential.
Ther Adv Med Oncol. 9:253–267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Rajawat J, Shukla N and Mishra DP:
Therapeutic targeting of poly(ADP-Ribose) polymerase-1 (PARP1) in
cancer: Current developments, therapeutic strategies, and future
opportunities. Med Res Rev. 37:1461–1491. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Vici P, Mariani L, Pizzuti L, Sergi D, Di
Lauro L, Vizza E, Tomao F, Tomao S, Mancini E, Vincenzoni C, et al:
Emerging biological treatments for uterine cervical carcinoma. J
Cancer. 5:86–97. 2014. View Article : Google Scholar : PubMed/NCBI
|