Introduction
Nasopharyngeal cancer (NPC) is one of the most
common cancer types in South China. In the early stages of the
disease, NPC is usually asymptomatic, with the majority of patients
being diagnosed in the middle or advanced stages. Radiotherapy is
the primary treatment for NPC; however, 40–60% of patients relapse
following definitive radiotherapy (1–5). Relapse
primarily occurs locally, in the neck region, or a combination of
the two. The majority of relapse cases appear between 2 and 3 years
after radiotherapy.
Relapse and metastasis of NPC are direct causes of
mortality. Radiological examination is the major method of
detecting relapse and metastasis. However, in order to be
visualized radiologically, lymph node lesions resulting from
relapse and metastasis must reach a maximum diameter of 0.8 cm,
according to the Union of International Cancer Control (6), at which point the chance of successful
treatment begins to decrease (7).
Preceding relapse and metastasis, tumour cells gain access to the
peripheral blood, where they are referred to as circulating tumour
cells (CTCs) (8). Tumour burden is
directly associated with the number of CTCs in the blood;
therefore, tumour status may be determined by CTC detection
(9).
Aptamers are oligonucleotide or peptide molecules,
which, through their unique secondary structure, bind to a specific
target molecule (10). Aptamers may
be screened using the Systematic Evolution of Ligands by
Exponential Enrichment technique, and due to ease of synthesis,
storage and transportation, have been employed in an extensive
number of applications (11).
Microfluidic chips contain complex fluids that are manipulated,
observed, detected and controlled at the micron level. In the
present study, the microfluidic reaction chamber of a chip was
modified for the incorporation of an array of bypass columns. This
design increased the area/volume ratio of the fluid environment,
thus increasing the reaction efficiency of the microfluidic chip
(12,13). Aptamers targeting NPC cell markers
were prepared in a preliminary study (14) and subsequently affixed to the inner
wall of the reaction chamber within the microfluidic chip. This
modified aptamer-bound microfluidic chip is capable of capturing
NPC cells from peripheral blood, for the non-invasive diagnosis of
NPC.
Materials and methods
Modelling of an array of bypass
columns in the microfluidic chip
The shape, arrangement and length of each major axis
of the bypass column were simulated using Fluent 6.3.26 software
(ANSYS, Inc., Canonsburg, PA, USA). Alterations in the magnitude of
shear force and flow rate were analysed in the microarray. The
columns were designed with a circular, elliptical (ratio of major
axis to minor axis, 10:7) or square form, and arranged in either an
aligned or staggered fashion. Flow rates between 10 and 2,000
µm/sec were employed. Multiple arrays of bypass columns were
simulated using the finite element method (FEM) (15). The microarray with optimal performance
was selected for preparation.
Preparation of the microfluidic
chip
The polydimethylsiloxane (PDMS) microfluidic chip
was manufactured by Suzhou Wenhao Chip Technology Co., Ltd.
(Suzhou, China). The SU-8 mould was prepared via a
photolithographic process (16,17), while
the PDMS array and cover slips were prepared by PDMS demoulding
technology. The typical final step in preparing a closed chip for
microfluidic experiments was accomplished using the bonding process
(18,19). The chip was composed of an inlet,
reaction chamber and an outlet; the reaction chamber was designed
with an array of circular bypass columns in a staggered
arrangement. The diameter of the bypass columns was 40 µm, and the
spacing between columns was 100 µm.
Modification of the inner wall of the
microfluidic chip
Biotin-labelled aptamers (Sangon Biotech Co., Ltd.,
Shanghai) were bound to the inner wall of the reaction chamber of
the microfluidic chip using a two-step approach, firstly by binding
the avidin (Thermo Fisher Scientific, Inc.) through physical
absorption to the inner wall of the reaction chamber, followed by
the immobilization of the biotin-labelled aptamers to the inner
wall of the reaction chamber using the biotin-avidin mechanism.
Avidin is a glycoprotein and each avidin molecule consists of four
subunits and each subunit can bind to one biotin molecule.
Immobilization of the aptamers to the inner wall of the reaction
chamber was determined using fluorescence microscopy
(magnification, ×400).
Aptamer synthesis and buffers
Aptamers were synthesized by Takara Biotechnology
Co., Ltd., (Dalian, China). The aptamer sequence is as follows:
Fluorescein
isothiocyanate-5′-ACCGACCGTGCTGGACTCTACCGCGCAGTGAGGTGAGTGGGGTAGGTTGTTACGTTTCCAGTATGAGCGAGCGTTGCG-3′-Biotin.
Buffers used in this study included elution buffer (DPBS containing
4.5 g/l glucose and 5 mM MgCl2), binding buffer (washing
buffer containing 0.1 mg/ml yeast tRNA and 1 mg bovine serum
albumin), and capture buffer (binding buffer and Histopaque-1119
from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at a 1:1
ratio).
Cell culture
The NPC cell line C666, used to screen aptamer in
the present study, human gastric cancer cell line SGC7901, human
colorectal cancer cell line HT29, human ovarian cancer cell line
SKOV3, human cervical cancer cell line Hela, and normal
nasopharyngeal epithelial cell line NP69 were preserved at our
laboratory. Keratinocyte-serum-free medium (SFM), 1640 medium, and
foetal bovine serum (FBS) were purchased from Gibco (Thermo Fisher
Scientific, Inc.). C666, SGC7901, HT29, A549, SKOV3 and HeLa cells
were cultured in 1640 medium containing 10% heat-inactivated FBS
and 100 U/ml penicillin-streptomycin. NP69 cells were cultured in
keratinocyte-SFM. All cells were washed twice with Dulbecco's
phosphate-buffered saline (DPBS) and treated with 0.25% trypsin.
Following trypsinization, the cells were either harvested or
routine passage was performed.
Cell capture experiment
Prior to cell capture, the concentration of the cell
suspension was adjusted to 106 cells/ml, and cells were
stained with Vybrant DiI or DiD dye (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Staining was performed at 37°C for 5 min
according to the manufacturer's protocol. The cells were rinsed
once with wash buffer and resuspended in capture buffer. The cell
concentration was adjusted to 106 cells/ml, and the
solution was kept on ice prior to use.
The microfluidic chip was treated with 1 mg/ml
avidin for 1 min and washed three times with binding buffer. After
incubating the chip in 30 µM of biotin-labelled aptamers for 1 min,
the chip was again washed three times with binding buffer. Finally,
1 ml of cell suspension (C666 and NP69) was prepared using capture
buffer, and injected into the microfluidic chip using a micro-pump.
To elute the target cells, the chip was washed three times with
binding buffer. To verify the capture efficiency of the modified
microfluidic chip, a flat microfluidic chip was constructed and the
capture of target cells for the two methods was compared by t-test
(n=4).
Separation of tumour cells from whole
blood
Whole blood samples (7.5 ml) were collected from
healthy volunteers and combined with an EDTA anticoagulant.
Physical absorption and aptamer immobilization were performed as
described above. Avidin were bound to the inner wall of the
reaction chamber by physical absorption, followed by the
immobilization of the biotin-labelled aptamers to the inner wall of
the reaction chamber. To prevent cell loss during trypsinisation,
NPC cells were treated with enzyme-free cell dissociation solution,
and subsequently added to the whole blood of the healthy
volunteers. The NPC cells from the whole blood samples were
captured by the microfluidic chip; the final concentrations of NPC
cells were 10, 100, 1,000, and 10,000 cells/ml, and each test was
performed in triplicate.
Capture of NPC cells
Whole blood samples containing a C666 cell
suspension or NP69, SGC7901, HT29, SKOV3 and Hela cells were
transported to the reaction chamber via the inlet of the
microfluidic device. The outlet of the reaction chamber was
connected to a cell harvester. To avoid introducing variation into
the whole blood samples, no buffer was added during cell
separation. A micro-magnetic stirrer was placed near the inlet and
outlet to prevent the cell aggregation and ensure uniform
distribution within the sample.
Statistical analysis
The shear force within different column types, fluid
flow rate and length of the major axis was analysed by independent
sample t-test for two-group comparison, and by analysis of variance
(ANOVA) using the least significant difference, Bonferroni and
Dunnett's T3 post hoc accordingly, for the comparison of more than
two groups. The purity of captured cells and the capture efficiency
at different lengths of the major axis and flow rate were
determined by Pearson correlation analysis. The difference in
capture efficiency between the ordinary and the modified
microfluidic chip was analysed by t-test. To confirm the
association of input and capture of microfluidic chip, the
correlation of cell number of input and capture were analysed by
Pearson correlation analysis. All data were anlyzed by SPSS 20.0
(IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate
a statistically significant.
Results
Effects of different parameters on
bypass column efficiency
Different arrays of bypass columns were simulated
using Fluent software. The impact of column shape and fluid flow
rate on the flow of NPC cells was examined. The fluid flow rate at
the inlet had a considerable impact on the shear force within the
micro-chamber. The flow rate was gradually increased, and the
maximum shear force in different arrays was compared. As the flow
rate near the inlet increased, the maximum shear force also
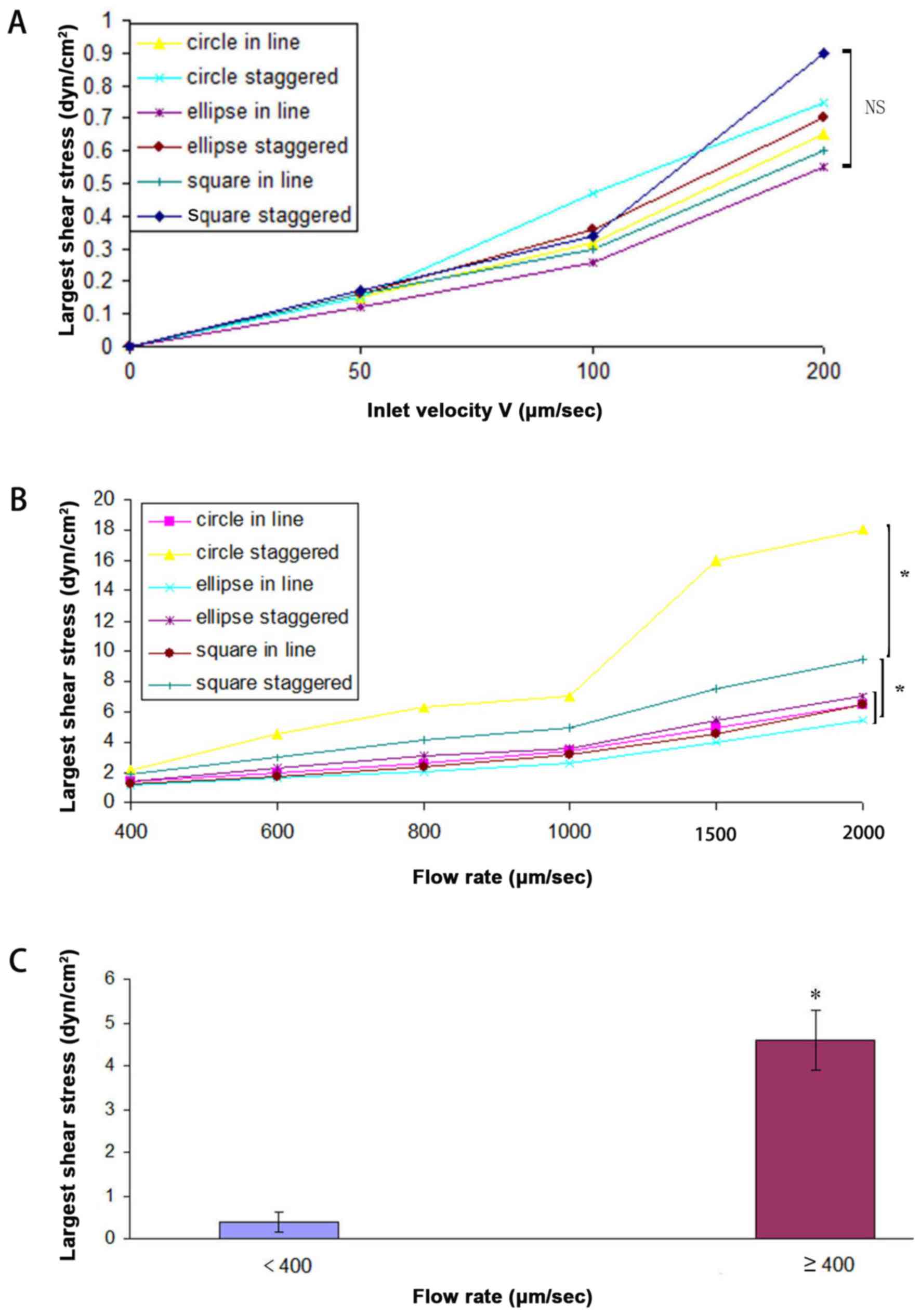
increased (Fig. 1). With a flow rate
<400 µm/sec (50, 100, 200 µm/sec), no significant difference was
observed in the maximum shear force across different arrays by
ANOVA (P>0.05; Fig. 1A). At a flow
rate ≥400 µm/sec (400, 600, 800, 1,000, 1,500 and 2,000 µm/sec),
the shear force significantly differed between circle staggered and
circle in line, circle staggered and ellipse in line, circle
staggered and ellipse staggered, circle staggered and square in
line, circle staggered and square staggered, square staggered and
circle in line, square staggered and ellipse in line, square
staggered and ellipse staggered, square staggered and square in
line, ellipse staggered and ellipse in line, and circle in line and
ellipse in line arrangements (Fig.
1B). Maximum shear force was observed for the circle columns in
a staggered arrangement, followed by the square columns in a
staggered arrangement. The shear force of the elliptical columns in
a staggered arrangement did not differ from that of the elliptical
columns in an aligned arrangement (as determined by ANOVA). As
determined by t-test (P<0.05), the shear force with flow rate
<400 µm/sec was lower compared with that observed for flow rate
≥400 µm/sec (Fig. 1C). Murthy et
al (20) determined the optimal
shear force for the absorption of cells near the micro-columns to
be <9.2 dyn/cm2. At a flow rate of 1,000 µm/sec, the
maximum shear force was <9.2 dyn/cm2; however, at
2,000 µm/sec, the maximum shear force exceeded 9.2
dyn/cm2 in circle staggered. Therefore, the smallest
flow rate of 50 µm/sec was selected and the effect of varying
column arrangement on the fluid flow pattern was determined.
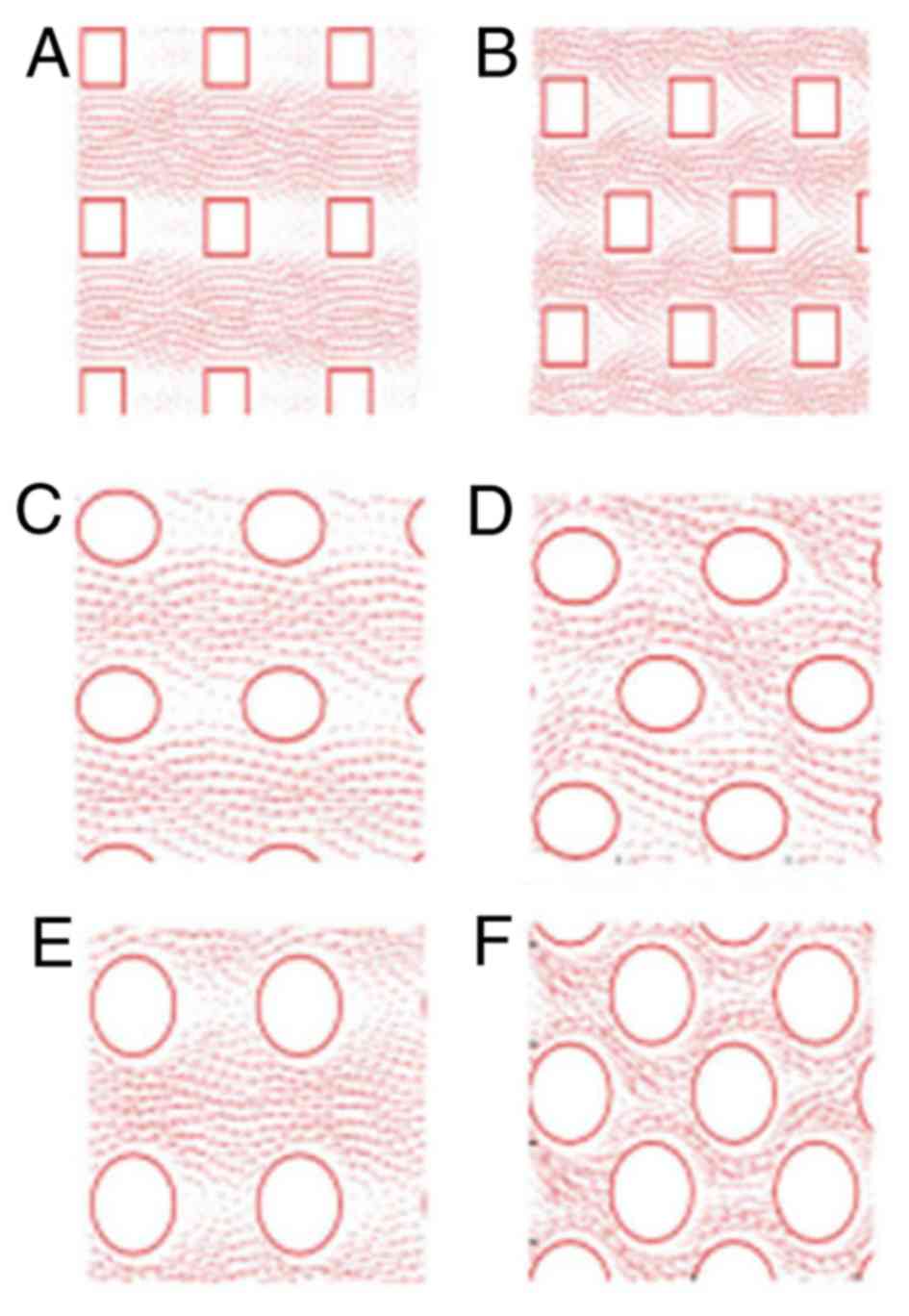
Fig. 2 illustrates that in an
staggered arrangement, the fluid flowed through the centre of the
array, and little fluid flowed around the columns. The majority of
the fluid was concentrated around the staggered column, the
so-called ‘flow around the column’ phenomenon (21). Therefore, the staggered arrangement
was superior to that of the aligned arrangement of columns for
fluid flow through the microchip. The major axis of the bypass
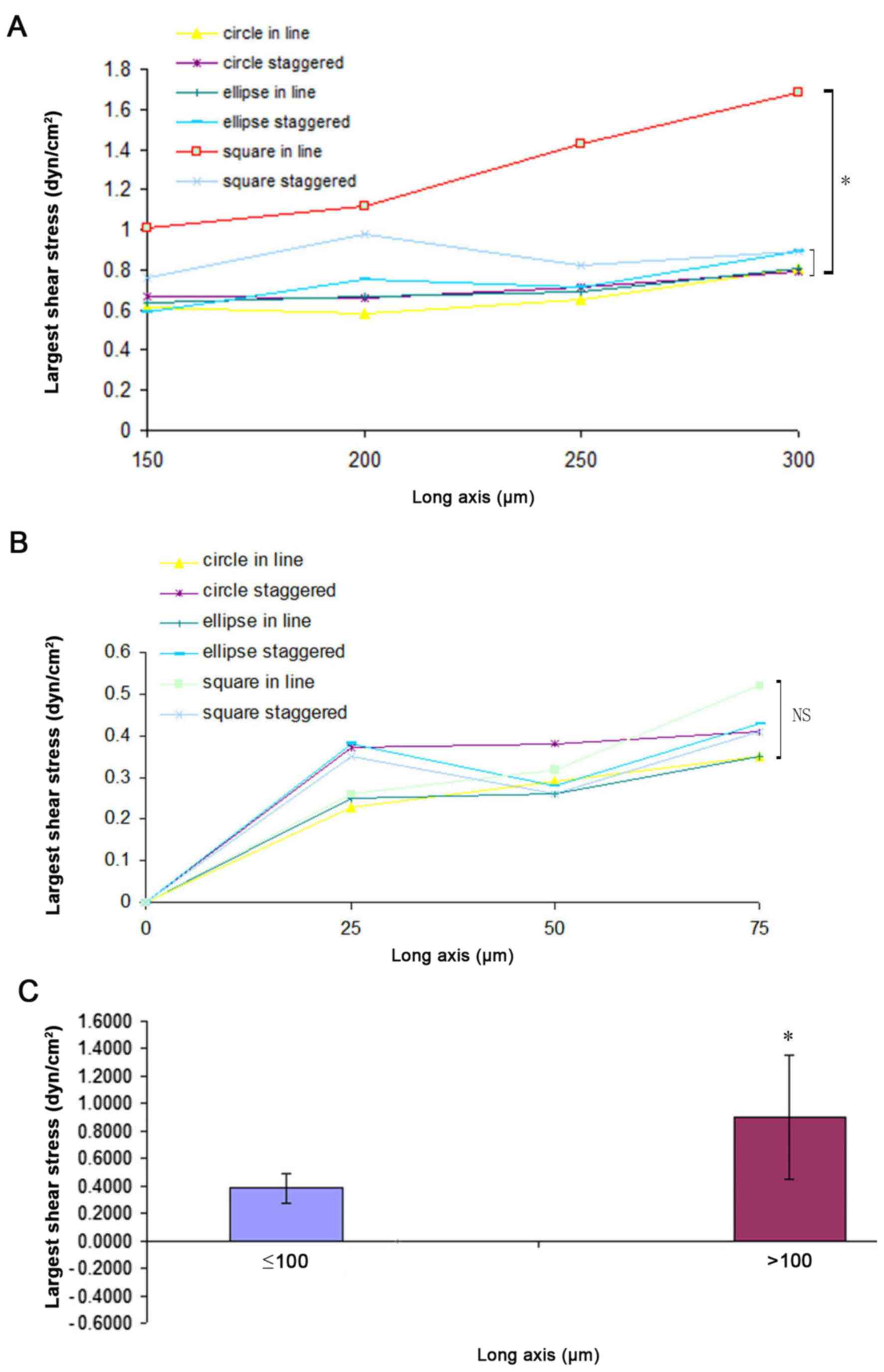
columns also influences shear force. At a fixed flow rate of 100
µm/sec, the effect of major axis length on shear force was
examined. Fig. 3 indicates that as
the length of the major axis increased, the shear force also
increased. When the length of the major axis was >100 µm (150,
200, 250 and 300 µm), the shear force of square in line was bigger
compared with the circle in line, ellipse in line, ellipse
staggered, square staggered and circle staggered (P<0.05;
Fig. 3A). The shear force of square
staggered was bigger compared with the circle in line and ellipse
in line arrangements by ANOVA (P<0.05). When the length of the
major axis was ≤100 µm, no significant difference was observed in
the maximum shear force across different arrays by ANOVA
(P>0.05; Fig. 3B). As determined
by t-test (P<0.05), the shear force with a major axis length
≤100 µm was lower compared with that observed for values >100 µm
(Fig. 3C). Thus, a major axis length
of between 25 and 100 µm was selected for the subsequent
experiments.
Separation of target cells by
microfluidics
Based on the results of the microfluidic chip
simulations, a range of 20–45 µm was selected as the major axis
length for the bypass columns; microfluidic chips were prepared
with varying lengths from within this range. The effect of major
axis length and inlet flow rate on the capture efficiency and
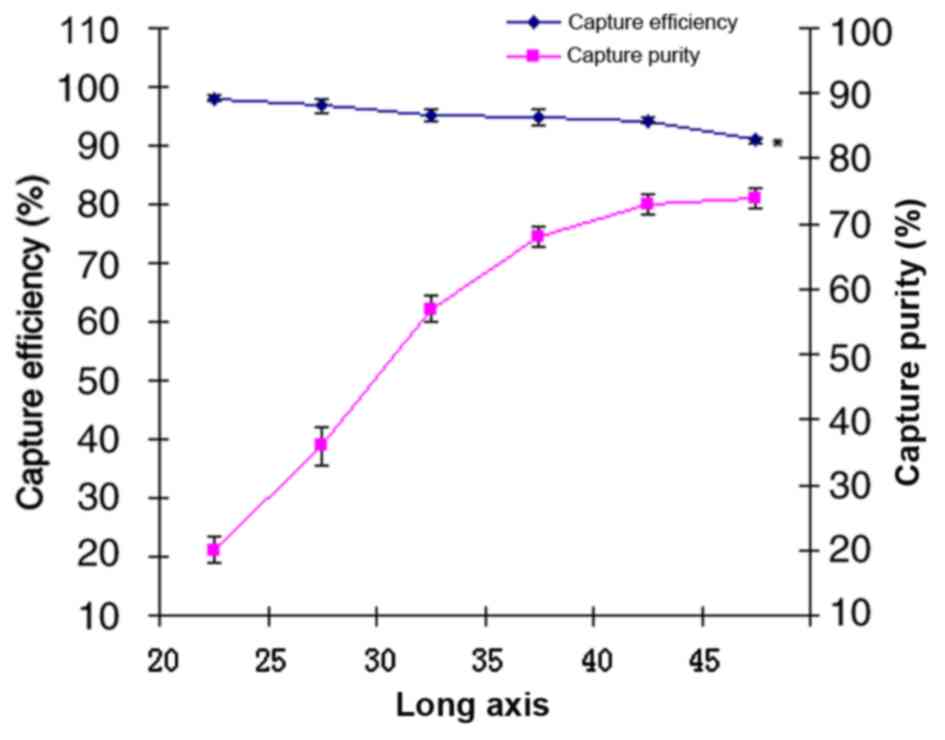
purity of target cells was examined. Fig.
4 illustrates that as the major axis length increased, the
purity of captured cells also increased, whilst the capture
efficiency decreased. Pearson correlation analysis indicated a
significant positive correlation between axis length and the purity
of the captured cells (R=0.94; P<0.01), a significant negative
correlation between axis length and the capture efficiency
(R=−0.95; P<0.01), and negative correlation between the capture
efficiency and the purity of captured (R=−0.86; P<0.05) at a
range of 20–45 µm axis length. With the increase of capture
efficiency, the purity of capture decreased therefore, the proper
long axis was maximum number of cells obtained from colligating
capture purity and capture efficiency (input cells × capture purity
× capture efficiency). When the long axis was 45 µm, the target
cells were captured the most. Therefore, the length of the major
axis was set at 45 µm and the effect of different flow rates on the
capture efficiency and purity of the captured cells were also
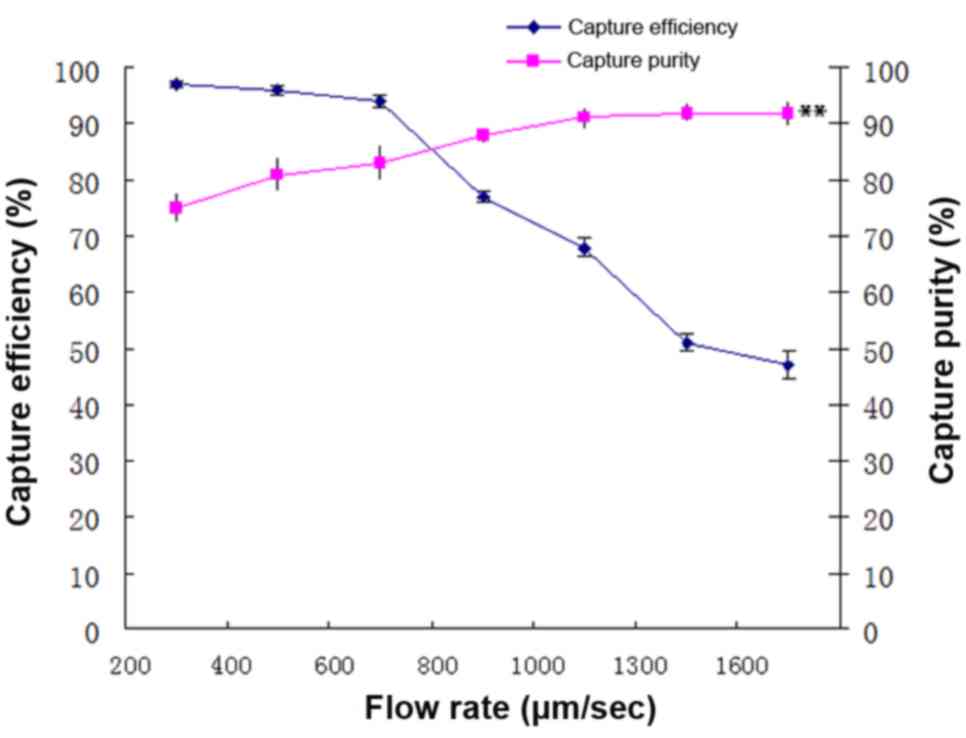
analysed. Fig. 5 illustrates that as
the flow rate increased the capture efficiency decreased, whilst
the purity of captured cells increased when the long axis was
fixed. A negative correlation was indicated between flow rate and
capture efficiency (R=−0.97; P<0.01), a positive correlation
between flow rate and capture purity (R=0.92; P<0.01), and a
negative correlation between capture efficiency and the capture
purity (R=−0.92; P<0.01). Therefore, the proper flow rate was
the maximum number of cells obtained from colligating capture
purity and the capture efficiency (input cells × capture purity ×
capture efficiency). At flow rates between 200 and 800 µm/sec, the
capture efficiency and purity of the captured cells were >70%.
The number of captured target cells was the highest at a flow rate
of 700 µm/sec; therefore, for subsequent experiments, a major axis
length of 45 µm, and a flow rate of 700 µm/sec were selected.
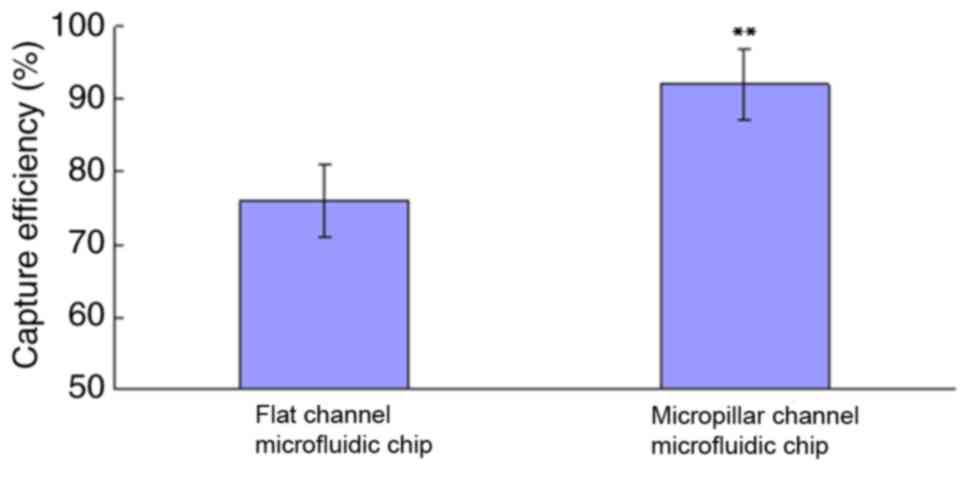
Fig. 6 compares the capture
efficiency between a conventional flat channel microfluidic chip
and modified version, the microfluidic chip reaction chamber with
an array of bypass columns defined as the micropillar channel
microfluidic chip. The t-test confirmed that the capture efficiency
of the modified microfluidic chip was considerably higher compared
with that of the conventional one (P<0.01).
To verify the enrichment capacity of microfluidic
chip for NPC cells, C666 cells, a nasopharyngeal cancer cell line
used to screen aptamer in the present study, were used as target
cells, and NP69, SGC7901, HT29, SKOV3 and Hela were selected as
controls. C666 cells were enriched using a microfluidic chip bound
with biotin-labelled aptamers. For 1 ml of cells, a 1:1,000 ratio
of target to control cells was sought, at a final concentration of
106 cells/ml. The two cell types were stained prior to
the experimentation; C666 cells were prestained with Vybrant DiI
dye (red), and NP69, SGC7901, HT29, SKOV3 and Hela were prestained
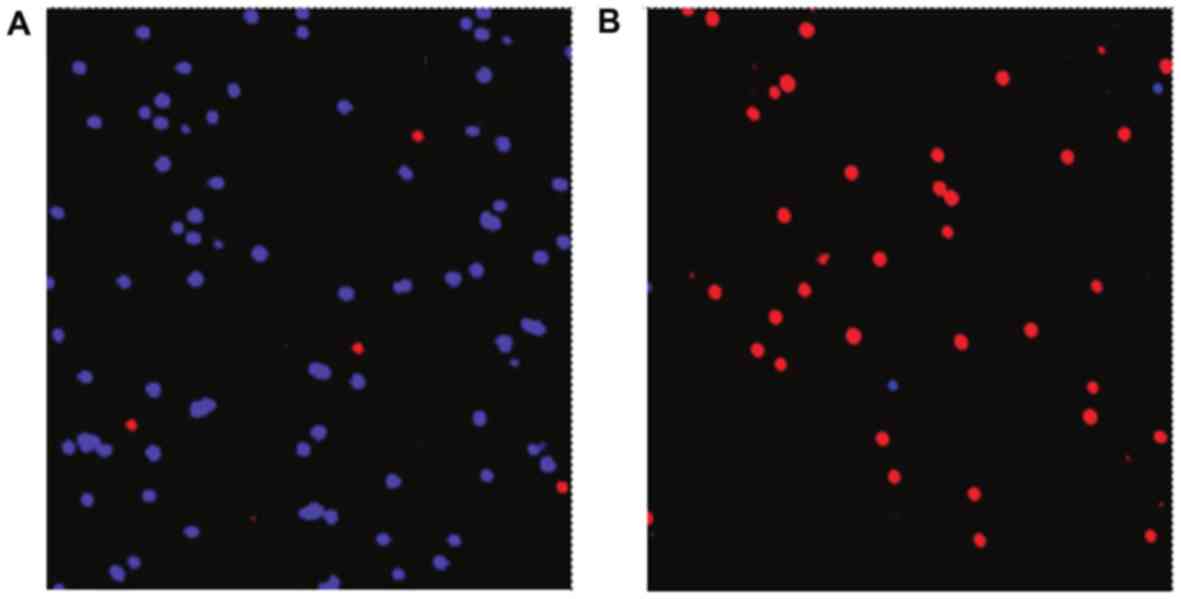
with Vybrant DiD dye (blue). Fig. 7
represents the cell mixture (target cell, C666; control cells,
NP69) prior to (Fig. 7A) and
following (Fig. 7B) separation of NPC
cells using the microfluidic chip. The modified aptamer-bound
microfluidic chip effectively enriched NPC cells with a capture
efficiency of 92%, the percentage of tumor cells isolated/total
tumor cells present.
Separation of NPC cells from whole
blood
The addition of cultured NPC cells into peripheral
blood samples was used as a model for CTCs in the peripheral blood
of tumour patients. Specifically, varying concentrations of C666
cells were added to 1 ml of peripheral blood, and NPC cells were
separated and enriched using the microfluidic chip. The capture
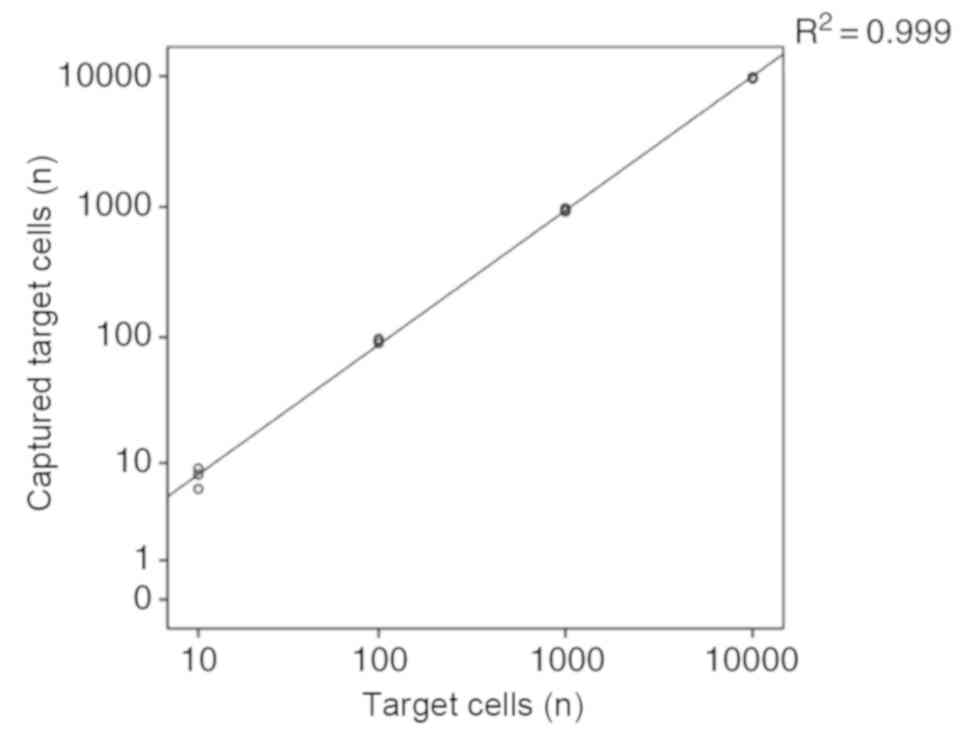
efficiency for NPC cells from blood samples was >90%. Fig. 8 indicates that the quantities of
captured cells ranged from 10–10,000 with varying concentrations of
total NPC cells, and the coefficient of correlation was 0.99
(calculated using linear regression).
Discussion
Liquid biopsy for tumours is currently a prevalent
topic in the field of tumour diagnosis and treatment. Tumour cells
in the peripheral blood of patients can be detected using different
techniques (22). Methods commonly
employed for the detection of CTCs include: i) Flow cytometry, used
for detecting DNA from eight CTCs in breast cancer; ii)
immune-magnetic bead separation; and iii) the CellSearch system
(Menarini Silicon Biosystems, Inc., Huntington Valley, PA, USA)
developed based on the principle of immune-magnetic bead-based
selection (23). The CellSearch
system is a semi-automatic CTC detector that binds the epithelial
cell adhesion molecule of CTCs to facilitate their separation from
peripheral blood cells. It is currently used to determine the
prognosis of patients with breast cancer. Additional methods
include membrane filtration, which separates tumour cells from the
peripheral blood on the basis of size and selected cell markers.
The aforementioned techniques (except for DNA detection) use
high-affinity antibodies to capture and count CTCs (24–26).
However, the number of available antibodies that are able to bind
to tumour cells or cell lines is limited, and off-target binding
remains a major challenge (27).
Specific preservation conditions are required to maintain the
functional activity of protein probes, (the principal limitation of
antibody-based capture), which restricts the application of these
techniques in tumour detection.
Aptamers not only exhibit higher affinity and
specificity compared with antibodies, but are also more easily
synthesized, preserved, transported and surface-modified. The use
of aptamers for capturing CTCs has been reported in recent years,
with capture efficiency varying between 70 and 98% (28,29). In
the present study, a microfluidic chip was modified by
incorporating an array of bypass columns into the reaction chamber
to increase the contact area between cells and the inner surface of
the chamber. Experiments indicated that the capture efficiency of
the modified microfluidic chip (90%) was higher compared with that
of a conventional microfluidic chip (78%). Additionally, the
modified aptamer-bound microfluidic chip targeting NPC cellsthat
was screened by preliminary research, successfully detected NPC
cells in the peripheral blood. The capture efficiency was 90%,
higher compared with the reported efficiency in existing studies
(30,31).
It was demonstrated that an aptamer-bound
microfluidic chip was able to separate NPC cells from whole blood
at a higher efficiency than is currently attainable, and neither
the aptamer-bound microfluidic chip nor the detected samples
required pre-treatment. The method may be performed quickly and
with an effective lower limit of 10 cells. The capture efficiency
of the aptamer-bound microfluidic chip was determined by
preliminary in vitro cell-based experiments, though the true
diagnostic value may be verified by clinical patient sample.
Additionally, the aptamer of the microfluidic chip may only
recognize the corresponding matched epitope, thus may not reliably
capture all types of cancer cells. Subsequent research may involve
samples from patients with nasopharyngeal cancer with mixed
aptamers to capture a greater range of cancer cell types. Despite
these limitations, the results of this study implicate far-reaching
clinical applications for the modified microfluidic chip.
Acknowledgements
The authors would like to thank Dr Xuesen Zou from
the Departments of Clinical Laboratory, Jiangxi Cancer Hospital for
simulation of various parameters of micropillar channel
microfluidic chip using Fluent 6.3.26 software in the study.
Funding
This study was supported by the National Natural
Science Foundation (grant no. 81360402), Jiangxi Provincial Nature
Fund (grant no. 20142BAB205058), Jiangxi Provincial Natural Science
Foundation (grant no. 20151BAB205027), and Jiangxi Province Science
and Technology Plan Projects (grant no. 20151BBG70131).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author by reasonable
request.
Authors' contributions
WXC, JGL, XSZ and WLC were responsible for the
conception and design of the present study. XHW, SYQ, YQZ, QMW,
JYL, WMX and CX were responsible for the experiment. WXC, WLC and
XHW assisted in data analysis and interpretation. All authors were
involved in the writing of the manuscript. All authors have read
and approved the final version of the manuscript.
Ethical approval and consent to
participate
The study was approved by the ethics committee of
Jiangxi Cancer Hospital, Nanchang, China. Written informed consent
was obtained from all volunteers.
Patient consent for publication
Written informed consent was obtained from all
volunteers.
Competing interests
The authors declare that they have no conflict of
interest.
References
|
1
|
Li JG, Yuan X, Zhang LL, Tang YQ, Liu L,
Chen XD, Gong XC, Wan GF, Liao YL, Ye JM and Ao F: A randomized
clinical trial comparing prophylactic upper versus whole-neck
irradiation in the treatment of patients with node-negative
nasopharyngeal carcinoma. Cancer. 119:3170–3176. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuo DY, Chang MH, Wang SY, Hsieh PY and
Shueng PW: Unusual axillary metastasis of recurrent nasopharyngeal
cancer: A case report. Medicine (Baltimore). 96:e68542017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang WY, Twu CW, Chen HH, Jiang RS, Wu CT,
Liang KL, Shih YT, Chen CC, Lin PJ, Liu YC and Lin JC: Long-term
survival analysis of nasopharyngeal carcinoma by plasma
Epstein-Barr virus DNA levels. Cancer. 119:963–970. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee AW, Lin JC and Ng WT: Current
management of nasopharyngeal cancer. Semin Radiat Oncol.
22:233–244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan C, Xu XH, Luo SW, Wang L, Sun M, Ni
LH, Xu L, Wang XL and Zeng G: Which neoadjuvant chemotherapy
regimen should be recommended for patients with advanced
nasopharyngeal carcinoma?: A network meta-analysis. Medicine
(Baltimore). 97:e119782018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
OuYang PY, You KY, Zhang LN, Xiao Y, Zhang
XM and Xie FY: External validity of a prognostic nomogram for
locoregionally advanced nasopharyngeal carcinoma based on the 8th
edition of the AJCC/UICC staging system: A retrospective cohort
study. Cancer Commun (Lond). 38:552018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren XY, Wen X, Li YQ, Zhang J, He QM, Yang
XJ, Tang XR, Wang YQ, Zhang PP, Chen XZ, et al: TIPE3
hypermethylation correlates with worse prognosis and promotes tumor
progression in nasopharyngeal carcinoma. J Exp Clin Cancer Res.
37:2272018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tu Q, Wu X, Le Rhun E, Blonski M, Wittwer
B, Taillandier L, De Carvalho Bittencourt M and Faure GC:
CellSearch technology applied to the detection and quantification
of tumor cells in CSF of patients with lung cancer leptomeningeal
metastasis. Lung Cancer. 90:352–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun B, Liu H, Wang S, Xiang J and Liu X:
Prognostic impact of circulating tumor cells in patients with
ampullary cancer. J Cell Physiol. 233:5014–5022. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bayat P, Nosrati R, Alibolandi M,
Rafatpanah H, Abnous K, Khedri M and Ramezani M: SELEX methods on
the road to protein targeting with nucleic acid aptamers.
Biochimie. 154:132–155. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zou Y, Duan N, Wu S, Shen M and Wang Z:
Selection, identification, and binding mechanism studies of an
ssDNA aptamer targeted to different stages of E. coli
O157:H7. J Agric Food Chem. 66:5677–5682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Filardi V: Carotid artery stenosis near a
bifurcation investigated by fluid dynamic analyses. Neuroradiol J.
26:439–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tian F, Cai L, Chang J, Li S, Liu C, Li T
and Sun J: Label-free isolation of rare tumor cells from untreated
whole blood by interfacial viscoelastic microfluidics. Lab Chip.
18:3436–3445. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen WX, Zhang KH, Zou XS, Chen YQ and Li
JG: Screening and identification of the nucleic acid aptamers in
nasopharyngeal carcinoma. Genet Mol Res. 12:6850–6857. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu C, Munglani G, Vogler H, Ndinyanka
Fabrice T, Shamsudhin N, Wittel FK, Ringli C, Grossniklaus U,
Herrmann HJ and Nelson BJ: Characterization of size-dependent
mechanical properties of tip-growing cells using a lab-on-chip
device. Lab Chip. 17:82–90. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Le Gac S, Cren-Olivé C, Rolando C and
Arscott S: A novel nib-like design for microfabricated nanospray
tips. J Am Soc Mass Spectrom. 15:409–412. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Benlarbi M, Blum LJ and Marquette CA:
SU-8-carbon composite as conductive photoresist for biochip
applications. Biosens Bioelectron. 38:220–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nuttall LP, Brossard FSF, Lennon SA, Reid
BPL, Wu J, Griffiths J and Taylor RA: Optical fabrication and
characterisation of SU-8 disk photonic waveguide heterostructure
cavities. Opt Express. 25:24615–24622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Altuna A, Menendez de la Prida L,
Bellistri E, Gabriel G, Guimerá A, Berganzo J, Villa R and
Fernández LJ: SU-8 based microprobes with integrated planar
electrodes for enhanced neural depth recording. Biosens
Bioelectron. 37:1–5. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Murthy SK, Sin A, Tompkins RG and Toner M:
Effect of flow and surface conditions on human lymphocyte isolation
using microfluidic chambers. Langmuir. 20:11649–11655. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oota-Ishigaki A, Masuzawa T and Nagayama
K: Analysis of the effect of the size of three-dimensional
micro-geometric structures on physical adhesion phenomena using
microprint technique. Int J Artif Organs. 41:277–283. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hille C and Pantel K: Prostate cancer:
Circulating tumour cells in prostate cancer. Nat Rev Urol.
15:265–266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Le UT, Bronsert P, Picardo F, Riethdorf S,
Haager B, Rylski B, Czerny M, Beyersdorf F, Wiesemann S, Pantel K,
et al: Intraoperative detection of circulating tumor cells in
pulmonary venous blood during metastasectomy for colorectal lung
metastases. Sci Rep. 8:87512018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schneck H, Gierke B, Uppenkamp F, Behrens
B, Niederacher D, Stoecklein NH, Templin MF, Pawlak M, Fehm T and
Neubauer H: Disseminated Cancer Cell Network (DCC Net) Duesseldorf:
EpCAM-independent enrichment of circulating tumor cells in
metastatic breast cancer. PLoS One. 10:e1445352015. View Article : Google Scholar
|
|
25
|
Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L,
Lee T, Lee EK, Reiss J, Lee YK, et al: Highly efficient capture of
circulating tumor cells by using nanostructured silicon substrates
with integrated chaotic micromixers. Angew Chem Int Ed Engl.
50:3084–3088. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yin C, Wang Y, Ji J, Cai B, Chen H, Yang
Z, Wang K, Luo C, Zhang W, Yuan C and Wang F: Molecular profiling
of pooled circulating tumor cells from prostate cancer patients
using a dual-antibody-functionalized microfluidic device. Anal
Chem. 90:3744–3751. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun C, Zhang R, Gao M and Zhang X: A rapid
and simple method for efficient capture and accurate discrimination
of circulating tumor cells using aptamer conjugated magnetic beads
and surface-enhanced Raman scattering imaging. Anal Bioanal Chem.
407:8883–8892. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiu WJ, Ling TK, Chiang HP, Lin HJ and
Huang CC: Monitoring cluster ions derived from aptamer-modified
gold nanofilms under laser desorption/ionization for the detection
of circulating tumor cells. ACS Appl Mater Interfaces. 7:8622–8630.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeng Z, Tung CH and Zu Y: A cancer
cell-activatable aptamer-reporter system for one-step assay of
circulating tumor cells. Mol Ther Nucleic Acids. 3:e1842014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wan Y, Liu Y, Allen PB, Asghar W, Mahmood
MA, Tan J, Duhon H, Kim YT, Ellington AD and Iqbal SM: Capture,
isolation and release of cancer cells with aptamer-functionalized
glass bead array. Lab Chip. 12:4693–4701. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Phillips JA, Xu Y, Xia Z, Fan ZH and Tan
W: Enrichment of cancer cells using aptamers immobilized on a
microfluidic channel. Anal Chem. 81:1033–1039. 2009. View Article : Google Scholar : PubMed/NCBI
|