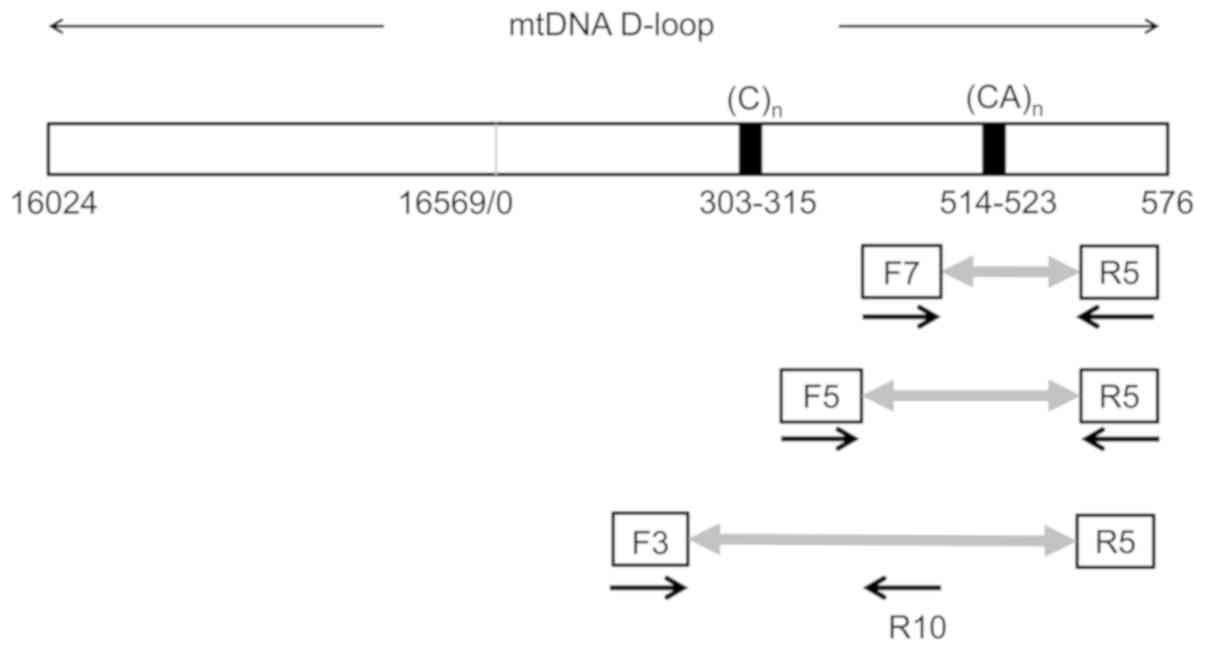
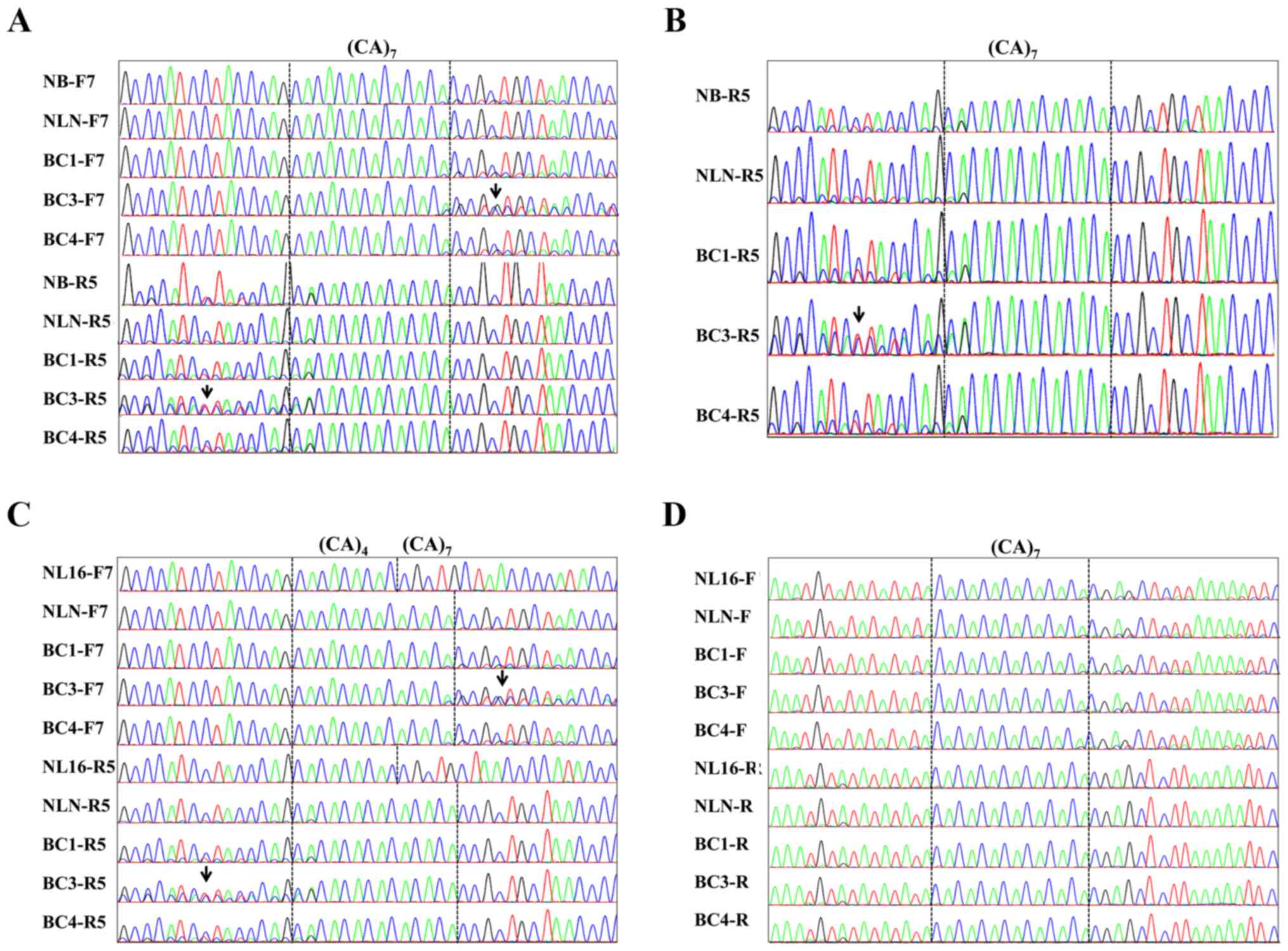
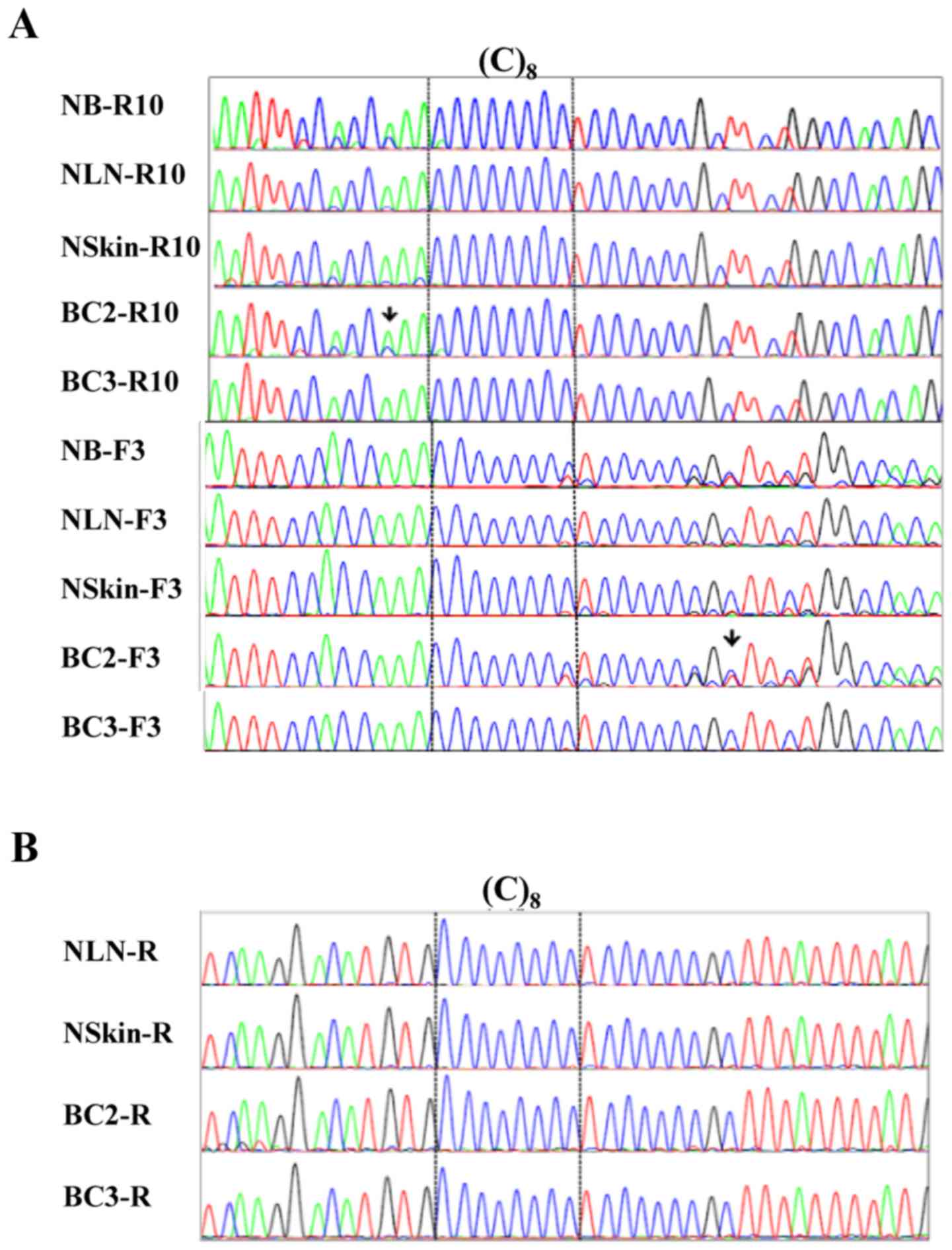
|
1
|
Wallace DC: Mitochondrial DNA sequence
variation in human evolution and disease. Proc Natl Acad Sci USA.
91:8739–8746. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanchez-Cespedes M, Parrella P, Nomoto S,
Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch
WM, et al: Identification of a mononucleotide repeat as a major
target for mitochondrial DNA alterations in human tumors. Cancer
Res. 61:7015–7019. 2001.PubMed/NCBI
|
|
3
|
Suzuki M, Toyooka S, Miyajima K, Iizasa T,
Fujisawa T, Bekele NB and Gazdar AF: Alterations in the
mitochondrial displacement loop in lung cancers. Clin Cancer Res.
9:5636–5641. 2003.PubMed/NCBI
|
|
4
|
Lee HC, Li SH, Lin JC, Wu CC, Yeh DC and
Wei YH: Somatic mutations in the D-loop and decrease in the copy
number of mitochondrial DNA in human hepatocellular carcinoma.
Mutat Res. 547:71–78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lievre A, Blons H, Houllier AM,
Laccourreye O, Brasnu D, Beaune P and Laurent-Puig P:
Clinicopathological significance of mitochondrial D-loop mutations
in head and neck carcinoma. Br J Cancer. 94:692–697. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen D and Zhan H: Study on the mutations
in the D-loop region of mitochondrial DNA in cervical carcinoma. J
Cancer Res Clin Oncol. 135:291–295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chatterjee A, Dasgupta S and Sidransky D:
Mitochondrial subversion in cancer. Cancer Prev Res (Phila).
4:638–654. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo W, Yang D, Xu H, Zhang Y, Huang J,
Yang Z, Chen X and Huang Z: Mutations in the D-loop region and
increased copy number of mitochondrial DNA in human laryngeal
squamous cell carcinoma. Mol Biol Rep. 40:13–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Geurts-Giele WR, Gathier GH, Atmodimedjo
PN, Dubbink HJ and Dinjens WN: Mitochondrial D310 mutation as
clonal marker for solid tumors. Virchows Arch. 467:595–602. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harfe BD and Jinks-Robertson S: DNA
mismatch repair and genetic instability. Annu Rev Genet.
34:359–399. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Naue J, Hörer S, Sänger T, Strobl C,
Hatzer-Grubwieser P, Parson W and Lutz-Bonengel S: Evidence for
frequent and tissue-specific sequence heteroplasmy in human
mitochondrial DNA. Mitochondrion. 20:82–94. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stewart JB and Chinnery PF: The dynamics
of mitochondrial DNA heteroplasmy: Implications for human health
and disease. Nat Rev Genet. 16:530–542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cline J, Braman JC and Hogrefe HH: PCR
fidelity of pfu DNA polymerase and other thermostable DNA
polymerases. Nucleic Acids Res. 24:3546–3551. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shinde D, Lai Y, Sun F and Arnheim N: Taq
DNA polymerase slippage mutation rates measured by PCR and
quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites.
Nucleic Acids Res. 31:974–980. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Einaga N, Yoshida A, Noda H, Suemitsu M,
Nakayama Y, Sakurada A, Kawaji Y, Yamaguchi H, Sasaki Y, Tokino T
and Esumi M: Assessment of the quality of DNA from various
formalin-fixed paraffin-embedded (FFPE) tissues and the use of this
DNA for next-generation sequencing (NGS) with no artifactual
mutation. PLoS One. 12:e01762802017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakayama Y, Yamaguchi H, Einaga N and
Esumi M: Pitfalls of DNA quantification using DNA-binding
fluorescent dyes and suggested solutions. PLoS One.
11:e01505282016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Simone D, Calabrese FM, Lang M, Gasparre G
and Attimonelli M: The reference human nuclear mitochondrial
sequences compilation validated and implemented on the UCSC genome
browser. Bmc Genomics. 12:5172011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Howell N and Smejkal CB: Persistent
heteroplasmy of a mutation in the human mtDNA control region:
Hypermutation as an apparent consequence of simple-repeat
expansion/contraction. Am J Hum Genet. 66:1589–1598. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alvi AJ, Rader JS, Broggini M, Latif F and
Maher ER: Microsatellite instability and mutational analysis of
transforming growth factor beta receptor type II gene (TGFBR2) in
sporadic ovarian cancer. Mol Pathol. 54:240–243. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maehara Y, Oda S and Sugimachi K: The
instability within: Problems in current analyses of microsatellite
instability. Mutat Res. 461:249–263. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Enomoto A, Esumi M, Yamashita K, Takagi K,
Takano S and Iwai S: Abnormal nucleotide repeat sequence in the
TGF-betaRII gene in hepatocellular carcinoma and in uninvolved
liver tissue. J Pathol. 195:349–354. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Longley MJ, Nguyen D, Kunkel TA and
Copeland WC: The fidelity of human DNA polymerase gamma with and
without exonucleolytic proofreading and the p55 accessory subunit.
J Biol Chem. 276:38555–38562. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mason PA, Matheson EC, Hall AG and
Lightowlers RN: Mismatch repair activity in mammalian mitochondria.
Nucleic Acids Res. 31:1052–1058. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kazak L, Reyes A and Holt IJ: Minimizing
the damage: Repair pathways keep mitochondrial DNA intact. Nat Rev
Mol Cell Biol. 13:659–671. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carelli V, Maresca A, Caporali L, Trifunov
S, Zanna C and Rugolo M: Mitochondria: Biogenesis and mitophagy
balance in segregation and clonal expansion of mitochondrial DNA
mutations. Int J Biochem Cell Biol. 63:21–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Larsson NG: Somatic mitochondrial DNA
mutations in mammalian aging. Annu Rev Biochem. 79:683–706. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Payne BA, Wilson IJ, Yu-Wai-Man P, Coxhead
J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M
and Chinnery PF: Universal heteroplasmy of human mitochondrial DNA.
Hum Mol Genet. 22:384–390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He Y, Wu J, Dressman DC, Iacobuzio-Donahue
C, Markowitz SD, Velculescu VE, Diaz LA Jr, Kinzler KW, Vogelstein
B and Papadopoulos N: Heteroplasmic mitochondrial DNA mutations in
normal and tumour cells. Nature. 464:610–614. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szulwach KE, Chen P, Wang X, Wang J,
Weaver LS, Gonzales ML, Sun G, Unger MA and Ramakrishnan R:
Single-cell genetic analysis using automated microfluidics to
resolve somatic mosaicism. PLoS One. 10:e01350072015. View Article : Google Scholar : PubMed/NCBI
|