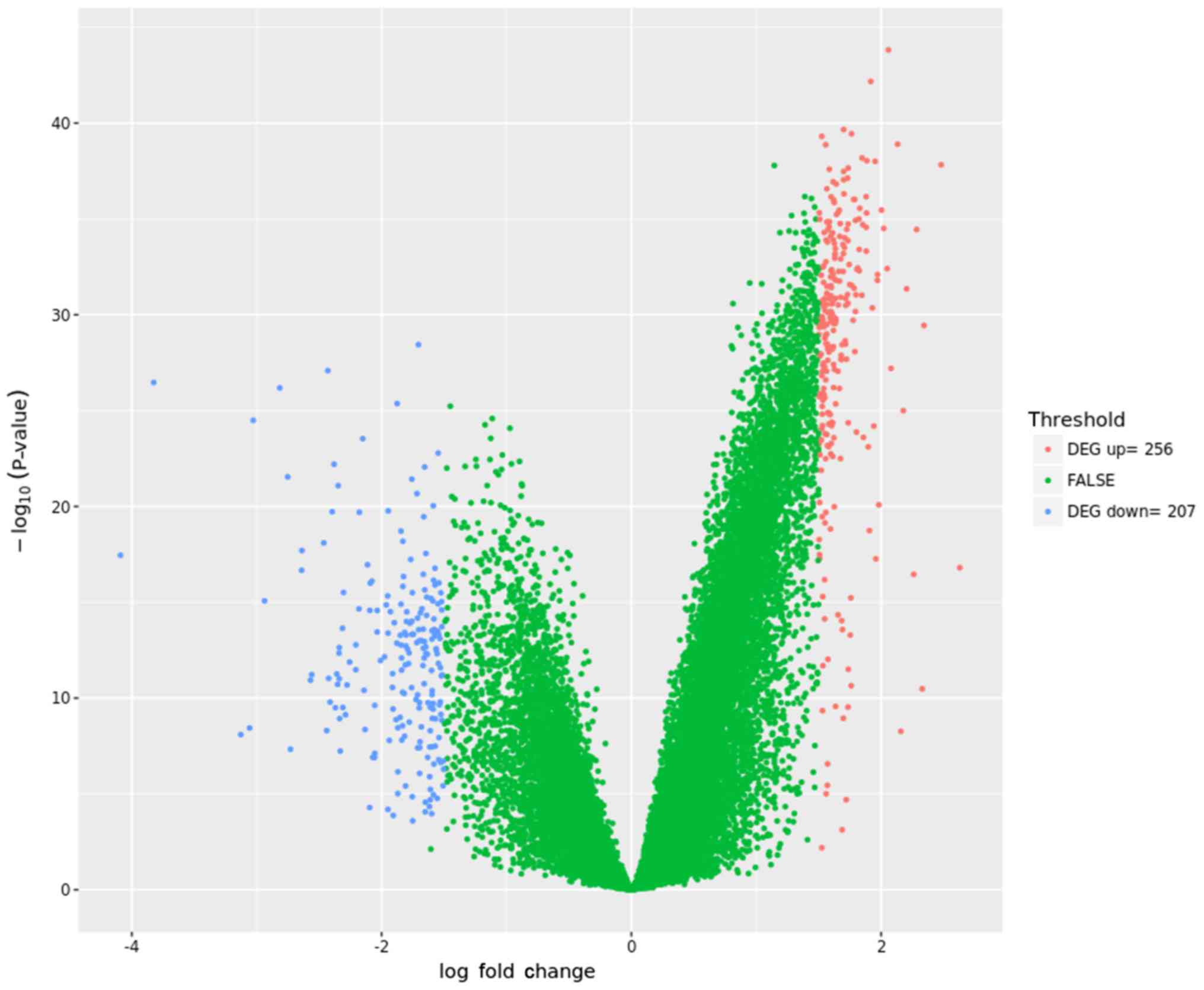
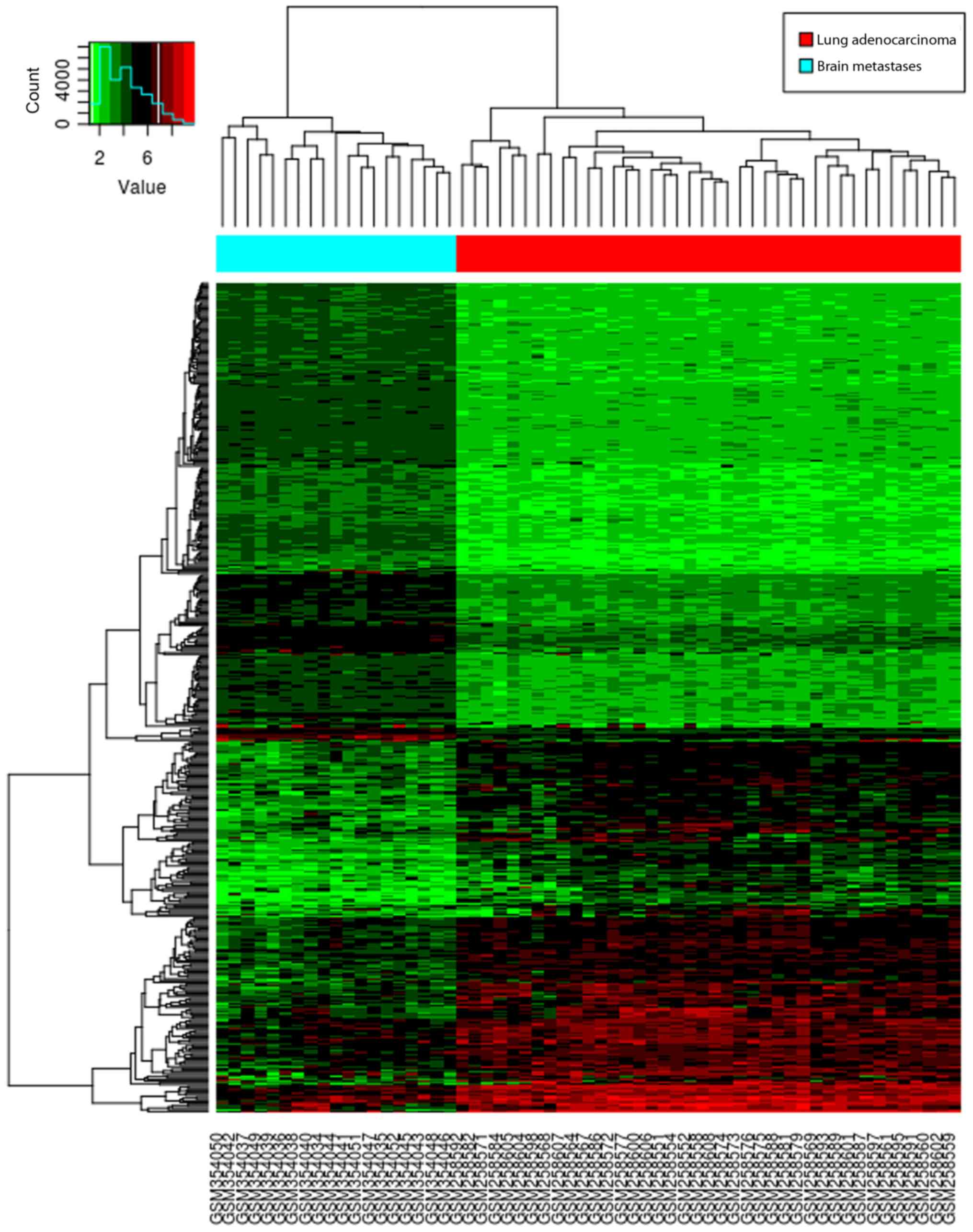
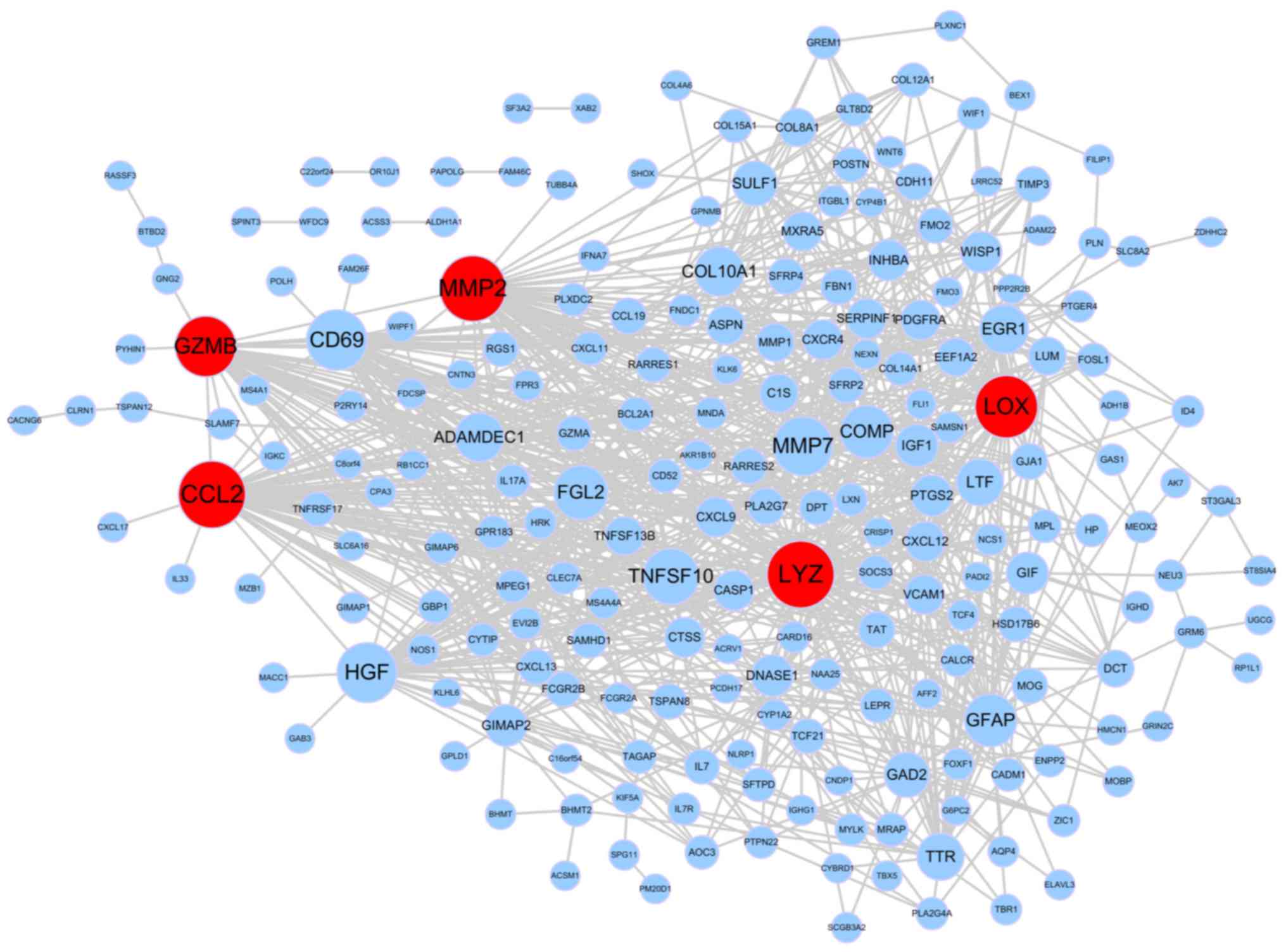
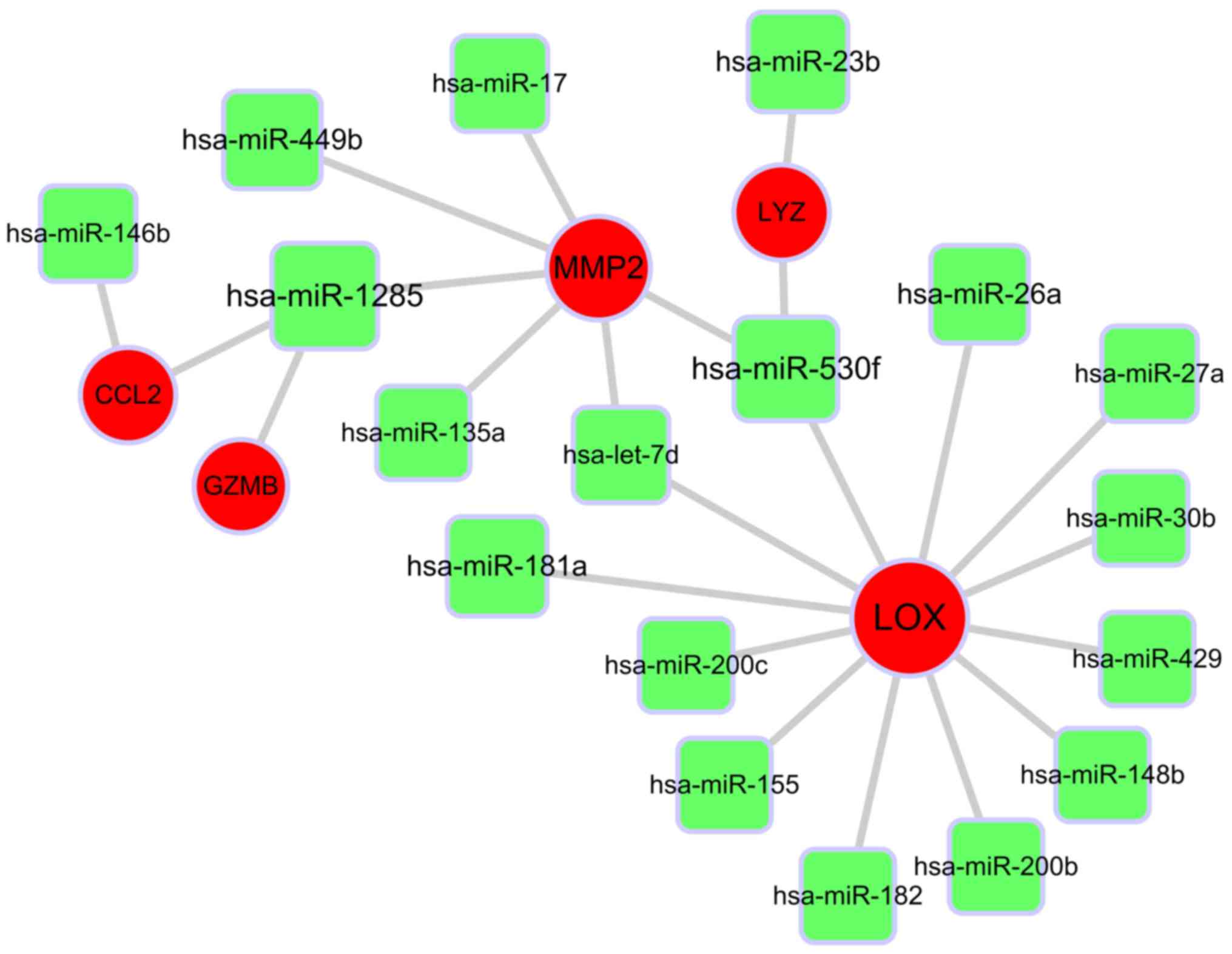
|
1
|
Nayak L, Lee EQ and Wen PY: Epidemiology
of brain metastases. Curr Oncol Rep. 14:48–54. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin NU and Winer EP: Brain metastases: The
HER2 paradigm. Clin Cancer Res. 13:1648–1655. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yau T, Swanton C, Chua S, Sue A, Walsh G,
Rostom A, Johnston SR, O'Brien ME and Smith IE: Incidence, pattern
and timing of brain metastases among patients with advanced breast
cancer treated with trastuzumab. Acta Oncol. 45:196–201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hubbs JL, Boyd JA, Hollis D, Chino JP,
Saynak M and Kelsey CR: Factors associated with the development of
brain metastases. Cancer. 116:5038–5046. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schuette W: Treatment of brain metastases
from lung cancer: Chemotherapy. Lung Cancer. 45 Suppl 2:S253–S257.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barnholtz-Sloan JS, Sloan AE, Davis FG,
Vigneau FD, Lai P and Sawaya RE: Incidence proportions of brain
metastases in patients diagnosed (1973 to 2001) in the metropolitan
detroit cancer surveillance system. J Clin Oncol. 22:2865–2872.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi AA, Digumarthy SR, Temel JS, Halpern
EF, Kuester LB and Aquino SL: Does initial staging or tumor
histology better identify asymptomatic brain metastases in patients
with non-small cell lung cancer? J Thorac Oncol. 1:205–210. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sundström JT, Minn H, Lertola KK and
Nordman E: Prognosis of patients treated for intracranial
metastases with whole-brain irradiation. Ann Med. 30:296–299. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Besse B, Le Moulec S, Mazières J,
Senellart H, Barlesi F, Chouaid C, Dansin E, Bérard H, Falchero L,
Gervais R, et al: Bevacizumab in patients with nonsquamous
non-small cell lung cancer and asymptomatic, untreated brain
metastases (BRAIN): A nonrandomized, phase II study. Clin Cancer
Res. 21:1896–1903. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nguyen DX, Chiang AC, Zhang XH, Kim JY,
Kris MG, Ladanyi M, Gerald WL and Massagué J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Valiente M, Obenauf AC, Jin X, Chen Q,
Zhang XH, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E and Massagué
J: Serpins promote cancer cell survival and vascular cooption in
brain metastasis. Cell. 156:1002–1016. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh M, Venugopal C, Tokar T, Brown KR,
McFarlane N, Bakhshinyan D, Vijayakumar T, Manoranjan B, Mahendram
S, Vora P, et al: RNAi screen identifies essential regulators of
human brain metastasis-initiating cells. Acta Neuropathol.
134:923–940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Castro D, Moreira M, Gouveia AM, Pozza DH
and De Mello RA: MicroRNAs in lung cancer. Oncotarget.
8:81679–81685. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hwang SJ, Lee HW, Kim HR, Song HJ, Lee DH,
Lee H, Shin CH, Joung JG, Kim DH, Joo KM and Kim HH: Overexpression
of microRNA-95-3p suppresses brain metastasis of lung
adenocarcinoma through downregulation of cyclin D1. Oncotarget.
6:20434–20448. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF
and Wang SF: MicroRNA-378 is associated with non-small cell lung
cancer brain metastasis by promoting cell migration, invasion and
tumor angiogenesis. Med Oncol. 29:1673–1680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao C, Xu Y, Zhang Y, Tan W, Xue J, Yang
Z, Zhang Y, Lu Y and Hu X: Downregulation of miR-145 contributes to
lung adenocarcinoma cell growth to form brain metastases. Oncol
Rep. 30:2027–2034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kikuchi T, Daigo Y, Ishikawa N, Katagiri
T, Tsunoda T, Yoshida S and Nakamura Y: Expression profiles of
metastatic brain tumor from lung adenocarcinomas on cDNA
microarray. Int J Oncol. 28:799–805. 2006.PubMed/NCBI
|
|
19
|
Kuner R, Muley T, Meister M, Ruschhaupt M,
Buness A, Xu EC, Schnabel P, Warth A, Poustka A, Sültmann H and
Hoffmann H: Global gene expression analysis reveals specific
patterns of cell junctions in non-small cell lung cancer subtypes.
Lung Cancer. 63:32–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lüke F, Blazquez R, Yamaci RF, Lu X,
Pregler B, Hannus S, Menhart K, Hellwig D, Wester HJ, Kropf S, et
al: Isolated metastasis of an EGFR-L858R-mutated NSCLC of the
meninges: The potential impact of CXCL12/CXCR4 axis in EGFRmut
NSCLC in diagnosis, follow-up and treatment. Oncotarget.
9:18844–18857. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C and Wong WH: Model-based analysis of
oligonucleotide arrays: Expression index computation and outlier
detection. Proc Natl Acad Sci USA. 98:31–36. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu G, Wang LG, Yan GR and He QY: DOSE: An
R/Bioconductor package for disease ontology semantic and enrichment
analysis. Bioinformatics. 31:608–609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahn T, Lee E, Huh N and Park T:
Personalized identification of altered pathways in cancer using
accumulated normal tissue data. Bioinformatics. 30:422–429. 2014.
View Article : Google Scholar
|
|
26
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun Y, Weng Y, Zhang Y, Yan X, Guo L, Wang
J, Song X, Yuan Y, Chang FY and Wang CL: Systematic expression
profiling analysis mines dys-regulated modules in active
tuberculosis based on re-weighted protein-protein interaction
network and attract algorithm. Microb Pathog. 107:48–53. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Goldenberg A, Wong KC and Zhang Z: A
probabilistic approach to explore human miRNA targetome by
integrating miRNA-overexpression data and sequence information.
Bioinformatics. 30:621–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Keeley EC, Mehrad B and Strieter RM: CXC
chemokines in cancer angiogenesis and metastases. Adv Cancer Res.
106:91–111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bachelder RE, Wendt MA and Mercurio AM:
Vascular endothelial growth factor promotes breast carcinoma
invasion in an autocrine manner by regulating the chemokine
receptor CXCR4. Cancer Res. 62:7203–7206. 2002.PubMed/NCBI
|
|
31
|
Akishima-Fukasawa Y, Nakanishi Y, Ino Y,
Moriya Y, Kanai Y and Hirohashi S: Prognostic significance of
CXCL12 expression in patients with colorectal carcinoma. Am J Clin
Pathol. 132:202–10; quiz 307. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fokas E, Steinbach JP and Rödel C: Biology
of brain metastases and novel targeted therapies: Time to translate
the research. Biochim Biophys Acta. 1835:61–75. 2013.PubMed/NCBI
|
|
33
|
Lee BC, Lee TH, Avraham S and Avraham HK:
Involvement of the chemokine receptor CXCR4 and its ligand stromal
cell-derived factor 1alpha in breast cancer cell migration through
human brain microvascular endothelial cells. Mol Cancer Res.
2:327–338. 2004.PubMed/NCBI
|
|
34
|
Hinton CV, Avraham S and Avraham HK: Role
of the CXCR4/CXCL12 signaling axis in breast cancer metastasis to
the brain. Clin Exp Metastasis. 27:97–105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hartmann TN, Burger JA, Glodek A, Fujii N
and Burger M: CXCR4 chemokine receptor and integrin signaling
co-operate in mediating adhesion and chemoresistance in small cell
lung cancer (SCLC) cells. Oncogene. 24:4462–4471. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salmaggi A, Maderna E, Calatozzolo C,
Gaviani P, Canazza A, Milanesi I, Silvani A, DiMeco F, Carbone A
and Pollo B: CXCL12, CXCR4 and CXCR7 Expression in brain
metastases. Cancer Biol Ther. 8:1608–1614. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fields GB: Interstitial collagen
catabolism. J Biol Chem. 288:8785–8793. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Woolley DE: Collagenolytic mechanisms in
tumor cell invasion. Cancer Metastasis Rev. 3:361–372. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liotta LA, Thorgeirsson UP and Garbisa S:
Role of collagenases in tumor cell invasion. Cancer Metastasis Rev.
1:277–288. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kudo-Saito C, Shirako H, Ohike M,
Tsukamoto N and Kawakami Y: CCL2 is critical for immunosuppression
to promote cancer metastasis. Clin Exp Metastasis. 30:393–405.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li X and Tai HH: Thromboxane A 2
receptor-mediated release of matrix metalloproteinase-1 (MMP-1)
induces expression of monocyte chemoattractant protein-1 (MCP-1) by
activation of protease-activated receptor 2 (PAR2) in A549 human
lung adenocarcinoma cells. Mol Carcinog. 53:659–666.
2014.PubMed/NCBI
|
|
42
|
Li X and Tai HH: Activation of thromboxane
A2 receptor (TP) increases the expression of monocyte
chemoattractant protein −1 (MCP-1)/chemokine (C-C motif) ligand 2
(CCL2) and recruits macrophages to promote invasion of lung cancer
cells. PLoS One. 8:e540732013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen W, Gao Q, Han S, Pan F and Fan W: The
CCL2/CCR2 axis enhances IL-6-induced epithelial-mesenchymal
transition by cooperatively activating STAT3-Twist signaling. Tumor
Biol. 36:973–981. 2015. View Article : Google Scholar
|
|
44
|
Lauer-Fields JL, Juska D and Fields GB:
Matrix metalloproteinases and collagen catabolism. Biopolymers.
66:19–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vu TH: Don't mess with the matrix. Nat
Genet. 28:202–203. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chakrabarti S and Patel KD: Matrix
metalloproteinase-2 (MMP-2) and MMP-9 in pulmonary pathology. Exp
Lung Res. 31:599–621. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu W, Xu H, Fang M, Wu X and Xu Y: MKL1
links epigenetic activation of MMP2 to ovarian cancer cell
migration and invasion. Biochem Biophys Res Commun. 487:500–508.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang TH, Hsia SM and Shieh TM: Lysyl
oxidase and the tumor microenvironment. Int J Mol Sci. 18:E622016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kasashima H, Yashiro M, Kinoshita H,
Fukuoka T, Morisaki T, Masuda G, Sakurai K, Kubo N, Ohira M and
Hirakawa K: Lysyl oxidase is associated with the
epithelial-mesenchymal transition of gastric cancer cells in
hypoxia. Gastric Cancer. 19:431–442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shih YH, Chang KW, Chen MY, Yu CC, Lin DJ,
Hsia SM, Huang HL and Shieh TM: Lysyl oxidase and enhancement of
cell proliferation and angiogenesis in oral squamous cell
carcinoma. Head Neck. 35:250–256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Osawa T, Ohga N, Akiyama K, Hida Y,
Kitayama K, Kawamoto T, Yamamoto K, Maishi N, Kondoh M, Onodera Y,
et al: Lysyl oxidase secreted by tumour endothelial cells promotes
angiogenesis and metastasis. Br J Cancer. 109:2237–2247. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wilgus ML, Borczuk AC, Stoopler M,
Ginsburg M, Gorenstein L, Sonett JR and Powell CA: Lysyl oxidase: A
lung adenocarcinoma biomarker of invasion and survival. Cancer.
117:2186–2191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rubio CA: The natural antimicrobial enzyme
lysozyme is up-regulated in gastrointestinal inflammatory
conditions. Pathogens. 3:73–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Serra C, Vizoso F, Alonso L, Rodríguez JC,
González LO, Fernández M, Lamelas ML, Sánchez LM, García-Muñiz JL,
Baltasar A and Medrano J: Expression and prognostic significance of
lysozyme in male breast cancer. Breast Cancer Res. 4:R162002.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Thomas DA and Massagué J: TGF-beta
directly targets cytotoxic T cell functions during tumor evasion of
immune surveillance. Cancer Cell. 8:369–380. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cao X, Cai SF, Fehniger TA, Song J,
Collins LI, Piwnica-Worms DR and Ley TJ: Granzyme B and perforin
are important for regulatory T cell-mediated suppression of tumor
clearance. Immunity. 27:635–646. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Smyth MJ, Street SE and Trapani JA:
Cutting edge: Granzymes A and B are not essential for
perforin-mediated tumor rejection. J Immunol. 171:515–518. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ramberg H, Alshbib A, Berge V, Svindland A
and Taskén KA: Regulation of PBX3 expression by androgen and Let-7d
in prostate cancer. Mol Cancer. 10:502011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Su B, Zhao W, Shi B, Zhang Z, Yu X, Xie F,
Guo Z, Zhang X, Liu J, Shen Q, et al: Let-7d suppresses growth,
metastasis, and tumor macrophage infiltration in renal cell
carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 13:2062014.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dai X and Cai Y: Down-regulation of
microRNA let-7d inhibits the proliferation and invasion of
trophoblast cells in preeclampsia. J Cell Biochem. 119:1141–1151.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Al-Shamy G and Sawaya R: Management of
brain metastases: The indispensable role of surgery. J Neurooncol.
92:275–282. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hoffman PC, Mauer AM and Vokes EE: Lung
cancer. Lancet. 355:479–485. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Suzuki M and Tarin D: Gene expression
profiling of human lymph node metastases and matched primary breast
carcinomas: Clinical implications. Mol Oncol. 1:172–180. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Johnson JD and Young B: Demographics of
brain metastasis. Neurosurg Clin N Am. 7:337–344. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Preusser M, Berghoff AS, Koller R,
Zielinski CC, Hainfellner JA, Liebmann-Reindl S, Popitsch N, Geier
CB, Streubel B and Birner P: Spectrum of gene mutations detected by
next generation exome sequencing in brain metastases of lung
adenocarcinoma. Eur J Cancer. 51:1803–1811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhou S, Liu P, Jiang W and Zhang H:
Identification of potential target genes associated with the effect
of propranolol on angiosarcoma via microarray analysis. Oncol Lett.
13:4267–4275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L and Li S: Cancer Genome Atlas Research Network, Xie W and
Yang D: lncRNA epigenetic landscape analysis identifies EPIC1 as an
oncogenic lncRNA that interacts with MYC and promotes cell-cycle
progression in cancer. Cancer Cell. 33:706–720.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li S, Li H, Xu Y and Lv X: Identification
of candidate biomarkers for epithelial ovarian cancer metastasis
using microarray data. Oncol Lett. 14:3967–3974. 2017. View Article : Google Scholar : PubMed/NCBI
|