Introduction
The incidence and mortality rates of colorectal
cancer (CRC) in the USA have declined between 1975 and 2006 due to
decreased smoking and red meat consumption (1). However, both the incidence and
mortality rates of CRC have rapidly increased in Asian countries in
the past three decades (2,3). Currently, CRC is the third most common
type of cancer worldwide, highlighting the importance of developing
novel anticancer methods (4).
MicroRNAs (miRs) are a family of non-coding RNAs
that can regulate gene expression predominantly by directly binding
to the 3′-untranslated region (3′-UTR) of target mRNAs (5). It is estimated that up to 33% of human
genes can be regulated by miRs (6).
Previously, the importance of miRs in regulating functions of
malignant cells, including cell proliferation, migration, invasion
and apoptosis, has been appreciated (7,8). Altered
expression levels of miRs have been identified in multiple types of
human tumor, including CRC, and miRs have been recognized as
biomarkers for tumor diagnosis and treatment (9–11).
miR-7-5p has been recognized as a tumor suppressor
in melanoma and breast cancer, as it is frequently downregulated in
these tumor types (12–14). Giles et al (12) reported that miR-7-5p expression was
reduced in metastatic melanoma-derived cells compared with primary
melanoma cells, and its effects on melanoma cell migration and
invasion was exerted partly via inhibition of insulin receptor
substrate 2 expression and oncogenic Akt signaling. In addition, it
has been identified that miR-7-5p is a potent inhibitor of melanoma
growth and metastasis by inactivation of the transcription factor
p65/nuclear factor-κB signaling pathway, which suggests that
miR-7-5p may serve a role in therapy for this disease (13). Furthermore, in vitro and in
vivo studies revealed that miR-7-5p overexpression could
inhibit breast cancer cell proliferation and induce cell apoptosis
by targeting REGγ (14). However, to
the best of our knowledge, the underlying mechanisms of miR-7-5p in
CRC progression remain unknown.
Krüppel-like factor 4 (KLF4) has been reported to
serve a critical role in cell differentiation and development
(15). Evidence has demonstrated
that KLF4 can function as either a tumor suppressor or an oncogene
in human tumors (16,17). Previous studies revealed that KLF4
expression was upregulated in CRC and could be regulated by miRs,
including miR-92a and miR-543 (18,19).
Given the importance of miR-7-5p and KLF4 in tumor initiation and
development, the current study investigated whether miR-7-5p could
regulate KLF4 in CRC. Furthermore, the effects of miR-7-5p and KLF4
expression levels on cell proliferation and migration were
examined.
Materials and methods
Patients and tumor tissues
Human CRC tumor tissues and adjacent non-tumor
tissues were obtained from 76 enrolled patients who received
surgical treatment between August 2009 and December 2011 at The No.
2 Hospital of Ningbo (Ningbo, China). All patients did not receive
any anticancer treatments prior to surgery. The tissue samples were
snap-frozen in liquid nitrogen and then stored at −80°C until
further use. The current study was approved by the Ethics Committee
of The No. 2 Hospital of Ningbo (Ningbo, China). Written informed
consent was obtained from all enrolled patients. The
clinicopathological features were collected and summarized in
Table I.
 | Table I.Associations of miR-7-5p expression
with the clinicopathological features of colorectal cancer. |
Table I.
Associations of miR-7-5p expression
with the clinicopathological features of colorectal cancer.
|
|
| microRNA-7-5p
expression, n |
|
|---|
|
|
|
|
|
|---|
| Variable | No. of cases | High | Low | P-value |
|---|
| Sex |
|
|
| 0.381 |
| Male | 42 | 15 | 27 |
|
|
Female | 37 | 14 | 23 |
|
| Age, years |
|
|
| 0.629 |
| ≥50 | 40 | 14 | 26 |
|
|
<50 | 39 | 15 | 24 |
|
| Tumor size, cm |
|
|
| 0.024 |
| ≥5 | 46 | 17 | 29 |
|
|
<5 | 33 | 12 | 21 |
|
| Tumor stage |
|
|
| 0.014 |
| I–II | 33 | 11 | 22 |
|
|
III–IV | 46 | 18 | 28 |
|
Cell culture
The human colon epithelial cell line HCEC 1CT and
the CRC cell lines HCT-116, SW480 and SW620 were purchased from the
Cell Bank of Chinese Academy of Sciences (Shanghai, China). All
cell lines were cultured in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 100 µg/ml streptomycin and ١٠٠ U/ml penicillin
(Beyotime Institute of Biotechnology, Haimen, China) in a 37°C
humidified incubator containing 5% CO2.
Cell transfection
The synthesized miR-7-5p mimic
(5′-CAACAAAUCACAGUCUGCCAUA-3′), inhibitor
(5′-UAUGGCAGACUGUGAUUUGUUG-3′) and negative control miR (NC-miR;
5′-ACCGCUAAUCAUACGAAUACAC-3′) were purchased from Chang Jing
Bio-Tech, Ltd. (Changsha, China). KLF4 small interfering (si)RNA
(si-KLF4; 5′-CCAGCCAGAAAGCACUACAAU-3′) and NC siRNA (NC-siR;
5′-AUGCAAUACCGCAGAACACCA-3′) were purchased from Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). The KLF4 plasmid, obtained from
OriGene Technologies, Inc. (Beijing, China), was also transfected
into cells with or without miR-7-5p mimic co-transfection.
Transfection was conducted using Lipofectamine® ٢٠٠٠
(Invitrogen; Thermo Fisher Scientific, Inc.) with ٥٠ nM miRNA, 50
nM siRNA, or 1 µg KLF4 plasmid, according to the manufacturer's
protocol. Cells were collected 48 h after transfection for
subsequent experiments.
Bioinformatics analysis
The TargetScan prediction software (version 7.1;
http://www.targetscan.org/vert_71/)
was used to predict the miRs that could bind to the 3′-UTR of
KLF4.
Luciferase activity assay
The wild-type (wt) or mutant (Mut) KLF4 3′-UTR was
inserted downstream of the firefly luciferase gene within a pmirGLO
plasmid (Promega Cooperation, Madison, WI, USA). CRC cells were
co-transfected with miR-7-5p mimic or NC-miR and wt or Mut KLF4
3′-UTR using Lipofectamine® ٢٠٠٠ (Invitrogen; Thermo
Fisher Scientific, Inc.). Relative luciferase activities were
measured using a Dual-Luciferase Reporter system (Promega
Cooperation) following transfection for 48 h, with firefly
luciferase activity used as the internal control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples and cell
lines using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. A
MicroRNA First-Strand Synthesis kit and a SYBR® Premix
Ex Taq™ II kit (both from Takara Biotechnology Co., Ltd., Dalian,
China) were employed to quantify the expression levels of miR-7-5p,
according to the manufacturer's protocols. The following
thermocycling conditions were used: 10 min at 95°C; 40 cycles of 1
min at 95°C; 2 min at 63°C; and 1 min at 72°C. U6 small nuclear
(sn)RNA was used as an endogenous control. The comparative cycle
threshold (2−ΔΔCq) method was used to quantify the
expression levels of miR-7-5p (20).
The primers used were as follows: miR-7-5p forward,
5′-ACACTCCAGCTGGGTGGAAGACTAGTGATTT-3′ and reverse,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAACAAA-3′; and U6 snRNA
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse
5′-AACGCTTCACGAATTTGCGT-3′.
Western blot analysis
Total protein was extracted from the tissue samples
and cell lines using radioimmunoprecipitation assay lysis buffer
(Beyotime Institute of Biotechnology). Protein concentration was
quantified using an Enhanced BCA Protein assay kit (Beyotime
Institute of Biotechnology). Subsequently, the proteins (50 µg)
were separated on 10% sodium dodecyl sulfate polyacrylamide gels
and transferred to polyvinylidene difluoride membranes. Following
blocking with 5% fat-free milk at 4°C for 4 h, the membranes were
incubated with the primary antibodies rabbit monoclonal anti-KLF4
(1:1,000; catalog no. ab215036) and rabbit monoclonal anti-β-actin
(1:1,000; catalog no. ab115777; both from Abcam, Cambridge, UK)
overnight at 4°C. Following washing with TBS and Tween-20, the
membranes were incubated with horseradish peroxidase-conjugated
goat anti-rabbit secondary antibodies (1:3,000; catalog no.
ab205718; Abcam) for 1 h at room temperature. The signals were
developed with an enhanced chemiluminescent kit (Beyotime Institute
of Biotechnology) and analyzed using ImageJ v.1.42 software
(National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assay
Cell proliferation rate was analyzed using Cell
Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology),
according to the manufacturer's protocol. Briefly, the cells were
seeded in 24-well plates at a density of 5×103
cells/well. Subsequently, 10 µl CCK-8 solution was added to each
well at 0, 24, 48 and 72 h, and the cells were further incubated
for 4 h at 37°C. The optical density was measured at 450 nm.
Cell migration assay
Cell migration rate was analyzed using a wound
healing assay. The cells were seeded in 6-well plates and cultured
until 80% confluence. Subsequently, a wound in the cell surface was
created using a 100-µl sterilized pipette tip. At 0 and 24 h after
the wound was created, images were obtained using a light
microscope (magnification, ×200) and analyzed using ImageJ v.1.42
software (National Institutes of Health).
Statistical analysis
Data are presented as the mean ± standard deviation.
All statistical analysis was conducted using GraphPad Prism 5
software (GraphPad Software Inc., La Jolla, CA, USA). Differences
between groups were analyzed using analysis of variance followed by
Tukey's test or the Student's t-test. Associations between miR-7-5p
and clinicopathological features were analyzed by Chi-square test.
Correlation between miR-7-5p and KLF4 expression levels was
analyzed by Spearman's correlation coefficient. Overall survival
was analyzed by a Kaplan-Meier curve and log-rank test. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-7-5p expression is reduced in CRC
tissues and cell lines
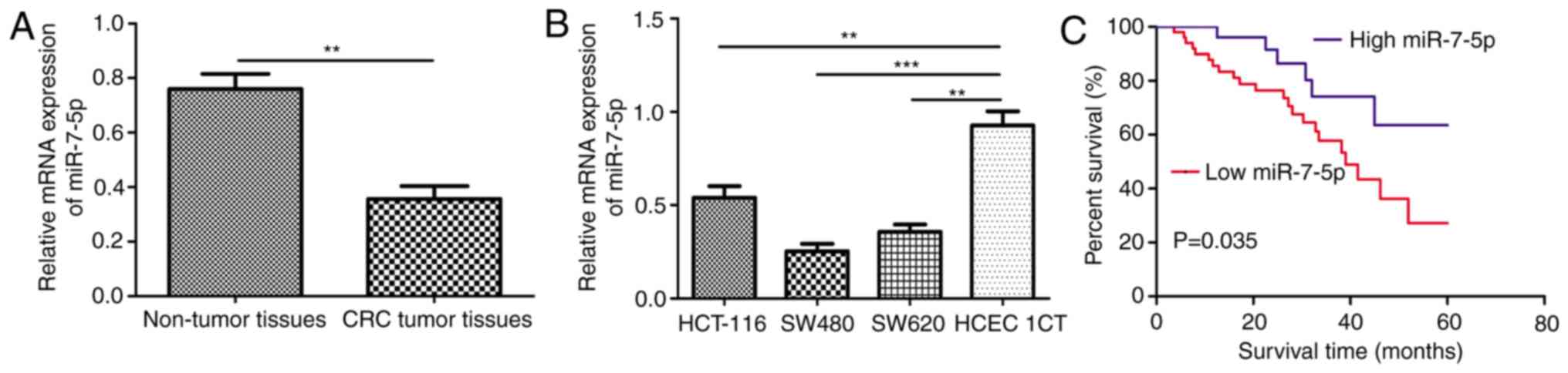
RT-qPCR demonstrated a significantly lower miR-7-5p
expression level in CRC tumor tissues compared with adjacent
non-tumor tissues (Fig. 1A).
Furthermore, miR-7-5p expression levels were examined in the human
colon epithelial cell line HCEC 1CT and CRC cell lines HCT-116,
SW480 and SW620. It was identified that the expression level of
miR-7-5p was significantly lower in HCT-116, SW480 and SW620 cells
compared with HCEC 1CT cells (Fig.
1B).
Clinical significance of miR-7-5p
expression in CRC
The enrolled patients were classified into high or
low miR-7-5p expression groups based on the expression level of
miR-7-5p. The 75th percentile of the 2−∆∆Cq values was
used as the cut-off point (0.41) for patients with high or low
miR-7-5p expression (21).
Subsequently, associations between miR-7-5p expression and
clinicopathological features were analyzed and it was revealed that
low miR-7-5p expression was significantly associated with tumor
size and tumor stage (22), however,
it was not associated with age and sex (Table I). Furthermore, analysis of miR-7-5p
expression on overall survival using Kaplan-Meier curve and
log-rank test revealed that low miR-7-5p expression predicts a poor
overall survival in patients with CRC compared with high expression
(Fig. 1C).
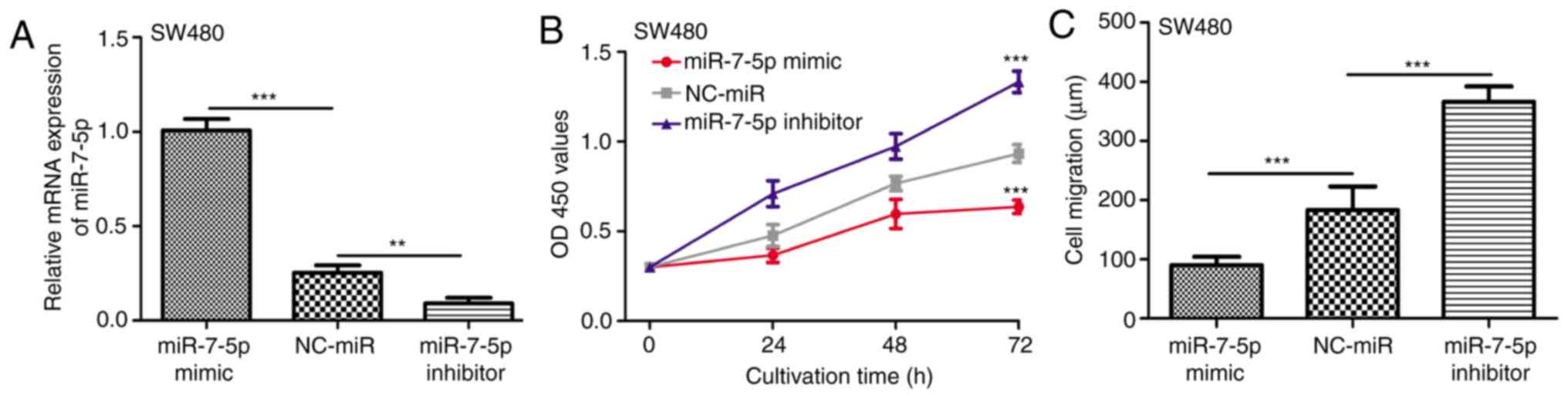
miR-7-5p inhibits CRC cell
proliferation and migration in vitro
The SW480 cell line exhibited the lowest miR-7-5p
expression level among the CRC cell lines investigated, therefore,
SW480 cells were selected for in vitro miR-7-5p biological
function analysis. The miR-7-5p mimic, miR-7-5p inhibitor and
NC-miR were used to regulate the expression of miR-7-5p in SW480
cells. It was confirmed that the expression level of miR-7-5p was
enhanced by miR-7-5p mimic and reduced by miR-7-5p inhibitor
(Fig. 2A). Subsequently, CCK-8 and
wound healing assays revealed that SW480 cells transfected with
miR-7-5p mimic exhibited significantly lower levels of cell
proliferation and migration compared with those transfected with
NC-miR (Fig. 2B and C). Furthermore,
downregulation of miR-7-5p in SW480 cells by miR-7-5p inhibitor
increased the levels of proliferation and migration compared with
the NC-miR group (Fig. 2B and
C).
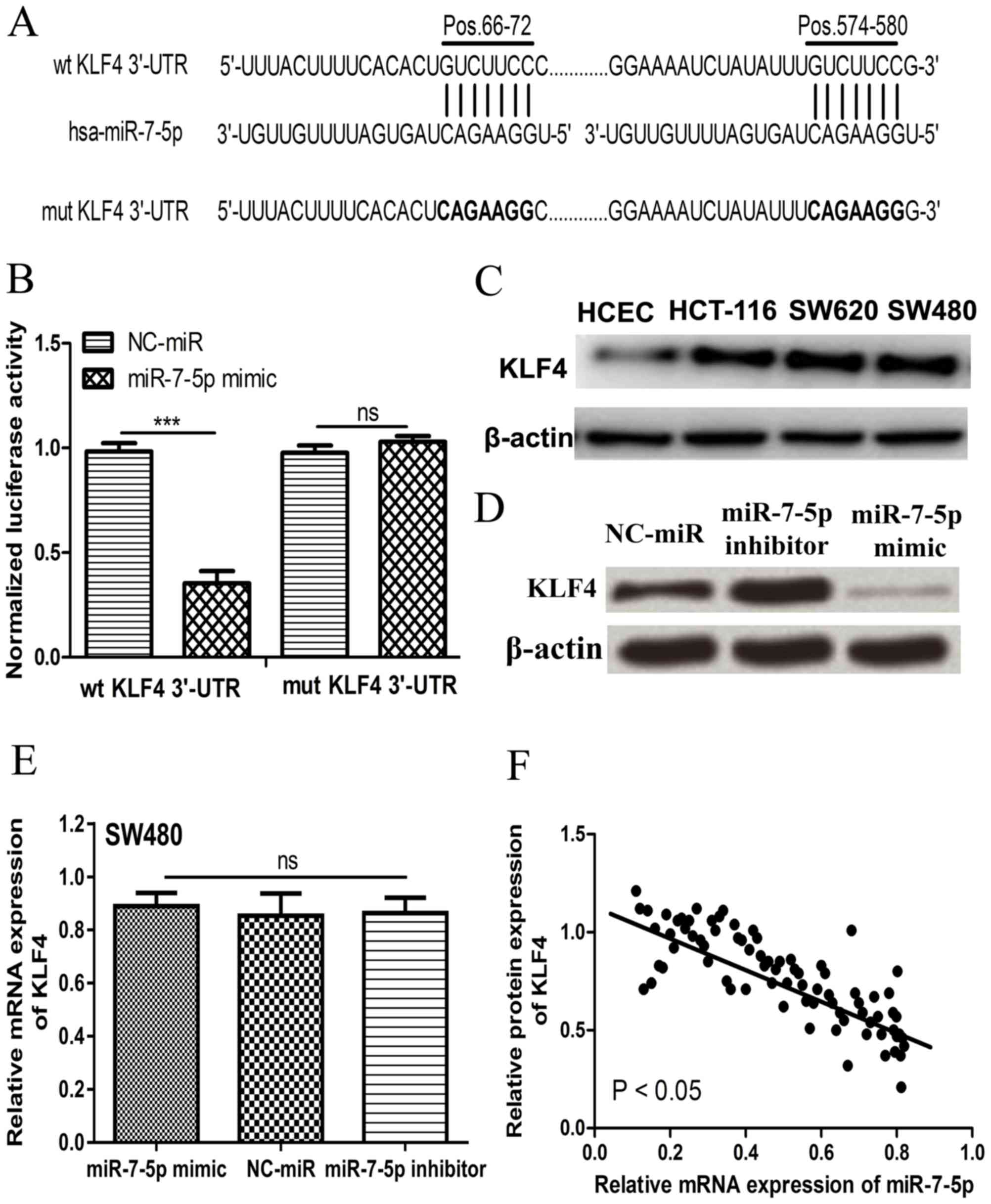
miR-7-5p directly targets KLF4 in
CRC
Analysis using TargetScan demonstrated that KLF4,
with two binding sites in its 3′-UTR, may be a target of miR-7-5p
(Fig. 3A). Luciferase activity
reporter assay was performed to confirm this prediction. It was
revealed that miR-7-5p mimic significantly inhibited the luciferase
activities of wt 3′-UTR of KLF4, however, it did not affect the
luciferase activity of Mut 3′-UTR of KLF4 (Fig. 3B). Furthermore, it was identified
that KLF4 expression was markedly higher in CRC cell lines compared
with the HCEC 1CT cell line (Fig.
3C). Additionally, the expression level of KLF4 was decreased
by miR-7-5p mimic but increased by miR-7-5p inhibitor in SW480
cells (Fig. 3D). However, miR-7-5p
mimic and miR-7-5p inhibitor did not affect the mRNA expression
level of KLF4, which suggests that miR-7-5p regulates KLF4
expression at the posttranscriptional phase (Fig. 3E). In addition, it was identified
that KLF4 protein expression was inversely correlated with miR-7-5p
expression in CRC tumor tissues (Fig.
3F).
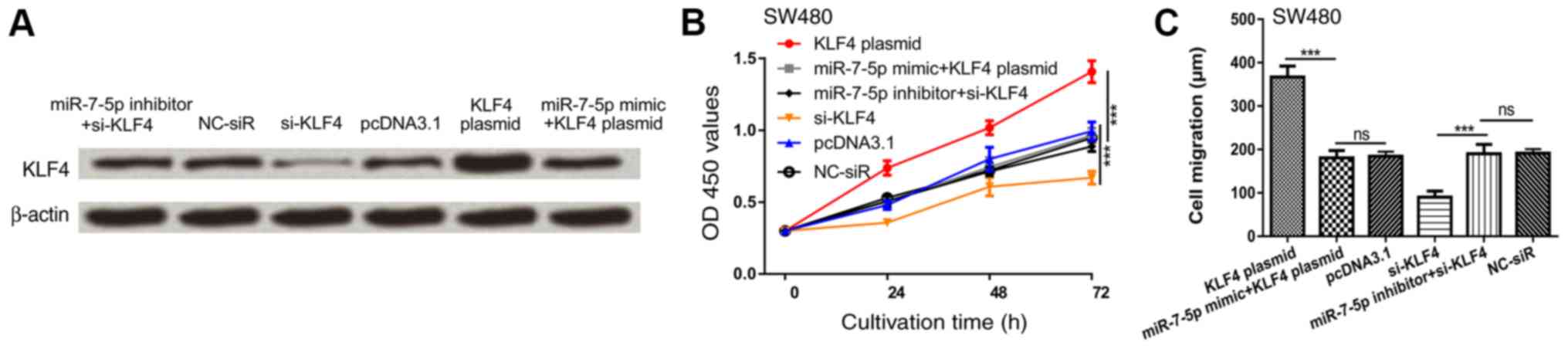
miR-7-5p inhibits CRC proliferation
and migration partly by regulating KLF4
To explore whether KLF4 serves as a mediator for the
suppressive role of miR-7-5p, si-KLF4 and KLF4 plasmid were
introduced into the SW480 cell line. As expected, KLF4 protein
expression was enhanced by KLF4 plasmid but reduced by si-KLF4
(Fig. 4A). The inhibitory effects of
miR-7-4p mimic on KLF4 expression could be reversed by KLF4 plasmid
and the stimulatory effects of miR-7-5p inhibitor on KLF4
expression could be reversed by si-KLF4 (Fig. 4A). Cell proliferation and migration
were enhanced following transfection with KLF4 plasmid but
decreased following transfection with si-KLF4 (Fig. 4B and C). KLF4 overexpression
significantly reversed the inhibition effects of miR-7-5p mimic on
SW480 cell proliferation and migration (Fig. 4B and C). Furthermore, downregulation
of KLF4 reversed the stimulation effects of miR-7-5p inhibitor on
SW480 cell proliferation and migration (Fig. 4B and C). These results indicated that
miR-7-5p inhibits CRC proliferation and migration in part via
regulating KLF4.
Discussion
KLF4 is a zinc-finger transcription factor that
functions as either a tumor suppressor or an oncogene in human
cancer (23). Previous studies have
identified that KLF4 serves a critical role in regulating malignant
cell behaviors by activating or repressing the expression of
downstream target genes (24–27).
Furthermore, accumulating evidence has suggested that KLF4
functions as an oncogene in CRC (18,19).
However, to the best of knowledge, the molecules that regulate KLF4
expression remain to be investigated.
A number of studies have demonstrated that miR-7-5p
expression is frequently downregulated in several types of tumor
(12–14). The present study demonstrated that
miR-7-5p expression was downregulated in CRC cell lines and
tissues, which, to the best of our knowledge, is the first study to
reveal the expression pattern of miR-7-5p in CRC. Furthermore, the
results demonstrated that patients with low miR-7-5p expression
exhibited a larger tumor size, advanced tumor stage and worse
5-year overall survival. These results suggest that low miR-7-5p
expression is associated with aggressive tumor behaviors.
Identifying the targets of miRs in tumors is crucial
for understanding the importance of miRs in tumor progression and
may provide promising antitumor therapeutic targets (18,19).
KLF4, whose expression is upregulated in CRC (18,19), was
predicted as a potential target of miR-7-5p. A luciferase activity
assay validated KLF4 as a direct target of miR-7-5p. Notably, it
was identified that KLF4 expression was inversely correlated with
miR-7-5p in CRC tumor tissues. It should be noted that abnormal
status of cell proliferation and cell migration are two major
characteristics of malignant cells (8). Therefore, the current study
investigated the effects of miR-7-5p on CRC cell proliferation and
cell migration in vitro. miR-7-5p overexpression
significantly inhibited cell proliferation and migration.
Furthermore, it was revealed that overexpression or downregulation
of KLF4 could partially restore the effects of miR-7-5p mimic or
inhibitor on cell behaviors. These findings demonstrated that
miR-7-5p regulates CRC cell proliferation and migration by
targeting KLF4.
In conclusion, the present study provided a novel
insight into the mechanisms underlying human CRC progression.
Decreased miR-7-5p expression was positively associated with CRC
progression by directly targeting the expression of KLF4.
Identification of this association may provide a novel therapeutic
approach for CRC.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YDX conceived and deigned the whole study. MJD and
YMX performed the study and were major contributors in writing the
manuscript. YDX reviewed the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
The No. 2 Hospital of Ningbo (Ningbo, China). Written informed
consent was obtained from all enrolled patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fedawa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hyodo I, Suzuki H, Takahashi K, Saito Y,
Tanaka S, Chiu HM, Kim NK, Li J, Lim R, Villalon A and Boku N:
Present status and perspectives of colorectal cancer in Asia:
Colorectal cancer working Group report in 30th Asia-Pacific Cancer
Conference. Jpn J Clin Oncol. 40 (Suppl 1):i38–i43. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung JJ, Lau JY, Young GP, Sano Y, Chiu
HM, Byeon JS, Yeoh KG, Goh KL, Sollano J, Rerknimitr R, et al: Asia
Pacific consensus recommendations for colorectal cancer screening.
Gut. 57:1166–1176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanahan D and Weinberg RA: Hallmarks of
Cancer: The Next Generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thomas J, Ohtsuka M, Pichler M and Ling H:
MicroRNAs: Clinical relevance in colorectal cancer. Int J Mol Sci.
16:28063–28076. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong YJ, Yu J and Ng SS: MicroRNA
dysregulation as a prognostic biomarker in colorectal cancer.
Cancer Manag Res. 6:405–422. 2014.PubMed/NCBI
|
|
12
|
Giles KM, Brown RA, Epis MR, Kalinowski FC
and Leedman PJ: miRNA-7-5p inhibits melanoma cell migration and
invasion. Biochem Biophys Res Commun. 430:706–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giles KM, Brown RA, Ganda C, Podgorny MJ,
Candy PA, Wintle LC, Richardson KL, Kalinowski FC, Stuart LM, Epis
MR, et al: microRNA-7-5p inhibits melanoma cell proliferation and
metastasis by suppressing RelA/NF-κB. Oncotarget. 7:31663–31680.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi Y, Luo X, Li P, Tan J, Wang X, Xiang T
and Ren G: miR-7-5p suppresses cell proliferation and induces
apoptosis of breast cancer cells mainly by targeting REGγ. Cancer
Lett. 358:27–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shields JM, Christy RJ and Yang VW:
Identification and characterization of a gene encoding a gut
enriched Kruppel-like factor expressed during growth arrest. J Biol
Chem. 271:20009–20017. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le
X, Yao J and Xie K: Loss of Kruppel-like factor 4 expression
contributes to Sp1 overexpression and human gastric cancer
development and progression. Clin Cancer Res. 12:6395–6402. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei D, Kanai M, Jia Z, Le X and Xie K:
Kruppel-like factor 4 induces p27Kip1 expression in and suppresses
the growth and metastasis of human pancreatic cancer cells. Cancer
Res. 68:4631–4639. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lv H, Zhang Z, Wang Y, Li C, Gong W and
Wang X: MicroRNA-92a promotes colorectal cancer cell growth and
migration by inhibiting KLF4. Oncol Res. 23:283–290. 2016.
View Article : Google Scholar
|
|
19
|
Zhai F, Cao C, Zhang L and Zhang J:
miR-543 promotes colorectal cancer proliferation and metastasis by
targeting KLF4. Oncotarget. 8:59246–59256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z,
Sun J, Tan FW, Ding DP, Xu XH, Zhou F, et al: microRNA-92a promotes
lymph node metastasis of human esophageal squamous cell carcinoma
via E-cadherin. J Biol Chem. 286:10725–10734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Edge SB, XXX BD, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Handbook. (7th).
2010.
|
|
23
|
Rowland BD and Peeper DS: KLF4, p21 and
context-dependent opposing forces in cancer. Nat Rev Cancer.
6:11–23. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu Q, Liu M, Zhang J, Xue L, Zhang G, Hu
C, Wang Z, He S, Chen L, Ma K, et al: Overexpression of KLF4
promotes cell senescence through microRNA-203-survivin-p21 pathway.
Oncotarget. 7:60290–60302. 2016.PubMed/NCBI
|
|
25
|
Zhang N, Zhang J, Shuai L, Zha L, He M,
Huang Z and Wang Z: Krüppel-like factor 4 negatively regulates
β-catenin expression and inhibits the proliferation, invasion and
metastasis of gastric cancer. Int J Oncol. 40:2038–2048.
2012.PubMed/NCBI
|
|
26
|
Hu W, Jia Y, Xiao X, Lv K, Chen Y, Wang L,
Luo X, Liu T, Li W, Li Y, et al: KLF4 downregulates hTERT
expression and telomerase activity to inhibit lung carcinoma
growth. Oncotarget. 7:52870–52887. 2016.PubMed/NCBI
|
|
27
|
Zhang W, Geiman DE, Shields JM, Dang DT,
Mahatan CS, Kaestner KH, Biggs JR, Kraft AS and Yang VW: The
gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates
the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J
Biol Chem. 275:18391–18398. 2000. View Article : Google Scholar : PubMed/NCBI
|