Introduction
Esophageal cancer (EC) remains the sixth most common
cause of cancer related death in the world despite recent progress
in multimodal therapy concepts involving neoadjuvant therapy and
standardized surgical approaches (1). EC is associated with high malignant
potential for local invasion and early dissemination resulting in
low overall-survival (OS) rates in patients even after curatively
intended surgery (2).
There are two major histological types of EC that
account for >90% of all malignant neoplasms in the esophagus:
squamous cell carcinoma and adenocarcinoma. The pathogenesis in
adenocarcinoma is generally considered to be driven by epithelial
metaplasia, often caused by acid reflux. In squamous cell
carcinoma, malignant transformation is predominantly associated
with alcohol intake and smoking (3).
The incidence of the particular subtypes has changed in the past
years with increase of adenocarcinoma and decrease of squamous cell
carcinoma in Western countries, mainly due to life style changes
(2). Due to the limited prognosis of
the majority of patients, identification of new biomarkers
predicting the individual prognosis and response to therapy are of
imperative need for allocation of individualized therapy
strategies.
The TP53 gene has been known as one of the
most important tumor suppressor genes, located on human chromosome
17p13.1. p53 plays a major role in tumorigenesis as it controls
cell growth, apoptosis and regulation of angiogenesis (4). Mutations in TP53 were found in
>50% of human cancers, which makes it one of the most mutated
genes in tumors (5). In esophageal
cancer, mutations have been described in frequencies between
40–70%, depending on the underlying cell type (4,6–8). As shown for several tumor entities,
inactivation of the TP53 tumor suppressor gene leads to the
development of malignant cell clones and accelerates the
carcinogenesis (9). Different
mechanisms of functional p53 inactivation have been described
including functionally relevant point mutations and gross
chromosomal alterations, mostly driven by chromosome 17p
deletions.
Previous studies reported inconclusive results on
whether p53 accumulation has a functional impact on progression of
EC (10–19). While some studies found associations
between the p53 expression level and tumor progression or
overall-survival, other studies were not able to confirm these
findings. Data on TP53 deletion status is available from
only one report including 40 patients suffering from esophageal
squamous cell carcinoma (20).
To further elucidate the clinical relevance of TP53
mutations and p53 expression as prognostic biomarkers in esophageal
cancer we took advantage of our preexisting tissue microarray (TMA)
containing nearly 700 esophageal cancer specimens with attached
clinical follow-up and clinico-pathological data.
Our findings demonstrate that strong p53
immunostaining is correlated with unfavorable prognosis in
esophageal cancer, while homozygous TP53 deletions represent
a catastrophic event leading to cell death.
Materials and methods
Patients
For this study, specimens from patients that had
undergone tumor resection in curative intent between 1992 and 2014
at the University Medical Center, Hamburg-Eppendorf were included.
Tissue samples from 691 patients were analyzed including 398
esophageal adenocarcinomas and 293 squamous cell carcinomas. All
data including patient sex, tumor histology, size, lymph node
metastasis and disease stage (UICC 7th edition) were obtained by
reviewing a combination of clinical and pathological records,
outpatient clinic medical records, epidemiological cancer
surveillance data bases and by communication with the patients and
their attending physicians. All resections were performed as
en-bloc esophagectomies with radical two field lymph node
dissection. Fifty patients underwent neoadjuvant therapy (AC n=30,
SCC n=20). Patients that died within 30 days due to postoperative
complications were not considered for survival analysis.
Informed consent was not required due to the
retrospective nature this study. Analysis of anonymized human
tissue samples by the treating physician (including the
pathologist) is permitted according to local laws (§12a
Hamburgisches Krankenhausgesetz). In addition, we obtained approval
for manufacturing and analyzing tissue microarrays made from tissue
samples from anonymized donors from our local review board, the
Ethics Commission of the Ärztekammer Hamburg (no. WF049/09).
TMA construction
The TMA was constructed as previously described
(21). In brief, tissue cores were
obtained from formalin-fixed paraffin-embedded (FFPE) tissue blocks
from patients with pathologically proven esophageal cancer.
Representative areas of the tumor were selected based on
hematoxylin-eosin staining. 691 tissue cylinders with a diameter of
0.6 mm were punched from the ‘donor’ tissue blocks using a
custom-made semi-automatic robotic precision instrument and placed
into one empty recipient paraffin block. The resulting TMA blocks
were used to produce 4 µm sections that were transferred to an
adhesive-coated slide system (Instrumedics Inc., Hackensack, NJ,
USA).
Immunohistochemistry
Freshly cut TMA sections were immunostained on one
day and in one experiment. Slides were deparaffinized and exposed
to heat-induced antigen retrieval for 5 min in an autoclave at
121°C in pH 7.8 Tris-EDTA-Citrate buffer. Primary antibody specific
for p53 (DO1, murine monoclonal IgG2a, Oncogene,
Cambridge, MA; USA; dilution 1:3,600) was applied at 37°C for 60
min. Bound antibody was then visualized using the EnVision Kit
(Dako, Glostrup, Denmark) according to the manufacturer's
directions.
Colon cancers with known p53 alterations served as
positive controls and normal prostate tissue as negative controls
on each TMA section. Nuclear p53 staining intensity was scored in a
four-step scale (0, 1+, 2+, 3+ staining intensity) and the
percentage of stained nuclei was estimated. A final
immunohistochemistry (IHC) result was assigned to each tumor as
described in earlier studies from our group (22). Negative: No staining at all or 1+/2+
staining in <10% of tumor cells, low: 1+ staining in ≥10% or 2+
staining in ≥10% but ≤70% of tumor cells or 3+ staining in ≤10% of
tumor cells, high: 2+ staining in >70% of tumor cells or 3+
staining in >10% of tumor cells. For calculation of results low
and high staining intensities were summed up as positive.
FISH
FISH was used to identify genomic TP53
deletions and translocations. A dual color FISH probe was
constructed from a spectrum green labeled BAC clone (BACs
RP11-89D11, RP11-404G1; Source Bioscience, Nottingham, UK) and a
commercial spectrum orange labeled centromere 17 (CEP17) reference
probe (no. 06J36-06; Abbott Molecular, Wiesbaden, Germany). Freshly
cut 4 µm TMA sections were used for dual color FISH. Before
hybridization, sections were deparaffinized and proteolytically
pretreated with a commercial kit (paraffin pretreatment reagent
kit; Abbott Molecular), followed by dehydration in 70, 80 and 96%
ethanol, air drying and denaturation for 10 min at 72°C in 70%
formamide-23 saline-sodium citrate (SSC) solution. Hybridization
was done overnight at 37°C in a humidified chamber, slides were
washed and counterstained with 0.2 mmol/l
40-6-diamidino-2-phenylindole in an antifade solution.
FISH Scoring
Each TMA spot was carefully evaluated and the
predominant TP53 signal counts were recorded for each FISH
probe. Tissue samples with missing tumor tissue as determined by
corresponding H&E slides were not considered for analysis. In
addition, tumor spots were excluded from analysis if there was
evidence for insufficient hybridization such as lack of TP53
signals in both tumor and peritumoral non-malignant tissue.
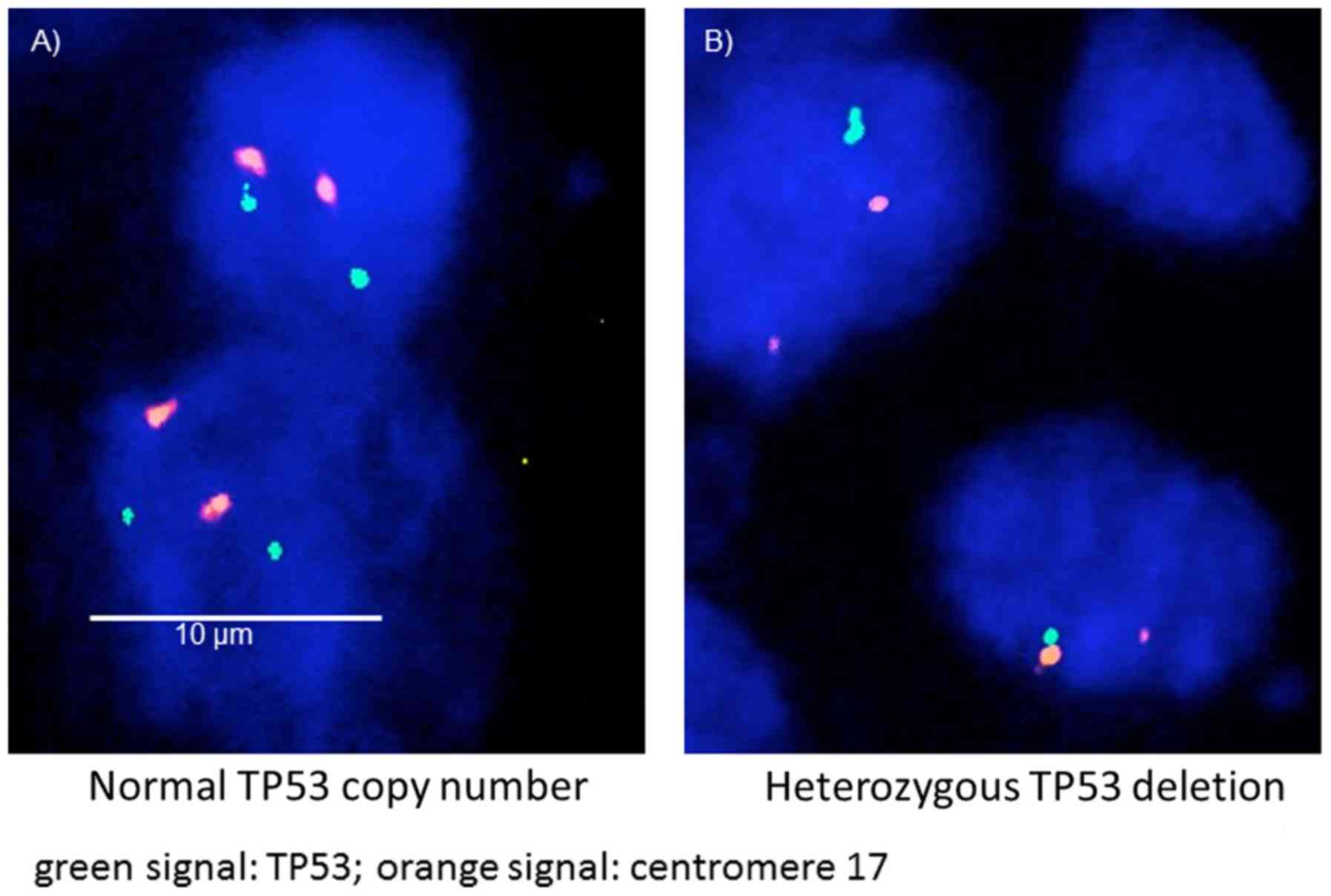
Deletion of TP53 was defined as presence of fewer
TP53 signals than centromere 17 probe signals (heterozygous
deletion) or complete absence of TP53 signals but presence
of at least one centromere 17 probe signal (homozygous deletion) in
≥60% of tumor nuclei. The aforementioned cut-off level was chosen
because a good correlation between FISH and array genomic
hybridization was already shown by our group for PTEN and
p53 in prostate cancer (22,23).
Statistical analysis
SPSS Statistics for Mac (version 17; SPSS, Inc.,
Chicago, IL, USA) was used for statistical analysis.
Interdependence between immunostaining and FISH results as well as
clinical data was calculated using the Chi-squared and Fisher's
exact tests and displayed by cross-tables. Group differences were
examined using the t-test. Survival curves were plotted using the
Kaplan-Meier method and analyzed using the log-rank test.
Univariate and multi-variate analyses were performed for prognostic
factors of recurrence-free and overall survival using the Cox
regression model. All tests were two-sided. P<0.05 was
considered to indicate a statistically significant difference.
Results
p53 IHC
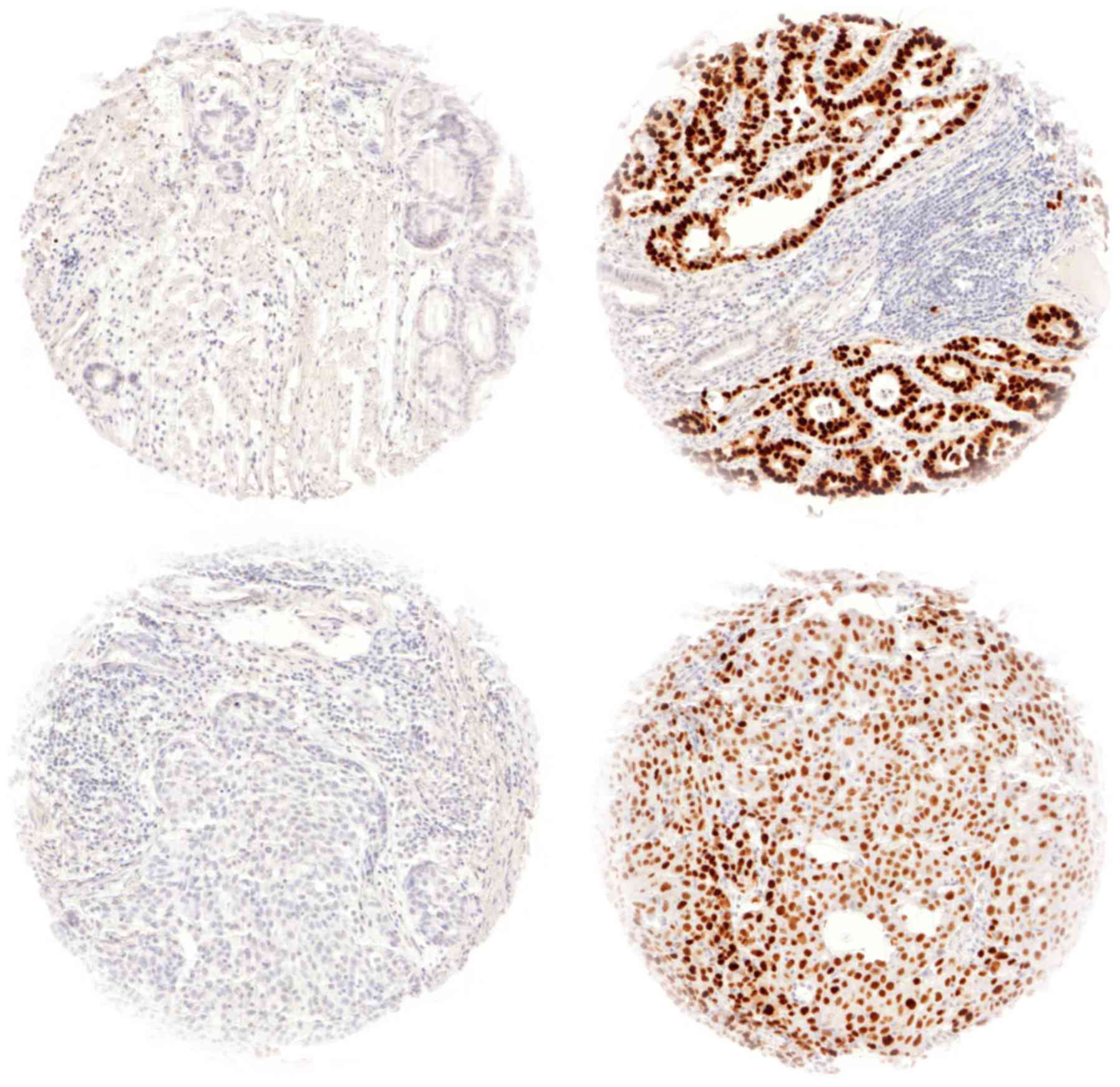
p53 immunostaining was interpretable in 574 (83.2%)
of 691 samples, in total 314 esophageal adenocarcinomas (AC) and
260 squamous cell carcinomas (SCC) were evaluable. Reasons for
non-informative cases included lack of tissue samples or absence of
unequivocal cancer tissue in the TMA spot. p53 positivity was seen
in 104 SCC (40.0%) and in 144 AC of the esophagus (45.9%).
Representative images of p53 immunostaining in AC and SCC are given
in Fig. 1. p53 positivity was
associated with tumor stage (P=0.019), UICC stage (P=0.004),
grading (P=0.027) and surgical resection margin status (P=0.006) in
esophageal AC (Table I), while no
associations between clinico-pathological data and p53
immunostaining in squamous cell carcinoma were revealed (Table II).
 | Table I.Association between p53
immunostaining and TP53 FISH with clinicopathological
parameters in adenocarcinoma. |
Table I.
Association between p53
immunostaining and TP53 FISH with clinicopathological
parameters in adenocarcinoma.
|
| p53 IHC result | TP53 FISH
result |
|---|
|
|
|
|
|---|
| Parameter | Evaluable cases
(n) | Negative (%) | Positive (%) | P-value | Evaluable cases
(n) | no del (%) | het del (%) | P-value |
|---|
| All tumors | 314 | 54.1 | 45.9 |
| 269 | 59.1 | 40.9 |
|
| Age, years |
|
|
| 0.645 |
|
|
| 0.031 |
|
≤65 | 107 | 52.3 | 47.7 |
| 91 | 89.2 | 10.8 |
|
|
>65 | 207 | 55.1 | 44.3 |
| 178 | 69.9 | 30.1 |
|
| Sex |
|
|
| 0.186 |
|
|
| 0.074 |
|
Male | 262 | 52.7 | 47.3 |
| 226 | 63.4 | 36.6 |
|
|
Female | 51 | 62.7 | 37.3 |
| 42 | 95.9 | 4.5 |
|
| Tumor stage |
|
|
| 0.019 |
|
|
| 0.195 |
|
pT1 | 61 | 70.5 | 29.5 |
| 40 | 92.2 | 7.8 |
|
|
pT2 | 33 | 60.6 | 39.4 |
| 26 | 95.9 | 4.1 |
|
|
pT3 | 96 | 49.0 | 51.0 |
| 156 | 72.8 | 27.2 |
|
|
pT4 | 22 | 45.4 | 54.6 |
| 21 | 98.1 | 1.9 |
|
| Lymph node
metastasis |
|
|
| 0.069 |
|
|
| 0.923 |
|
pN0 | 96 | 64.6 | 35.4 |
| 79 | 88.4 | 11.6 |
|
|
pN1 | 51 | 53.0 | 47.0 |
| 50 | 92.5 | 7.5 |
|
|
pN2 | 81 | 51.9 | 48.1 |
| 65 | 89.2 | 10.8 |
|
|
pN3 | 84 | 45.2 | 54.8 |
| 74 | 88.8 | 11.2 |
|
| Distant
metastasis |
|
|
| 0.475 |
|
|
| 0.974 |
| M0 | 277 | 54.9 | 45.1 |
| 237 | 63.9 | 36.1 |
|
| M1 | 37 | 48.6 | 51.4 |
| 32 | 95.2 | 4.8 |
|
| UICC stage |
|
|
| 0.004 |
|
|
| 0.977 |
| I | 62 | 72.6 | 27.4 |
| 52 | 92.5 | 7.5 |
|
| II | 41 | 61.0 | 39.0 |
| 33 | 95.1 | 4.9 |
|
|
III | 173 | 48.0 | 52.0 |
| 151 | 76.3 | 23.7 |
|
| IV | 34 | 44.1 | 55.9 |
| 30 | 95.5 | 4.5 |
|
| Grading |
|
|
| 0.027 |
|
|
| 0.922 |
| G1 | 15 | 86.7 | 13.3 |
| 12 | 98.5 | 1.5 |
|
| G2 | 113 | 54.9 | 45.1 |
| 98 | 84.2 | 15.8 |
|
| G3 | 175 | 50.9 | 49.1 |
| 152 | 76.2 | 23.8 |
|
| G4 | 6 | 83.3 | 16.7 |
| 3 | 99.6 | 0.4 |
|
| Surgical resection
margin |
|
|
| 0.006 |
|
|
| 0.029 |
| R0 | 228 | 59.2 | 40.8 |
| 174 | 72.7 | 27.3 |
|
| R1 | 78 | 38.5 | 61.5 |
| 63 | 86.4 | 13.6 |
|
| R2 | 3 | 66.7 | 33.3 |
| 3 | 99.6 | 0.4 |
|
 | Table II.Association of p53 immunostaining and
TP53 fluorescence in situ hybridization with
clinicopathological parameters in squamous cell carcinoma. |
Table II.
Association of p53 immunostaining and
TP53 fluorescence in situ hybridization with
clinicopathological parameters in squamous cell carcinoma.
|
| p53 IHC result | TP53 FISH
result |
|---|
|
|
|
|
|---|
| Parameter | Evaluable cases
(n) | Negative (%) | Positive (%) | P-value | Evaluable cases
(n) | no del (%) | het del (%) | P-value |
|---|
| All tumors | 260 | 60.0 | 40.0 |
| 237 | 80.6 | 19.4 |
|
| Age, years |
|
|
| 0.678 |
|
|
| 0.11 |
|
<65 | 101 | 58.4 | 41.6 |
| 94 | 90.3 | 9.7 |
|
|
>65 | 159 | 61.0 | 39.0 |
| 143 | 90.3 | 9.7 |
|
| Sex |
|
|
| 0.568 |
|
|
| 0.945 |
|
Male | 190 | 61.1 | 38.9 |
| 171 | 86.1 | 13.9 |
|
|
Female | 70 | 57.1 | 42.9 |
| 66 | 94.5 | 5.5 |
|
| Tumor stage |
|
|
| 0.694 |
|
|
| 0.028 |
|
pT1 | 43 | 67.4 | 32.6 |
| 42 | 97.9 | 2.1 |
|
|
pT2 | 55 | 56.4 | 43.6 |
| 46 | 98.3 | 1.7 |
|
|
pT3 | 144 | 59.7 | 40.3 |
| 134 | 85.2 | 14.8 |
|
|
pT4 | 18 | 55.6 | 44.4 |
| 15 | 99.2 | 0.8 |
|
| Lymph node
metastasis |
|
|
| 0.276 |
|
|
| 0.009 |
|
pN0 | 123 | 53.7 | 46.3 |
| 115 | 92.4 | 7.6 |
|
|
pN1 | 58 | 63.8 | 36.2 |
| 53 | 95.8 | 4.2 |
|
|
pN2 | 52 | 67.3 | 32.7 |
| 49 | 95.8 | 4.2 |
|
|
pN3 | 26 | 65.4 | 34.6 |
| 20 | 96.6 | 3.4 |
|
| Distant
metastasis |
|
|
| 0.873 |
|
|
| 0.62 |
| M0 | 211 | 59.7 | 40.3 |
| 195 | 83.5 | 16.5 |
|
| M1 | 49 | 61.2 | 38.8 |
| 42 | 97.0 | 3.0 |
|
| UICC stage |
|
|
| 0.903 |
|
|
| 0.074 |
| I | 60 | 13.1 | 10.0 |
| 6 | 97.5 | 2.5 |
|
| II | 60 | 14.6 | 8.5 |
| 9 | 96.2 | 3.8 |
|
|
III | 91 | 20.8 | 14.2 |
| 23 | 90.3 | 9.7 |
|
| IV | 49 | 11.2 | 7.3 |
| 8 | 96.6 | 3.4 |
|
| Grading |
|
|
| 0.63 |
|
|
| 0.81 |
| G1 | 3 | 0.4 | 0.8 |
| 4 | 99.6 | 0.4 |
|
| G2 | 163 | 38.1 | 24.6 |
| 151 | 88.6 | 11.4 |
|
| G3 | 93 | 21.5 | 14.2 |
| 81 | 92.8 | 7.2 |
|
| Surgical resection
margin |
|
|
| 0.84 |
|
|
| 0.54 |
| R0 | 192 | 43.8 | 30.0 |
| 179 | 86.4 | 13.6 |
|
| R1 | 52 | 12.7 | 7.3 |
| 45 | 95.3 | 4.7 |
|
| R2 | 14 | 3.1 | 2.3 |
| 12 | 98.7 | 1.3 |
|
TP53 FISH analysis
FISH was interpretable for TP53 deletions in
269 (67.6%) samples in esophageal adenocarcinoma and 237 (80.9%)
samples in squamous cell carcinoma. Non-informative cases were
caused by inefficient hybridization, missing tissue spots or
absence of representative tumor tissue on the TMA spot.
Heterozygous TP53 deletions were detectable in 110 samples
(40.9%) in EC and 46 samples (19.4%) in SCC. TP53 deletions
were associated with age (P=0.031) and surgical resection margin
(P=0.029) in adenocarcinoma (Table
I). For squamous cell carcinoma, associations were detected
between heterozygous TP53 deletions and tumor stage
(P=0.028) and presence of lymph node metastasis (P=0.009) (Table II). Not one single TMA spot revealed
cells with homozygous TP53 deletions.
Combined effect of p53 immunostaining
and TP53 deletions
Data on both p53 immunostaining and TP53 FISH
were available from 244 adenocarcinomas and 223 squamous cell
carcinomas. A correlation between high p53 immunostaining and
TP53 deletion could not be revealed for AC (P=0.643) while
in SCC an association (P=0.044) was seen. In adenocarcinoma, 54
(21.8%) tumors showed heterozygous TP53 deletion accompanied
by high p53 expression which was also displayed by 25 SCC (11.2%).
However, this finding was merely associated with resection margin
status (P=0.013) in esophageal AC.
Survival analysis
In total, 283 patients with adenocarcinoma and 231
with squamous cell carcinoma were available for survival analysis.
High-level p53 immunostaining was linked to shortened
overall-survival (OS) in both esophageal AC and SCC as analyzed by
Kaplan-Meier (P=0.021 and P=0.013, respectively; Fig. 2A and D). TP53 deletions were
not associated with OS irrespective of the histological type
(P=0.973 (AC) and P=0.099 (SCC); Fig. 2B
and E). Combination of p53 expression and TP53 deletion
status did not improve the prognostic power compared to p53 IHC
alone (Fig. 2C and F).
Multivariate analysis
For both histological types, UICC stage and a
complete surgical resection (R0) proved to be independent
prognostic markers as analyzed by multivariate cox-regression
model. Furthermore, strong p53 immunostaining was also
independently associated with OS in SCC (Table III).
 | Table III.Multivariate analysis of p53
immunostaining in esophageal cancer. |
Table III.
Multivariate analysis of p53
immunostaining in esophageal cancer.
|
| Adenocarcinoma | Squamous cell
carcinoma |
|---|
|
|
|
|
|---|
|
|
| 95% confidence
interval |
|
| 95% confidence
interval |
|
|---|
|
|
|
|
|
|
|
|
|---|
|
|
| Parameter | HR | Lower | Upper | P-value | HR | Lower | Upper | P-value |
|---|
| Sex
(male/female) | 0.741 | 0.462 | 1.187 | 0.212 | 0.815 | 0.562 | 1.184 | 0.284 |
| Age group
(<65/>65 years) | 1.414 | 0.965 | 2.072 | 0.076 | 0.920 | 0.663 | 1.277 | 0.620 |
| UICC Stage | 2.232 | 1.626 | 3.065 | <0.001 | 1.288 | 1.019 | 1.629 | 0.034 |
| Distant
metastasis | 1.095 | 0.581 | 2.063 | 0.780 | 1.001 | 0.579 | 1.730 | 0.998 |
| Surgical resection
margin | 1.677 | 1.152 | 2.440 | 0.007 | 1.396 | 1.061 | 1.836 | 0.017 |
| Grading | 1.219 | 0.874 | 1.700 | 0.243 | 1.131 | 0.829 | 1.543 | 0.438 |
| p53
immunostaining | 1.186 | 0.829 | 1.698 | 0.351 | 1.459 | 1.054 | 2.022 | 0.023 |
| TP53
FISH | 0.853 | 0.593 | 1.227 | 0.391 | 1.095 | 0.745 | 1.610 | 0.643 |
Discussion
Our study shows that p53 overexpression is linked to
shortened overall survival in patients suffering from EC. To our
knowledge, this is the study includes the largest number of cases
for EC and SCC with corresponding survival data. Furthermore, we
correlated p53 IHC and TP53 FISH analysis. As follows in the
discussion, previous studies either present inconclusive results or
include only few patients.
In our analysis p53 alterations are present in
40–45% of esophageal cancer, irrespective of the underlying
histological type. Genome studies conducted either with array
comparative genome hybridization (CGH) or whole genome sequencing
repeatedly revealed TP53 mutations as the most common gene
mutation in EC with reported mutation rates between 40–70%
(4,7,24). The
variance of the reported mutation frequencies is in concordance to
immunohistochemical confirmation studies for SSC and the results
revealed from our study (19,25–28).
In contrast, data from large patient cohorts concerning p53
expression in esophageal adenocarcinoma are hardly available. A
recent review article by Chen and colleagues analyzed biomarkers in
esophageal cancer and found only 2 studies focusing on p53
expression in esophageal adenocarcinoma with 97 patients included
in the meta-analysis (14). These
studies also showed p53 protein alterations in 40–65%, although the
case load is too small to draw general conclusions. Our group has
previously shown that the applied IHC protocols, for prostate
cancer, reveal high concordance of IHC data with sequencing
results, arguing for a strong validity of our IHC protocol
(22).
p53 is physiologically expressed by numerous cell
types and plays a major role in cell cycle control. It is
considered one of the most important tumor suppressor genes
(9). p53 mutations have been found
in multiple cancers with large variances in the mutation frequency,
depending on the observed tumor entity (29).
In our study heterozygous TP53 deletions were
found in 41% of AC and 19% of SCC. Only little data on TP53
gene deletions have yet been reported. Recently, a study on 40
patients suffering from esophageal squamous cell carcinoma revealed
heterozygous deletion of TP53 in 22 patients (55%). A
correlation with clinical data could not be established in this
study (20). The deletion rate is
somewhat higher than it is in our data, which is partially caused
by our stringent criteria for defining TP53 deletions. These
were applied to avoid false deletion calling due to truncation of
the nuclei during tissue sectioning. Most samples with TP53
deletion had >80% cells with fewer TP53 fluorescence
signals than centromere 17 signals. In contrast to other studies,
we did not use automated systems for detection of particular gene
deletions which is another possibility for deviating deletion rates
compared to previous published reports (20). As already shown in prostate cancer on
a TMA with >11,000 patient samples, 100% concordance between
array-CGH detected deletions and FISH could be achieved for
PTEN and TP53 deletions using these criteria
(22,23,30).
Therefore, differences in the deletion rate are explained by
different protocols calling more deletions as present within one
patient sample. The deletion rate of 19% supports the assumption
that TP53 deletions have only minor significance as a
pathway for p53 inactivation in esophageal SCC. In line with the
results revealed by our study, the impact of p53 mutations is of
higher importance with respect to tumor progression than the
heterozygous gene deletion of chromosome 17p13. These findings also
match a Northern blot analysis on expression of the p53 gene in
esophageal tumorigenesis. RNA was extracted from tumor, Barrett's
epithelium, and histologically normal esophageal mucosa. p53 was
found to be overexpressed in cancerous or metaplastic tissue in
comparison to normal tissue. Thus, the authors concluded that p53
is implicated in the progression of Barrett's epithelium to
invasive cancer (31).
By analyzing p53 immunostaining and TP53
deletions, we were able to correlate p53 IHC and TP53
deletion status with corresponding clinical data of our patient
cohort. Patients with p53 overexpression in IHC presented with
shortened OS and unfavorable clinic-pathological data, meaning that
p53 mutations must have an influence on tumor biology. This effect
is also paralleled by a significant correlation between advanced
tumor stages and high p53 expression levels in AC (Table I: Tumor stage (P=0.019), UICC stage
(P=0.004), grading (P=0.027) and surgical resection margin status
(P=0.006)). Furthermore, the multivariate analysis of p53
immunostaining in a setting with various established prognostic
factors for esophageal cancer, such as UICC stage, distant
metastasis, resection margin and grading revealed that p53
accumulation is an independent prognostic marker in SCC. In
contrast, in our analysis, heterozygous TP53 deletions do
not affect the OS, neither in AC nor in SCC. These results confirm
the findings of former studies in EC and other tumor entities that
mono-allelic protein expression is sufficient for regular cell
cycle control mediated by p53 (29).
Our data show, that mono-allelic TP53 deletion cannot be
considered a major driver for tumor progression in esophageal
cancer while there is no mutation in the remaining TP53 allele.
Since the poor outcome of patients with strong p53
immunostaining was independent of the TP53 deletion status, it is
tempting to speculate that these cancers may at least carry
dominant negative mutations with complete inactivation of wild-type
p53 protein through complex formation with mutant p53 protein. This
mechanism is known to lead to massive nuclear accumulation of
inactive p53 complexes composed of mutated and non-mutated p53
protein (29). While IHC reveals
both mutated and non-mutated p53, cells with accumulation of
inactive p53 complexes are the ones that will predominantly show
strong staining. Thus, strong p53 immunostaining in our samples
indicates an accumulation of unfunctional p53.
Alterations of p53 have been found in virtually
every region of the protein but only a handful of the most
frequently occurring mutations haven been studied in depth for
their contribution to cancer progression (32). In EC, the majority of patients
present with TP53 exon single nucleotide mutations
(approximately 70%) while frameshift mutations are much less
frequent. Only 15% of cases have been reported to have insertions
or deletions as found by genome wide sequencing and PCR exon
analysis (6,32). Numerous in vitro and in
vivo studies confirmed the ability of mutant p53 to drive
enhanced cancer invasion and motility, with evidence that mutant
p53 can enhance signaling through receptors such as transforming
growth factor β (TGFβ) or epidermal growth factor receptor (EGFR).
Additionally, although mutated p53 has generally lost the ability
to bind p53 DNA binding regions in target gene promoters, various
p53 mutants can bind directly to DNA with some degree of
selectivity and may thereby control the transcription of certain
genes (33,34). Furthermore, there is increasing
evidence that mutated p53 has an inhibitory effect on transcription
factors, such as TAp63, which is of regulative importance for
numerous miRNA with important roles in cancer invasion and
progression (35,36).
In contrast to other cancers, such as prostate
cancer, the combination of gene deletion and mutation status does
not increase the malignant potential in esophageal cancer (22). In esophageal adenocarcinoma only 53
(21%) patients showed TP53 deletion combined with strong p53
immunostaining, the rate being even lower in SCC with only 11%.
These low frequencies suggest that biallelic inactivation, which is
associated with a total loss of functional p53, is a catastrophic
cellular event related with a high level of apoptosis. Studies
using transgenic mouse models carrying heterozygous TP53
deletions show larger and more mammalian tumors than animals with
two wild type TP53 alleles (37). Moreover, the results from animal
studies confirm the findings in our study, as these results
indicate that loss of both TP53 alleles is not a
prerequisite for tumor formation and that mere reduction in p53
levels may be sufficient to promote tumorigenesis.
Other studies have examined serum levels of p53
antibodies and their correlation to outcome. Shimada et al
(38) reported on 28 patients with
SCC that were positive for serum p53 antibodies out of a cohort of
105 cancers. These patients' survival was significantly worse than
that of seronegative patients. To our knowledge, no correlation
between serum p53 antibody levels and p53 expression in cancer
tissue has yet been described for esophageal cancer. Our study does
not provide any data on this issue since serum p53 antibodies were
not examined in our cohort.
Seeing that cancers with strong p53 expression are
correlated with poor survival independent of the established
clinic-pathological prognostic markers supports the notion that
these patients might profit from neoadjuvant and adjuvant
therapeutic regimes in a multimodal setting. Tumor size, nodal
status and resection margin appear to not be the only relevant
markers for prediction of survival in EC and therefore the search
for prognostic biomarkers is warranted even in this highly
malignant disease with poor overall survival even in less advanced
stages.
In summary, the results of our large-scale TMA
analysis in esophageal adeno- and squamous cell carcinoma show that
different types of p53 alterations characterize subgroups of
patients with different outcomes. Strong p53 expression is
correlated with unfavorable prognosis in esophageal cancer and
represents an independent prognosticator in SCC. Furthermore,
homozygous TP53 deletions are catastrophic cellular events
related with a high level of apoptosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NM, SN, MK, RS and CHM conceived and designed the
study, and acquired, analyzed and interpreted the data. SS, AH, EB,
FJ, WW, AQ, MB, KG and MT were involved in drafting the manuscript,
made substantial contributions to conception, analysis and
interpretation of data and revised it critically for important
intellectual content. JI, GS and FG made substantial contributions
to study design, acquisition of data and gave final approval of the
version to be published.
Ethics approval and consent to
participate
Informed consent was not required due to the
retrospective nature this study. Approval for manufacturing and
analyzing tissue microarrays made from the tissue samples of
anonymized donors was obtained from our local review board, the
Ethics Commission of the Ärztekammer Hamburg (no. WF049/09).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
van Hagen P, Hulshof MC, van Lanschot JJ,
Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ,
Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al: Preoperative
chemoradiotherapy for esophageal or junctional cancer. N Engl J
Med. 366:2074–2084. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rustgi AK and El-Serag HB: Esophageal
carcinoma. N Engl J Med. 371:2499–2509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bian YS, Osterheld MC, Bosman FT,
Benhattar J and Fontolliet C: p53 gene mutation and protein
accumulation during neoplastic progression in Barrett's esophagus.
Mod Pathol. 14:397–403. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dulak AM, Stojanov P, Peng S, Lawrence MS,
Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, et
al: Exome and whole-genome sequencing of esophageal adenocarcinoma
identifies recurrent driver events and mutational complexity. Nat
Genet. 45:478–486. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L,
Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al: Genomic and
molecular characterization of esophageal squamous cell carcinoma.
Nat Genet. 46:467–473. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y,
Liu Z, Zhan Q, Liu Y, Yu D, et al: Genome-wide association study
identifies three new susceptibility loci for esophageal
squamous-cell carcinoma in Chinese populations. Nat Genet.
43:679–684. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Muller PA and Vousden KH: p53 mutations in
cancer. Nat Cell Biol. 15:2–8. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang K, Chen L, Zhang J, Wu Z, Lan L,
Wang L, Lu B and Liu Y: Elevated p53 expression levels correlate
with tumor progression and poor prognosis in patients exhibiting
esophageal squamous cell carcinoma. Oncol Lett. 8:1441–1446. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murata A, Baba Y, Watanabe M, Shigaki H,
Miyake K, Karashima R, Imamura Y, Ida S, Ishimoto T, Iwagami S, et
al: p53 immunohistochemical expression and patient prognosis in
esophageal squamous cell carcinoma. Med Oncol. 30:7282013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yao W, Qin X, Qi B, Lu J, Guo L, Liu F,
Liu S and Zhao B: Association of p53 expression with prognosis in
patients with esophageal squamous cell carcinoma. Int J Clin Exp
Pathol. 7:7158–7163. 2014.PubMed/NCBI
|
|
13
|
Blanchard P, Quero L, Pacault V,
Schlageter MH, Baruch-Hennequin V and Hennequin C: Prognostic
significance of anti-p53 and anti-KRas circulating antibodies in
esophageal cancer patients treated with chemoradiotherapy. BMC
Cancer. 12:1192012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen M, Huang J, Zhu Z, Zhang J and Li K:
Systematic review and meta-analysis of tumor biomarkers in
predicting prognosis in esophageal cancer. BMC Cancer. 13:5392013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dey B, Raphael V, Khonglah Y and Lynrah
KG: Immunohistochemical analysis of P53 and PRB in esophageal
squamous cell carcinoma. J Clin Diagn Res. 8:FC01–FC03.
2014.PubMed/NCBI
|
|
16
|
Madani K, Zhao R, Lim HJ and Casson AG:
Prognostic value of p53 mutations in oesophageal adenocarcinoma:
Final results of a 15-year prospective study. Eur J Cardiothorac
Surg. 37:1427–1432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pühringer-Oppermann F, Stahl M, Keller G
and Sarbia M: Lack of prognostic impact of p53 gene mutation and
p53 phosphorylation at serine 15 in multimodally treated
adenocarcinomas of the gastroesophageal junction. J Cancer Res Clin
Oncol. 132:433–438. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang ZB, Peng XZ, Chen SS, Ning FL, Du CJ,
Wang K, Ma W and Cheng YF: High p53 and MAP1 light chain 3A
co-expression predicts poor prognosis in patients with esophageal
squamous cell carcinoma. Mol Med Rep. 8:41–46. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu XL, Zheng WH, Tao KY, Li XX, Xu WZ,
Wang Y, Zhu SM and Mao WM: p53 is an independent prognostic factor
in operable esophageal squamous cell carcinoma: A large-scale study
with a long follow-up. Med Oncol. 31:2572014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Niyaz M, Turghun A, Ping ZH, Zhu Z,
Sheyhedin I, Ren C and Awut I: TP53 gene deletion in esophageal
cancer tissues of patients and its clinical significance. Mol Med
Rep. 7:122–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mirlacher M and Simon R: Recipient block
TMA technique. Methods Mol Biol. 664:37–44. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kluth M, Harasimowicz S, Burkhardt L,
Grupp K, Krohn A, Prien K, Gjoni J, Haß T, Galal R, Graefen M, et
al: Clinical significance of different types of p53 gene alteration
in surgically treated prostate cancer. Int J Cancer. 135:1369–1380.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krohn A, Diedler T, Burkhardt L, Mayer PS,
De Silva C, Meyer-Kornblum M, Kötschau D, Tennstedt P, Huang J,
Gerhäuser C, et al: Genomic deletion of PTEN is associated with
tumor progression and early PSA recurrence in ERG fusion-positive
and fusion-negative prostate cancer. Am J Pathol. 181:401–412.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shang L, Liu HJ, Hao JJ, Jiang YY, Shi F,
Zhang Y, Cai Y, Xu X, Jia XM, Zhan QM and Wang MR: A panel of
overexpressed proteins for prognosis in esophageal squamous cell
carcinoma. PLoS One. 9:e1110452014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saemi N, Khoshnevis J, Akbari ME, Meysamie
A, Korourian A, Gholizadeh B, Larijani L, Moradi A, Baikpour M,
Baikpour M and Zham H: Evaluating the correlation between the
survival rate of patients with esophageal squamous cell carcinoma
and expression of p53 and cyclin D1 biomarkers along with other
prognostic factors. J Gastrointest Cancer. 49:35–40. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang L, Yu X, Li J, Zhang Z, Hou J and Li
F: Prognostic significance of p53 expression in patients with
esophageal cancer: A meta-analysis. BMC Cancer. 16:3732016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Z, Wang P, Gao Y and He J: The high
expression instead of mutation of p53 is predictive of overall
survival in patients with esophageal squamous-cell carcinoma: A
meta-analysis. Cancer Med. 6:54–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Muller PA and Vousden KH: Mutant p53 in
cancer: New functions and therapeutic opportunities. Cancer Cell.
25:304–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Steurer S, Mayer PS, Adam M, Krohn A, Koop
C, Ospina-Klinck D, Tehrani AA, Simon R, Tennstedt P, Graefen M, et
al: TMPRSS2-ERG fusions are strongly linked to young patient age in
low-grade prostate cancer. Eur Urol. 66:978–981. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sorsdahl K, Casson AG, Troster M, Van
Meyel D, Inculet R and Chambers AF: p53 and ras gene expression in
human esophageal cancer and Barrett's epithelium: A prospective
study. Cancer Detect Prev. 18:179–185. 1994.PubMed/NCBI
|
|
32
|
Leroy B, Fournier JL, Ishioka C, Monti P,
Inga A, Fronza G and Soussi T: The TP53 website: An integrative
resource centre for the TP53 mutation database and TP53 mutant
analysis. Nucleic Acids Res 41 (Database Issue). D962–D969. 2013.
View Article : Google Scholar
|
|
33
|
Brázdová M, Navrátilová L, Tichý V,
Němcová K, Lexa M, Hrstka R, Pečinka P, Adámik M, Vojtesek B,
Paleček E, et al: Preferential binding of hot spot mutant p53
proteins to supercoiled DNA in vitro and in cells. PLoS One.
8:e595672013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weisz L, Oren M and Rotter V:
Transcription regulation by mutant p53. Oncogene. 26:2202–2211.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dong P, Karaayvaz M, Jia N, Kaneuchi M,
Hamada J, Watari H, Sudo S, Ju J and Sakuragi N: Mutant p53
gain-of-function induces epithelial-mesenchymal transition through
modulation of the miR-130b-ZEB1 axis. Oncogene. 32:3286–3295. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tucci P, Agostini M, Grespi F, Markert EK,
Terrinoni A, Vousden KH, Muller PA, Dötsch V, Kehrloesser S, Sayan
BS, et al: Loss of p63 and its microRNA-205 target results in
enhanced cell migration and metastasis in prostate cancer. Proc
Natl Acad Sci USA. 109:15312–15317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Doyle B, Morton JP, Delaney DW, Ridgway
RA, Wilkins JA and Sansom OJ: p53 mutation and loss have different
effects on tumourigenesis in a novel mouse model of pleomorphic
rhabdomyosarcoma. J Pathol. 222:129–137. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shimada H, Nabeya Y, Okazumi S, Matsubara
H, Funami Y, Shiratori T, Hayashi H, Takeda A and Ochiai T:
Prognostic significance of serum p53 antibody in patients with
esophageal squamous cell carcinoma. Surgery. 132:41–47. 2002.
View Article : Google Scholar : PubMed/NCBI
|