Introduction
Ovarian cancer (OC) is the most common gynecologic
malignancy and remains the leading cause of cancer-related
mortality worldwide (1). Despite
advances in chemotherapy and surgical treatment, the 5 year
survival rate of OC is ~30% due to its frequent recurrence
(2). Negative outcome of OC are
mainly due to asymptomatic stages, rapid metastasis and
chemotherapy resistance (3).
Although the understanding of OC is constantly progressing, its
underlying molecular mechanisms remain unclear. Most patients with
OC have developed metastases by the time of their first diagnosis
(4). There is therefore an urgent
need to identify potential prognostic biomarkers for the prediction
of OC clinical outcomes.
Circular RNAs (circRNAs) are produced from
back-splicing of precursor mRNAs and represent a novel class of
endogenous noncoding RNAs (5). These
mRNAs have been considered for decades as abnormal splicing
products of RNAs due to their low expression levels. Alongside the
development of high-throughput sequencing and bioinformatics
technologies, recent studies have confirmed that circRNAs are
abundant, stable and conserved in mammalian cells (6–8). The
involvement of circRNAs in cancer pathology is therefore
intensively studied. It has been reported that circRNAs negatively
modulate micro (mi)RNA expression by harboring their binding sites,
and subsequently further affect the levels of downstream mRNA
(9). For example, the circRNA of
ciRS-7 sequence works as a competing endogenous RNA that sponges
the miRNA miR-7, which leads to an increase in miR-7 targets
expression in various types of cancer (10). Furthermore, RNA-sequencing analyses
revealed that circRNAs are enriched in cancer cell-derived exosomes
(11). A recent study reported that
a large amount of long RNA species, including circRNAs, are
detected in human blood-derived exosomes (12). These studies indicate that circRNAs
may serve as potential biomarkers in cancer diagnosis and help
monitor cancer progression. Although circRNAs have roles in
oncogenesis and tumor progression, their role in OC remains
unclear.
The present study aimed to identify potential
circRNA signatures that could predict the survival of patients with
OC. A re-annotation strategy was performed to evaluate the
expression level of circRNAs based on microarray datasets obtained
from the Gene Expression Omnibus (GEO) database. In the GSE9891
dataset, a signature comprising six circRNAs associated with
survival was identified. An expression-based risk score model was
designed to extrapolate the prognostic efficacy of this signature.
The risk score model was further validated in GSE63885 and GSE26193
datasets. The results demonstrated that the signature was
significantly associated with patient overall survival (OS),
progression-free survival (PFS) and disease-free survival (DFS).
Further analysis revealed that this signature was more sensitive
and specific than the existing clinical and other molecular
signatures in predicting survival. In conclusion, the present study
demonstrated that the six-circRNA signature may serve as a
potential prognostic biomarker of OC.
Materials and methods
The expression dataset of OC
Three independent OC datasets (Affymetrix
HG-U133_Plus_2.0 array platform) GSE26193 (n=107), GSE9891 (n=278)
and GSE63885 (n=75), were downloaded from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/).
Re-annotation of circRNAs from
microarray dataset
A re-annotation strategy was performed to identify
the circRNAs from microarray dataset (13). The circRNA transcripts were
downloaded from circBase (hg19; http://www.circbase.org/) (14). The SeqMap tool was used to map probes
to circRNA transcripts (15). In the
mapping procedure, mismatches were not allowed in mapping probes to
circRNA transcripts. Probes that were uniquely mapped to circRNAs
were retained. To further purify probes that were specific to
circRNAs, probes that also mapped to other transcripts were
excluded based on GENCODE (v19) (https://www.gencodegenes.org/) annotation and RefSeq
(GRCH37) database (https://www.ncbi.nlm.nih.gov/refseq/). With regards to
probes that mapped the same circRNA, the arithmetic mean expression
value was used. A total of 630 circRNAs were eventually identified
from the microarray data.
Clinical information of patients with
OC
Clinical characteristics of patients with OC were
derived from series matrix file or supporting information of the
corresponding dataset available in the GEO database. Patients with
OC from GSE9891 were randomly assigned into two groups as training
(n=139) and validating (n=139) datasets. There were no significant
differences in the clinical characteristics between the two groups
(P>0.05). Detailed clinical characteristics obtained from these
datasets are presented in Table
I.
 | Table I.Clinical characteristics of different
OC datasets. |
Table I.
Clinical characteristics of different
OC datasets.
|
| Number of patients
with OC |
|
|---|
|
|
|
|
|---|
| Characteristic | Training set
(n=139) | Validating set
(n=139) | GSE26193 (n=107) | GSE63885 (n=75) | P-values |
|---|
| Stage |
|
|
|
| 0.94a |
| I | 11 | 13 | 21 | 0 |
|
| II | 9 | 8 | 10 | 2 |
|
| III | 107 | 107 | 59 | 63 |
|
| IV | 12 | 10 | 17 | 10 |
|
| Age, years |
|
|
|
| 0.34b |
| Mean ±
SD | 60.24±10.47 | 59.04±10.68 | NA | NA |
|
|
Range | 23–80 | 22–80 | NA | NA |
|
| Histological
grade |
|
|
|
| 0.73a |
| G1 | 10 | 9 | 1 | 0 |
|
| G2 | 51 | 44 | 33 | 9 |
|
| G3 | 77 | 84 | 67 | 48 |
|
| G4 | 0 | 0 | 0 | 18 |
|
| Malignancy |
|
|
|
| 1a |
|
Malignant | 9 | 9 | NA | NA |
|
| Low
malignant potential | 130 | 130 | NA | NA |
|
| Recurrence |
|
|
|
| 0.90a |
|
Yes | 94 | 96 | 80 | 70 |
|
| No | 45 | 43 | 27 | 5 |
|
| Survival,
months |
|
|
|
| 0.47b |
| Mean ±
SD | 30.83±26.61 | 32.84±18.69 | 49.92±39.60 | 42.80±28.89 |
|
|
Range | 0–214 | 0–113 | 0–243 | 3–136 |
|
|
State |
|
|
|
| 0.63a |
|
Survival | 85 | 80 | 31 | 9 |
|
|
Mortality | 54 | 59 | 76 | 66 |
|
Statistical analysis
Student's t-test was used to determine the
difference of age and survival time between training and validating
groups. χ2 and Fisher's exact tests were used to
determine the difference of stage, grade, malignancy, recurrence
and survival status between training and validating groups.
Univariate and multivariate Cox regression analyses were performed
to evaluate the association between survival and circRNAs
expression in each OC cohort. The random survival forests variable
hunting (RSFVH) algorithm was carried out to select important
predictors (16). This strategy has
been used in a previous study to identify prognostic lncRNAs in OC
(17). The risk score for each
patient with OC was calculated according to the linear combination
of the expression values weighted by the coefficient from
univariate Cox regression analysis as follows:
RiskScore=∑i=1nβiExp(ci),(1)
where βi is the Cox regression
coefficient of a circRNA and n is the number of circRNAs regulated
by the same TF. Exp(ci) is the expression value
of circRNA i in the corresponding patient. The median risk
score was used as the cutoff point to divide the patients into high
and low risk groups. Kaplan-Meier (KM) survival curves were plotted
for patients in different risk groups, and statistical significance
was assessed by the log-rank test (P<0.05). A time-dependent
receiver operating characteristic (ROC) curve analysis was
performed using an R package named survivalROC (https://cran.r-project.org/web/packages/survivalROC/).
The function roc.KM.calc returns the true positive (TP) and false
positive (FP) values at the time point of interest. The sensitivity
is calculated as TP/(TP + false negative), and the specificity is
calculated as true negative (TN)/(TN+FP). All analyses were
performed based on R framework (v3.4; http://www.r-project.org/).
Functional analysis
Pearson correlation coefficients were used to
evaluate the co-expression association between circRNAs and mRNAs
[correlation coefficients >0 and false discovery rate (FDR)
<0.05]. For each circRNA, the co-expressed mRNAs were used to
perform gene set functional enrichment analysis. The R package
named clusterProfiler was used to predict biological functions of
circRNAs based on the gene sets of co-expressed mRNAs (18). Gene Ontology (GO) terms of
‘Biological Process’ and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathways with FDR <0.05 were considered to be
significantly enriched.
Results
Identification of potential prognostic
circRNAs
To identify potential prognostic circRNAs, 278
patients with OC from GSE9891 were randomly assigned into two
groups as training (n=139) and validating (n=139) datasets
(Table I). There were no significant
differences between the two groups in the clinical characteristics
(P>0.05). A re-annotation strategy was performed to evaluate the
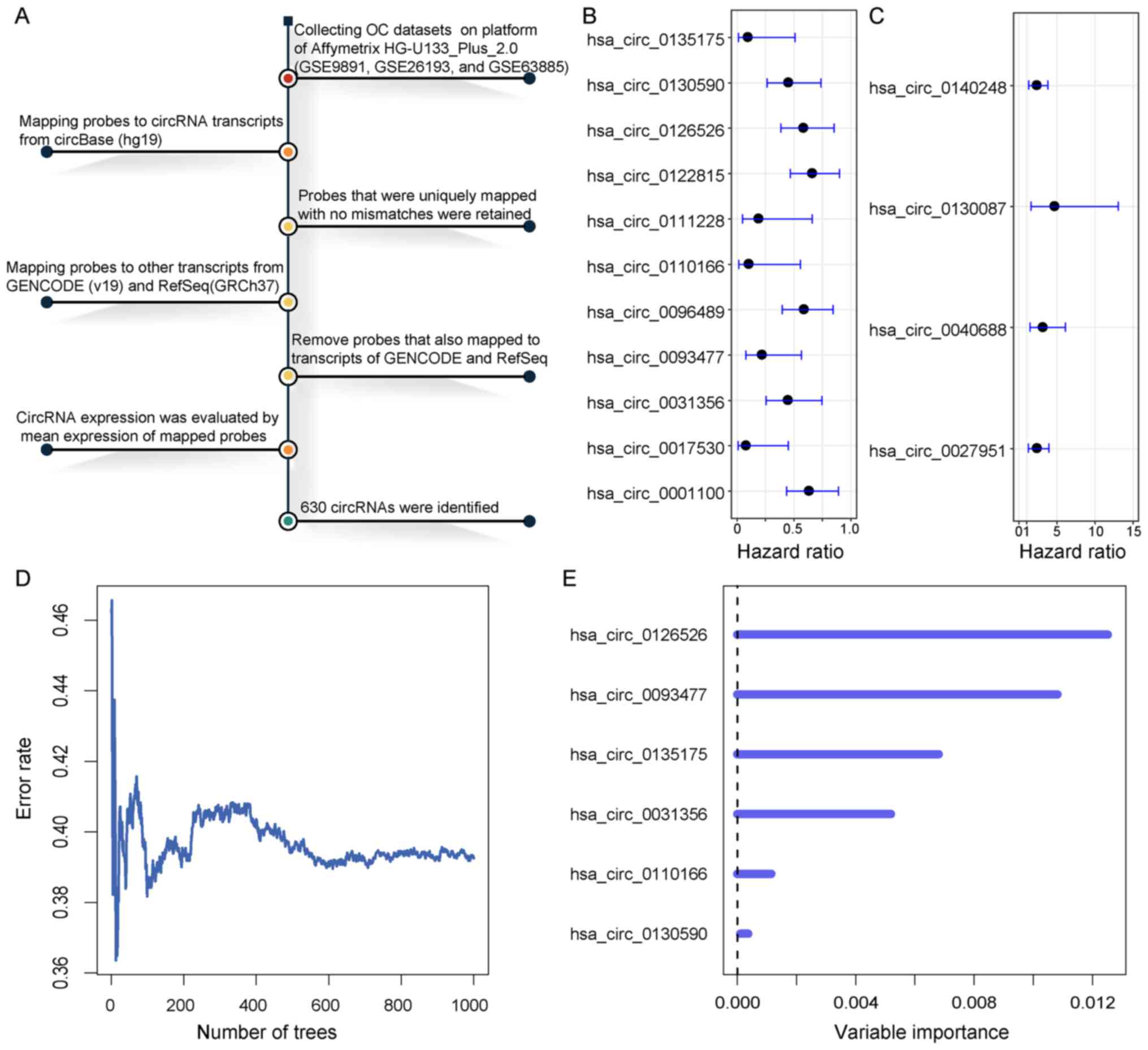
expression of circRNAs from microarray dataset (Fig. 1A). A total of 630 circRNAs were
identified from the microarray data. The univariate Cox regression
analysis was performed for circRNAs expression data in the training
dataset. A set of 15 circRNAs were significantly associated with
patient OS at a threshold of P<0.01, of which 11 were considered
as protective factors with hazard ratio (HR) values of 0–1
(Fig. 1B); however, four circRNAs
were considered as risk factors for OC with HR values >1
(Fig. 1C). To provide a smaller set
of circRNAs with more predictive efficacy, the RSFVH method
(16) was used to select the
circRNAs highly associated with OS. Subsequently, a panel of six
circRNAs (circ_0031356, 0093477, 0110166, 0126526, 0130590 and
0135175) was identified (Fig. 1D and
E). These six circRNAs had negative coefficients, which
suggested that lower expressions were associated with poor OS
(Fig. 1B). These circRNAs
represented protective factors with HR values of 0–1.
A six-circRNA signature predicts OS in
the training and validating datasets
A KM survival analysis was performed for the six
circRNAs based on their expression values in the training dataset.
The six circRNAs presented significant differences in patient OS
between high and low risk groups (log-rank test P<0.05; data not
shown). To improve the predictive efficacy of the six-circRNA
signature, a risk score formula was developed by integrating the
expression values and corresponding coefficients derived from Cox
regression analysis. A total of 139 training patients were assigned
into a high (n=70) and a low risk group (n=69) by using the median
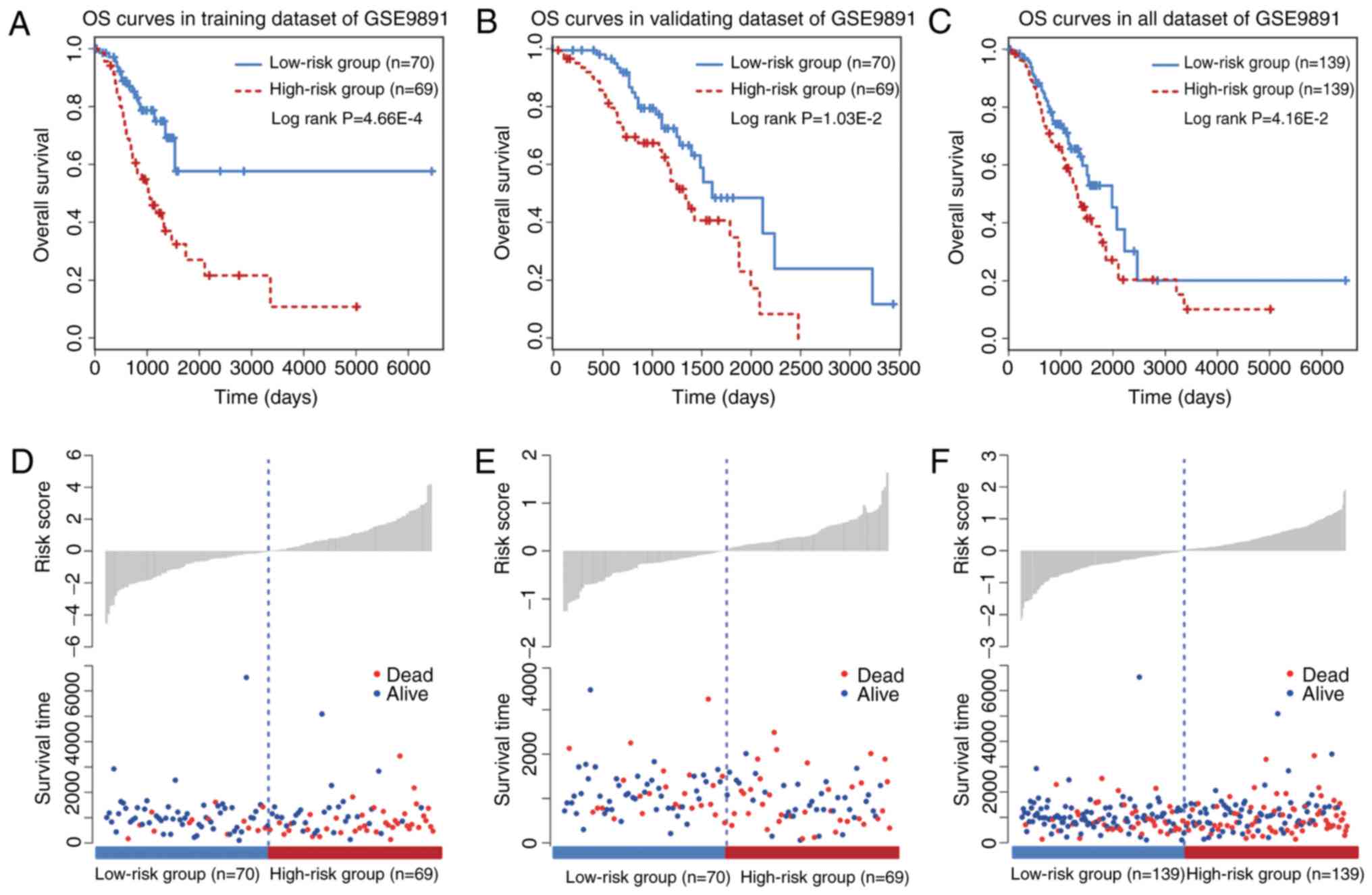
risk score as a cutoff point (P=4.66×10−4; Fig. 2A). Furthermore, the prognostic
efficiency of the six-circRNA signature was investigated in the
validating dataset using the same risk score threshold from the
training set. Based on this strategy, the validating patients were
also significantly divided into high (n=70) and low risk groups
(n=69; P=1.03×10−2; Fig.
2B). KM survival curves for the six-circRNA signature in the
278 patients with OC of GSE9891 are presented in Fig. 2C. By using the median risk score, the
six-circRNA signature significantly divided all the OC patients of
GSE9891 into high (n=139) and low risk groups (n=139)
(P=4.16×10−2). Distribution of patient risk scores and
survival status are presented in Fig.
2D-F. Patients in the low and high risk groups tended to have
different survival rates.
Validation of the six-circRNA
signature in other independent cohorts
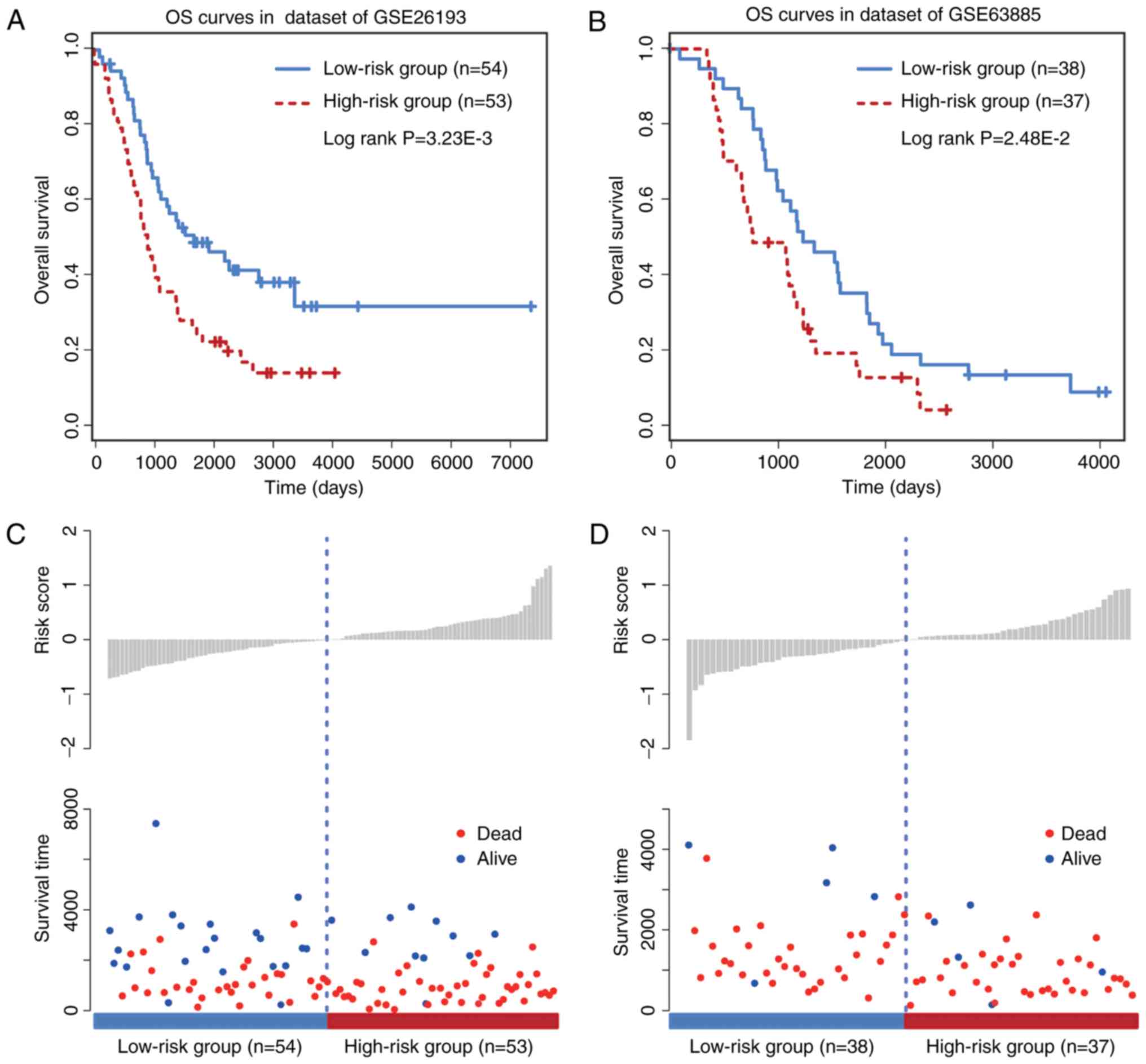
To further evaluate the prognostic efficacy of the
six-circRNA signature, a survival analysis was performed in the two
other independent OC datasets GSE26193 (n=107) and GSE63885 (n=75)
by using the same model construction as for the GSE9891 dataset.
Patients in these independent datasets were given risk scores and
classified into high or low risk groups. The six-circRNA signature
was significantly associated with prognosis in GSE26193 (HR=2.48,
95% CI=1.43–4.28, P=1.17×10−3) and GSE63885 (HR=2.63,
95% CI=1.40–4.96, P=2.28×10−3) datasets. In addition,
significant association between the risk score and OS was observed
in the two independent datasets (Fig. 3A
and B). The distribution of patient risk scores and survival
status is presented in Fig. 3C and
D. Patients with higher risk scores were prone to have shorter
survival time, whereas patients with lower risk scores tended to
have longer survival time. These observations were consistent with
findings obtained from the training and validating datasets.
Prognostic performance of six-circRNA
signature on PFS/DFS
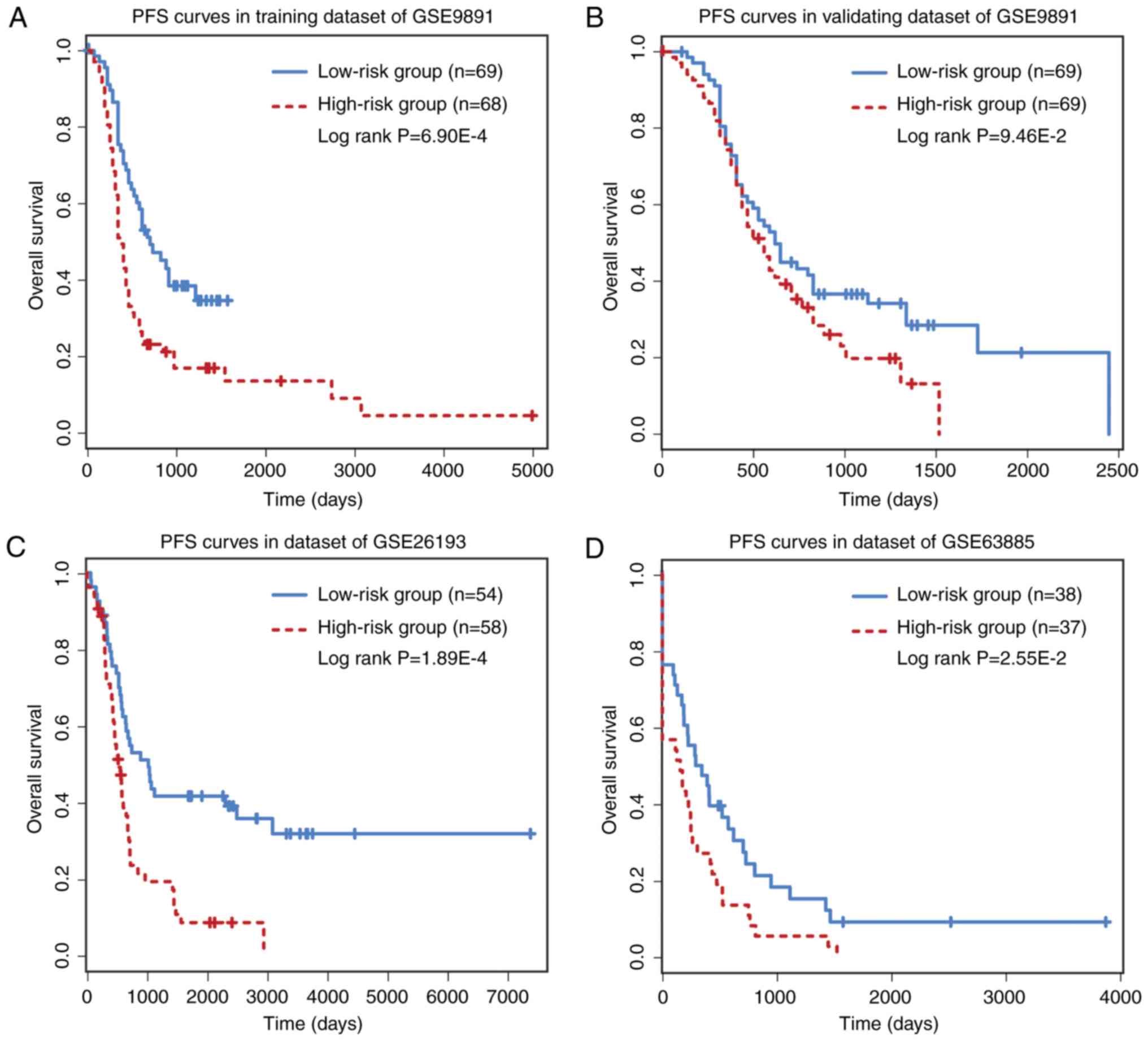
The prognostic performance of the six-circRNA
signature on PFS and DFS was analyzed. PFS analysis was made on
GSE9891 (n=275) and GSE26193 (n=107) datasets, while DFS analysis
was performed on GSE63885 (n=75) dataset. The six-circRNA signature
was significantly associated with patient PFS in the GSE9891
(HR=1.76, 95% CI=1.16–2.68, P=7.58×10−3) and GSE26193
(HR=2.17, 95% CI=1.30–3.61, P=2.83×10−3) datasets. In
GSE63885, the six-circRNA signature was significantly associated
with patient DFS (HR=2.76, 95% CI=1.38–5.54,
P=4.13×10−3). In addition, a significant association
between risk scores and PFS/DFS was observed in the three datasets
(Fig. 4). In PFS analysis, the
six-circRNA signature successfully divide patients from the GSE9891
and GSE26193 datasets into high and low risk groups, respectively
(Fig. 4A-C). In DFS analysis, the
six-circRNA signature could also significantly separate GSE63885
patients into high and low risk groups (Fig. 4D).
Independence of six-circRNA signature
from clinical characteristics
To evaluate whether the six-circRNA signature was
associated with other clinical variables, multivariate Cox
regression analyses were performed in each OC cohort. The
six-circRNA signature and other clinical and pathological
variables, including age at diagnosis, International Federation of
Gynecologists and Obstetricians (FIGO) stage and tumor grade, were
analyzed as covariables (Table II).
Following multivariate Cox regression analysis, the six-circRNA
signature was significantly associated with patient survival in
GSE9891 (P=1.08×10−4), GSE26193 (P=5.28×10−3)
and GSE63885 (P=4.04×10−3). Furthermore, a
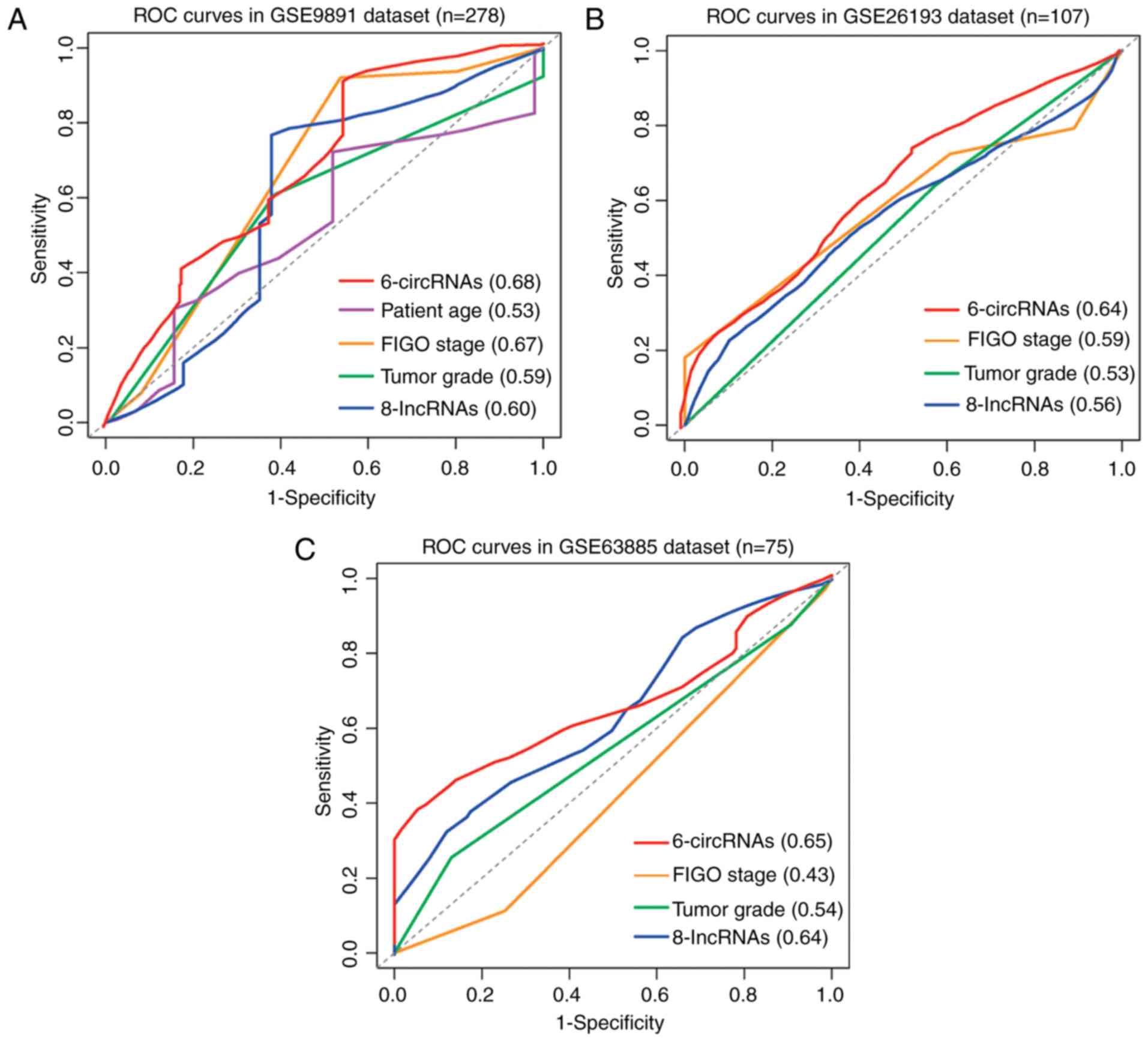
time-dependent ROC analysis was performed to assess the sensitivity
and specificity of OS prediction between the six-circRNA signature
and other clinical and molecular variables, including a panel of
eight lncRNAs obtained from a previous study (19). The median survival time was used as a
cutoff point to identify positive/negative cases. Following
comparison of the area under the curve (AUC) values of the ROC
curves, the predictive value of the six-circRNA signature was
higher than other clinical and molecular variables in different OC
datasets (Fig. 5A-C). In GSE9891,
the six-circRNA signature reached the highest AUC value of 0.68.
The AUC value of FIGO stage variable reached 0.67 in GSE9891;
however, it was only 0.59 and 0.43 in GSE26193 and GSE63885
datasets, respectively. These results demonstrated that the
six-circRNA signature was more sensitive and specific than the
existing clinical and molecular signatures in predicting the
survival of patients with OC.
 | Table II.Univariate and multivariate Cox
regression analyses of the six-circRNA signature and other clinical
variables. |
Table II.
Univariate and multivariate Cox
regression analyses of the six-circRNA signature and other clinical
variables.
| A, GSE9891
(Training set) |
|---|
|
|---|
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Grade
(G1/G2/G3) | 1.154
(0.741–1.797) |
5.26×10−01 | 0.679
(0.408–1.130) |
1.36×10−01 |
| Stage
(I/II/III/IV) | 3.047
(1.787–5.193) |
4.24×10−05 | 3.529
(1.939–6.422) |
3.67×10−05 |
| Age | 1.020
(0.990–1.051) |
1.83×10−01 | 1.023
(0.993–1.053) |
1.41×10−01 |
| Six-circRNA
signature | 1.509
(1.260–1.806) |
7.46×10−06 | 1.488
(1.242–1.784) |
1.69×10−05 |
|
| B, GSE9891
(Validating set) |
|
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|
|
|
|
|
Variables | HR (95%
CI) | P-value | HR (95%
CI) | P-value |
| Grade
(G1/G2/G3) | 1.618
(1.015–2.578) |
4.31×10−02 | 1.264
(0.772–2.068) |
3.52×10−01 |
| Stage
(I/II/III/IV) | 1.568
(0.939–2.619) |
8.55×10−02 | 1.716
(0.939–3.136) |
7.90×10−02 |
| Age | 1.031
(1.004–1.059) |
2.18×10−02 | 1.032
(1.005–1.059) |
2.03×10−02 |
| Six-circRNA
signature | 2.202
(1.357–3.575) |
1.40×10−03 | 2.110
(1.267–3.513) |
4.10×10−03 |
|
| C, GSE26193
(n=107) |
|
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|
|
|
|
|
Variables | HR (95%
CI) | P-value | HR (95%
CI) | P-value |
|
| Grade
(G1/G2/G3) | 1.207
(0.829–1.759) |
3.26×10−01 | 0.819
(0.537–1.247) |
3.51×10−01 |
| Stage
(I/II/III/IV) | 2.057
(1.546–2.738) |
7.54×10−07 | 2.212
(1.588–3.083) |
2.72×10−06 |
| Six-circRNA
signature | 2.475
(1.432–4.279) |
1.17×10−03 | 2.134
(1.253–3.635) |
5.28×10−03 |
|
| D, GSE63885
(n=75) |
|
|
| Univariate Cox
analysis | Multivariate Cox
analysis |
|
|
|
|
|
Variables | HR (95%
CI) | P-value | HR (95%
CI) | P-value |
|
| Grade
(G2/G3/G4) | 1.707
(1.129–2.582) |
1.13×10−02 | 1.639
(1.043–2.575) |
3.20×10−02 |
| Stage
(II/III/IV) | 2.315
(1.239–4.323) |
8.45×10−03 | 2.018
(1.087–3.748) |
2.62×10−02 |
| Six-circRNA
signature | 2.630
(1.396–4.957) |
2.78×10−03 | 2.643
(1.363–5.128) |
4.04×10−03 |
Function prediction of six-circRNA
signature
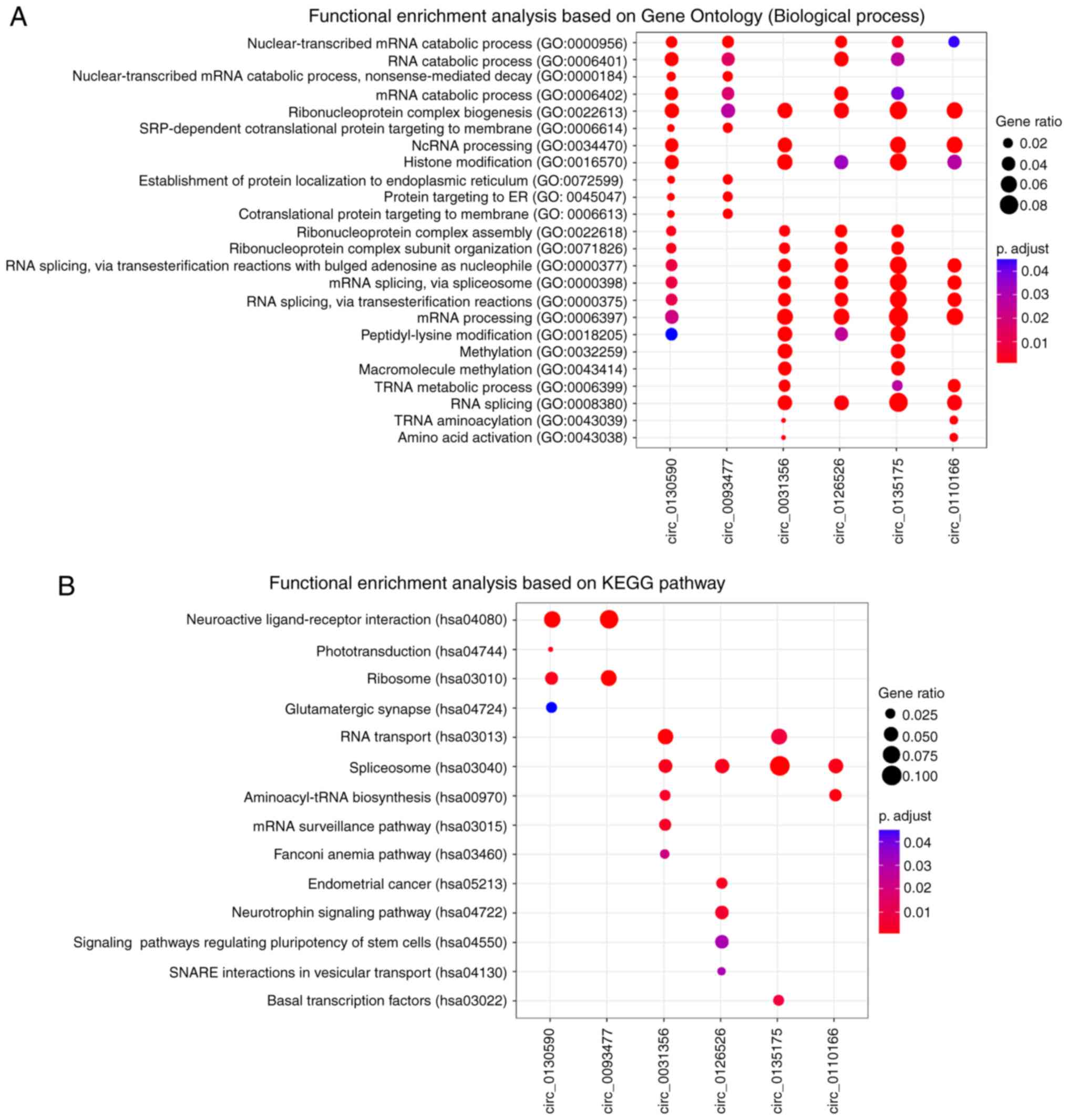
To explore the potential functional roles of these
six prognostic circRNAs, a ‘guilt-by-association’ analysis
(20) was performed to identify
co-expressed circRNA-mRNA pairs on GSE9891 dataset, which has the
largest samples size. For the six circRNAs, functional enrichment
analysis was performed based on their co-expressed mRNAs by using
clusterProfiler R package (18). The
results from GO enrichment analysis revealed that the six circRNAs
were significantly enriched in 24 GO terms based on ‘biological
processes’ ontology (Fig. 6A). These
related GO terms could be organized into different functional
clusters, including ‘RNA catabolic processes’, ‘ribonucleoprotein
complex biogenesis’, ‘histone modification’ and ‘methylation’. KEGG
pathway enrichment analysis revealed that circ_0130590 and 0093477
were significantly enriched in ‘neuroactive ligand-receptor
interaction’ and ‘ribosome’ pathways (Fig. 6B). Circ_0031356, 0126526, 0135175 and
0110166 were significantly enriched in ‘spliceosome pathway’.
‘Endometrial cancer’ and certain other signaling pathways were
enriched by circ_0126526. These results suggested that the six
circRNAs may participate in numerous biological processes involved
in OC tumorigenesis.
Discussion
In the present study, a re-annotation strategy was
performed to evaluate circRNAs expression from microarray dataset
of OC. Following univariate Cox regression analysis, 15 circRNAs
were significantly associated with patients OS. Amongst these
prognostic candidates, 11 circRNAs were determined as protective
factors with HR values of 0–1, while 4 circRNAs were determined as
risk factors with HR values >1. In addition, the RSFVH method
revealed that a panel of six circRNAs (circ_0031356, circ_0093477,
circ_0110166, circ_0126526, circ_0130590 and circ_0135175) was
associated with patient OS. Subsequently, an expression-based risk
score model was constructed to extrapolate the prognostic efficacy
of these circRNAs. The results revealed that the six-circRNA
signature was significantly associated with patient survival in the
training and validating datasets of GSE9891. The risk score model
was further validated in the independent cohorts of GSE26193 and
GSE63885. Furthermore, PFS analysis was performed on GSE9891 and
GSE26193 dataset, while DFS analysis was performed on GSE63885. The
six-circRNA signature was significantly associated with patient
PFS/DFS in GSE9891, GSE26193 and GSE63885 datasets. In addition,
consensus cluster analysis (21) was
performed on GSE9891 and GSE26193 datasets by considering the
expression correlation of the six-circRNA signature. The clustering
results revealed that patients in GSE9891 and GSE26193 could be
clustered into different risk groups, which was consistent with the
results found by using risk scores of the six-circRNA signature
(data not shown).
To further evaluate the independence of the
six-circRNA signature, multivariate Cox regression analyses were
performed based on the six-circRNA signature and other clinical and
pathological variables. Results demonstrated that the six-circRNA
signature was significantly associated with patient survival in
GSE9891, GSE26193 and GSE63885 datasets. In addition, certain
clinical variables, including patient age, stage and tumor grade,
were significantly associated with OS in different OC cohorts
(Table II). Additional
stratification analyses according to these factors therefore
require further investigation. With regards to OC stage, patients
from different OC datasets were divided into early (I/II) and late
(III/IV) stage subgroups. Results revealed that the six-circRNA
signature significantly subdivided patients in late stage subgroups
but not in early stage groups (data not shown). With regards to OC
age at diagnosis, patients from validating GSE9891 dataset were
stratified into younger and older groups according to the age
median value. Results revealed that the six-circRNA signature
significantly subdivided patients at different age levels (data not
shown). With regards to OC tumor grade, patients in the GSE63885
dataset were stratified into low (G1/G2) and high (G3/G4) grade
subgroups; however, P-values were not significant with regards to
OC survival analysis in the two subgroups (data not shown). This
last finding may be due to unknown confounding factors or the small
size of each subgroup.
In the present study, only 630 circRNAs were
identified from the microarray dataset; the re-annotation strategy
of circRNA expression from the microarray dataset may not have been
able to cover all circRNA transcripts. By considering this
limitation, it is possible that certain potential signatures may
have been ignored. For example, blood or exosome-derived biomarkers
are crucial for the prediction of cancer survival (22,23).
However, validation of the prognostic biomarkers from peripheral
blood or exosome populations requires expression dataset tested
previously in the human circulation system and well-annotated
follow up information. The follow up procedure may take 3–5 years
(24). The acquisition of fast
growing RNA-sequencing datasets may allow the discovery of novel
noncoding signatures associated with survival. Future work will
consider blood or exosome datasets to validate circRNA
biomarkers.
In conclusion, a re-annotation strategy was
performed to identify circRNA expressions from microarray dataset.
Following the design of a risk score model, a panel of six circRNAs
was significantly associated with OS, PFS and DFS in patients with
OC. In addition, the present study revealed that the six-circRNA
signature was independent of other clinical characteristics and
more powerful than other molecular signatures. The present study
highlighted some novel, potentially powerful prognostic markers for
OC.
Acknowledgements
Not applicable.
Funding
This study was supported by The National Natural
Science Foundation of China (grant no. 81772780).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GZ conceived, designed and supervised the study. YH,
QG, LS, CK and YC carried out data processing and experimental
analysis. GZ, YH and QG drafted the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors confirm that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rustin G, van der Burg M, Griffin C, Qian
W and Swart AM: Early versus delayed treatment of relapsed ovarian
cancer. Lancet. 377:380–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bowtell DD: The genesis and evolution of
high-grade serous ovarian cancer. Nat Rev Cancer. 10:803–808. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li C, Bonazzoli E, Bellone S, Choi J, Dong
W, Menderes G, Altwerger G, Han C, Manzano A, Bianchi A, et al:
Mutational landscape of primary, metastatic, and recurrent ovarian
cancer reveals c-MYC gains as potential target for BET inhibitors.
Proc Natl Acad Sci USA. 116:619–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kumar L, Shamsuzzam A, Haque R, Baghel T
and Nazir A: Circular RNAs: The emerging class of non-coding RNAs
and their potential role in human neurodegenerative diseases. Mol
Neurobiol. 54:7224–7234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shang Q, Yang Z, Jia R and Ge S: The novel
roles of circRNAs in human cancer. Mol Cancer. 18:62019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chekulaeva M and Rajewsky N: Roles of long
noncoding RNAs and circular RNAs in translation. Cold Spring Harb
Perspect Biol. a0326802018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du Z, Fei T, Verhaak RG, Su Z, Zhang Y,
Brown M, Chen Y and Liu XS: Integrative genomic analyses reveal
clinically relevant long noncoding RNAs in human cancer. Nat Struct
Mol Biol. 20:908–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Glazar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang H and Wong WH: SeqMap: Mapping
massive amount of oligonucleotides to the genome. Bioinformatics.
24:2395–2396. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishwaran H and Kogalur UB: Consistency of
random survival forests. Stat Probab Lett. 80:1056–1064. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhan X, Dong C, Liu G, Li Y and Liu L:
Panel of seven long noncoding RNA as a candidate prognostic
biomarker for ovarian cancer. Onco Targets Ther. 10:2805–2813.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao
H, Zhong Z and Sun J: Comprehensive analysis of lncRNA expression
profiles reveals a novel lncRNA signature to discriminate
nonequivalent outcomes in patients with ovarian cancer. Oncotarget.
7:32433–32448. 2016.PubMed/NCBI
|
|
20
|
Li ZC, Huang MH, Zhong WQ, Liu ZQ, Xie Y,
Dai Z and Zou XY: Identification of drug-target interaction from
interactome network with ‘guilt-by-association’ principle and
topology features. Bioinformatics. 32:1057–1064. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wilkerson MD and Hayes DN:
ConsensusClusterPlus: A class discovery tool with confidence
assessments and item tracking. Bioinformatics. 26:1572–1573. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Panagopoulou M, Karaglani M,
Balgkouranidou I, Biziota E, Koukaki T, Karamitrousis E, Nena E,
Tsamardinos I, Kolios G, Lianidou E, et al: Circulating cell-free
DNA in breast cancer: Size profiling, levels, and methylation
patterns lead to prognostic and predictive classifiers. Oncogene.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Varkaris A, Katsiampoura A, Davis JS, Shah
N, Lam M, Frias RL, Ivan C, Shimizu M, Morris J, Menter D, et al:
Circulating inflammation signature predicts overall survival and
relapse-free survival in metastatic colorectal cancer. Br J Cancer;
2019, View Article : Google Scholar
|
|
24
|
Huelsmann E, Zighelboim I, Ahmed A and
Dewdney S: The role of neoadjuvant chemotherapy in the management
of patients with advanced stage ovarian cancer: Survey results from
members of the society of gynecologic oncologists, a 5-year
follow-up. Gynecol Oncol Rep. 20:47–50. 2017. View Article : Google Scholar : PubMed/NCBI
|