Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common form of cancer (1) and is the
third most common cause of cancer-associated mortality worldwide
(2). Hepatitis B virus (HBV)
infection is one of the important etiological factors that can lead
to liver damage. It was estimated that ~5% of the world population
are infected with HBV, especially in China (3). A large proportion of patients with
chronic HBV infection may develop necroinflammatory liver disease
in the order of increasing severity from sustained liver injury to
cirrhosis, hepatic failure and HCC (4). Early diagnosis and treatment are vital
for improving the overall survival rate of patients with HCC. The
detection of serum a fetoprotein (AFP) may aid the diagnosis of
HCC, but its sensitivity is low, especially at the early stage of
HCC. Therefore, it is critical to find novel non-invasive serum
biomarkers for the independent or combined diagnosis of HCC.
MicroRNA (miRNA), a type of small non-coding single
stranded RNA of 18–25 nucleotides in length, can interact with the
target genes to suppress their expression by promoting the
degradation of target genes or inhibiting translation (5,6). miRNAs
have a wide range of functions and are involved in a variety of
biological processes, including cell proliferation, growth,
differentiation, apoptosis, metabolism and signaling pathway
transduction (7,8). They may also serve as important
clinical biomarkers for the diagnosis and prognosis of multiple
types of cancer (9–11). miRNA-122 is one of the key biomarkers
of the miRNAs and constitutes ~52% of the total hepatic miRNome
(12). It is the most abundant
liver-specific miRNA, which has notable anti-inflammatory effects
and is involved in a variety of liver functions (13,14). It
has been reported that miRNA-122 exhibited dysregulated expression
in HCC tissues, serum and cell lines (15,16);
however, contradictory conclusions regarding the differential
expression and prognosis of miRNA-122 have been reported (17). In addition, the numerous functions of
miRNA-122 in HCC and other types of cancer remain to be further
investigated.
In the present study, the clinical value and
associated function of miRNA-122 in various types of cancer,
particularly in HCC were systematically analyzed. Firstly, the
expression levels of miRNA-122 in patients with HCC, benign and
healthy groups were analyzed and the significance of miRNA-122 and
AFP in the differential diagnosis of HCC was evaluated. Secondly,
the analysis on the differential expression and survival rate of
miRNA-122 in a variety of common cancer types using miRNA-seq data
from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) was performed. Thirdly,
the target genes of miRNA-122 were comprehensively predicted for
further functional enrichment analysis. Finally, the associated
research progress of miRNA-122 was systematically summarized. The
content of the present study was progressive and may improve
current understanding of miRNA-122 from a macroscopic and
comprehensive perspective.
Materials and methods
Serum sample collection
The present study was approved by the Institutional
Review Board of the First Affiliated Hospital of Guangxi Medical
University (Guangxi, China) and written informed consent was
obtained from all individuals in this research project. A total of
150 individuals were enrolled from September 2016 to September
2017, including patients with HCC (50 cases), patients with benign
lesions (50 cases) and healthy controls (50 cases). Patent
characteristics are presented in table
I. HCC patients were diagnosed by serum AFP concentration
analysis and abdominal ultrasonography; diagnoses were confirmed by
histopathological examination. In addition, all the serum samples
of HCC patients were collected prior to surgical resection. All the
serum samples were collected, centrifuged at 2,000 × g for 5 min at
room temperature and stored as 500-µl aliquots at −80°C prior to
experimental use.
 | Table I.Clinical characteristics of the
included individuals. |
Table I.
Clinical characteristics of the
included individuals.
|
Characteristics | HCC | Hepatitis B | Cirrhosis | Healthy | P-value |
|---|
| Mean age ± SD,
years | 48.6±11.9 | 42.9±14.4 | 48.0±11.0 | 47.2±11.7 | >0.05 |
| Sex |
|
|
|
|
|
| Male, n
(%) | 35 (70) | 17 (68) | 18 (72) | 35 (70) | >0.05 |
| Female,
n (%) | 15 (30) | 8 (32) | 7 (28) | 15 (30) |
|
| AFP, ng/ml |
10,676.3±20,568.7 | 32.4±74.3 | 20.5±47.5 | 2.7±1.6 | <0.001 |
| Cirrhosis |
|
|
|
|
|
| Yes, n
(%) | 38 (76) | 0 (0) | 25 (100) | 0 (0) |
|
| No, n
(%) | 12 (24) | 25 (100) | 0 (0) | 50 (100) |
|
| Hepatitis B |
|
|
|
|
|
| Yes, n
(%) | 42 (84) | 25 (100) | 7 (28) | 0 (0) |
|
| No, n
(%) | 8 (16) | 0 (0) | 18 (72) | 50 (100) |
|
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) experiment
Total RNA of serum samples was extracted using the
Serum/Plasma miRNA Extraction and Isolation kit DP503. This kit is
a newly-developed products specifically for serum and plasma miRNA
extraction. Total RNA was reverse transcribed using the miRcute
Enhanced miRNA cDNA First Strand Synthesis kit (KR211). All the
reagents were obtained from Tiangen Biotech Co., Ltd. (Beijing,
China) and all operations were conducted in strict accordance with
the manufacturer's protocol. The temperature protocol was as
follows: 60 min for miRNA plus A tail reaction and reverse
transcription reaction at 42°C; 3 min for enzyme inactivation
reaction at 95°C. The RT-qPCR experiment was performed using the
reagent of SYBR-Green FP411. The thermocycling conditions of the
RT-qPCR were as follows: 15 min for initial template denaturation
at 95°C, following 45 cycles of PCR template denaturation for 20
sec at 94°C; annealing and extension for 34 sec at 60°C. The
melting/dissociation curve stage was produced following RT-qPCR.
The 7500 Real-Time PCR Instrument from Thermo Fisher Scientific,
Inc. (Waltham, MA, USA) was used in the detection. U6 was set as an
internal reference. The primer of U6: Forward:
5′-AGCGGGAAATCGTGCGTGACA-3′, reverse: 5′-GTGGACTTGGGAGAGGACTGG-3′.
The primer of miRNA-122: Forward: 5′-GCGAAAGCATTTGCCAAGAA-3′,
reverse: 5′-CATCACAGACCTGTTATTGC-3′. Calculation of the relative
miRNA expression levels was based on 2−ΔΔCq (18).
Statistical analysis
The values in tables were reported as the mean ±
standard deviation or number percentage, unless otherwise stated.
The receiver operating characteristic curve, area under curve
(AUC), sensitivity, specificity, positive predictive value and
negative predictive value (NPV) were used to analyze diagnostic
accuracy. Youden index=sensitivity-(1-specificity). When the Youden
index is at its maximum, the corresponding detection value can be
used as the cutoff (19). Student's
t-test or one-way analysis of variance (ANOVA) was used to analyze
continuous variables where appropriate in this study. Student's
t-test was used for the analysis of differential expression between
two groups and the one-way ANOVA was used for the analysis of
differential expression among more than two groups. Bonferroni's
post hoc test was used following the ANOVA. The χ2 test
or Fisher's exact test was used for the comparisons of differences
in the categorical data between groups. A two-tailed P≤0.05 was
considered to indicate a statistically significant difference. The
aforementioned analyses were performed using IBM SPSS 20.0 (IBM,
Corps., Armonk, NY, USA). The scatter plot was produced using
GraphPad Prism 5 software (Graphpad Software, Inc., La Jolla, CA,
USA).
Differential expression and survival
analysis of miRNA-122 in TCGA and literature summary in PubMed
At present, a number of studies have demonstrated
that miRNAs or other genes frequently exhibit diversity as clinical
markers of cancer. The diagnosis and prognosis of a certain
biomarker in clinical pan-cancer is also a popular research model
(20,21). Therefore, data of TCGA was thoroughly
used to analyze the expression levels of miRNA-122 in a variety of
common tumors in this study. Data mining is one of the hot topics
in bioinformatics research. The statistical results are more
scientific and accurate than simple difference analysis. TCGA is
currently a popular database for cancer research. It is a project
initiated by the National Cancer Institute and the National Human
Genome Research Institute in 2005. It has provided a large amount
of genomic data and associated clinical data for researchers in
basic and translational medicine for cancer. In this study,
analysis of differential expression was performed on the basis that
cancer tissues and para-cancerous tissues must be present
simultaneously. In TCGA database, the number of tumor tissues are
greater than the normal tissues. Certain types of tumor do not
contain normal tissue samples. Therefore, the criteria that
adjacent normal tissue samples ≥3 was set. The ‘edgeR’ package of R
(version 3.3.3; http://www.r-project.org) was used for differential
expression analysis based on a False Discovery Rate <0.05 and
the absolute value of log2 (fold change) ≥1. In
addition, the overall survival analysis of cancers with
differentially expressed miRNA-122 was performed using the
‘survival’ package of R (http://bioconductor.org/packages/3.6/bioc/). The
present study also searched for relevant references by searching
the keyword ‘miRNA-122’ or ‘microRNA-122’ or ‘miR-122’ in PubMed
(https://www.ncbi.nlm.nih.gov/pubmed),
and summarized the expression and prognosis of miRNA-122 in various
types of cancer.
Target gene prediction and functional
enrichment analysis
The target gene prediction software available for
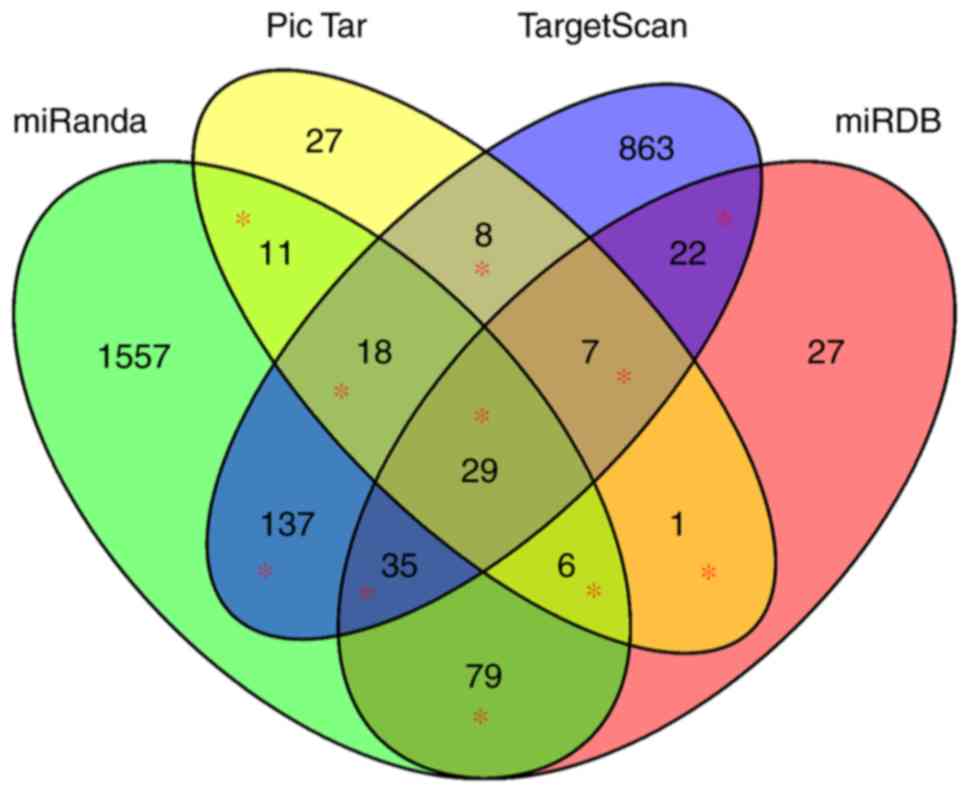
miRNA is multifarious. In the present study, four classical and
common sets of software were selected to predict the target gene of
miRNA-122, including miRanda (http://www.microrna.org/) (22), Pic Tar (http://pictar.mdc-berlin.de/) (23), miRDB (http://www.mirdb.org/) (24) and TargetScanHuman (http://www.targetscan.org/vert_71/) (25), respectively. Comparing the target
genes of the four types of software, the intersection of any two
predictor softwares were selected as candidate target genes for
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway functional enrichment analyses. The Venn diagram
demonstrating intersections were constructed using the
‘VennDiagram’ package of R. Furthermore, the GO and KEGG pathway
analyses are the most common functional enrichment analyses and
were performed using the online software, Database for Annotation,
Visualization and Integrated Discovery (DAVID), version 6.8
(https://david.ncifcrf.gov/) (26). The selection criteria for functional
items were in accordance with P≤0.05 and count number ≥2.
Results
Clinical characteristics
The clinical information and laboratory biomarker
analysis of the different groups are presented in Table I. For statistical analysis, all the
participants were divided into three groups: Including HCC (50
cases), benign (HBV and cirrhosis, 50 cases) and healthy (50
cases). There was no statistically significant difference between
the age and gender of the studied groups as they were matched with
each other. However, there was a significant difference between the
different groups regarding the clinical biomarker AFP (P<0.001
for all comparisons).
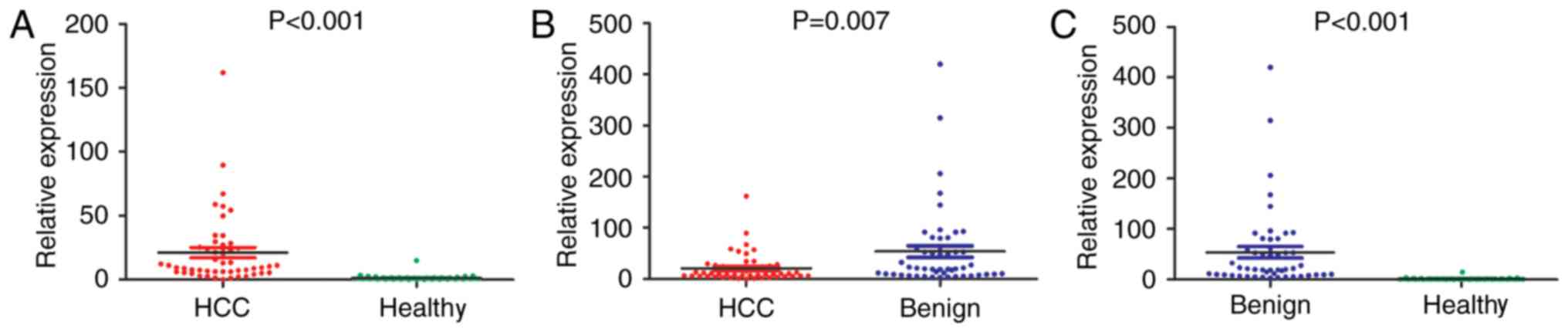
Different expression of miRNA-122
among three groups
The expression levels of miRNA-122 were detected
among the HCC, benign and healthy groups. The results indicated
that there was a significant difference in the expression levels of
miRNA-122 among the three groups (Fig.
1). Compared with the healthy group, the HCC and benign groups
exhibited significantly increased expression levels of miRNA-122
(P<0.001). In addition, the expression levels of miRNA-122
between the HCC and benign groups were compared, and it was
demonstrated that the benign group exhibited significantly higher
expression levels of miRNA-122 compared with the HCC group
(P=0.007). Overall, the expression levels of miRNA-122 in the
healthy, HCC and benign groups demonstrated a stepwise increase
(healthy group < HCC group < benign group); there was a
statistically significant difference between the groups
(P<0.001; Fig. 1C). Therefore,
miRNA-122 can be used not only for the identification of HCC and
normal controls, but also for the identification of HCC and benign
lesions. There is also a study demonstrating that the serum level
of miRNA-122 in patients with hepatitis is increased compared with
in HCC patients (27). The
experiments of the present study confirmed this.
Subgroup analysis of miRNA-122 in
individuals with HBV
In the study of the clinical markers of HCC, it is
particularly important to analyze individuals with hepatitis B.
Therefore, subgroup analysis (HBV-positive and -negative groups) of
the expression levels in individuals with hepatitis B was performed
in the present study. The results are presented in Table II. There was no statistically
significant difference in the expression levels of miRNA-122
between the positive and negative HBV groups. However, the
concentration of AFP was significantly associated with HBV
infection in the HCC (P=0.03) and HCC + Benign group (P=0.003). In
HBV-infected individuals, AFP concentrations were increased
compared with HBV negative patients.
 | Table II.Subgroup analysis of miRNA-122 in
HBV-associated individuals. |
Table II.
Subgroup analysis of miRNA-122 in
HBV-associated individuals.
|
| HCC | Benign | HCC + benign |
|---|
|
|
|
|
|
|---|
| Measurement | HBV+
(n=42) | HBV−
(n=8) | P-value | HBV+
(n=32) | HBV−
(n=18) | P-value | HBV+
(n=74) | HBV−
(n=26) | P-value |
|---|
| miRNA-122, mean ±
SD | 23.5±29.6 | 9.5±8.3 | 0.199 | 64.3±93.7 | 35.2±29.5 | 0.215 | 28.1±68.6 | 27.3±27.7 | 0.324 |
| AFP, ng/ml |
12,588.2±21,920.6 | 639.1±1,254.2 | 0.030 | 33.0±75.0 | 14.7±26.4 | 0.634 | 139.5±17,646.9 | 206.8±753.3 | 0.003 |
Diagnostic accuracy of miRNA-122, AFP
and miRNA-122 + AFP
In the previous differential expression analysis, it
was demonstrated that the miRNA-122 expression levels were
significantly different between the HCC vs. healthy groups and the
HCC vs. benign groups (Fig. 1).
Additionally, AFP has also been recognized as a serological
biomarker for the diagnosis of HCC. Therefore, the diagnostic value
of assessing miRNA-122, AFP and miRNA-122 + AFP in HCC was
investigated in the present study (Table III). The diagnostic sensitivity of
miRNA-122 (sensitivity, 90%) was increased compared with AFP
(sensitivity, 86%) and the combination of the two biomarkers could
further improve the diagnostic accuracy between the HCC and healthy
groups (AUC, 0.980; 95% CI, 0.958–1.000). However, the diagnostic
sensitivity and specificity of miRNA-122 were poor between HCC and
benign groups (sensitivity, 64%; specificity, 62%).
 | Table III.Diagnostic value of miRNA-122, AFP
and miRNA-122 + AFP. |
Table III.
Diagnostic value of miRNA-122, AFP
and miRNA-122 + AFP.
|
| HCC vs.
Healthy | HCC vs. Benign |
|---|
|
|
|
|
|---|
| Parameter | miR-122 | AFP | miR-122 + AFP | miR-122 | AFP | miR-122 + AFP |
|---|
| Sensitivity, % | 90.0 | 86.0 | 90.0 | 64.0 | 86.0 | 86.0 |
| Specificity, % | 94.0 | 100.0 | 100.0 | 62.0 | 80.0 | 80.0 |
| AUC (95% CI) | 0.954 | 0.961 | 0.980 | 0.667 | 0.878 | 0.876 |
|
| (0.907–1.000) | (0.927–0.996) | (0.958–1.000) | (0.562–0.772) | (0.811–0.944) | (0.808–0.944) |
| PPV, % | 94.0 | 1.0 | 1.0 | 63.0 | 81.0 | 81.0 |
| NPV, % | 90.0 | 88.0 | 91.0 | 63.0 | 85.0 | 85.0 |
| Cutoff | 2.80 | 8.78 |
| 14.29 | 8.59 |
|
Differential expression and survival
analysis of miRNA-122 by pan-cancer analysis of TCGA database
It is an indisputable fact that miRNA-122 is highly
expressed in HCC. However, the expression of miRNA-122 in other
types of tumor requires further investigation. In the present
study, the differential expression of miRNA-122 was analyzed in a
variety of common tumors using the publicly available data of TCGA
database. Following strict screening, 17 types of cancer were
selected to analyze the differential expression of miRNA-122
between cancerous and adjacent normal tissues; the results are
presented in Table IV. The
expression levels of miRNA-122 were significantly upregulated in
eight types of cancer, including colorectal carcinoma, breast,
kidney and lung cancers, prostate adenocarcinoma, stomach
adenocarcinoma, thyroid carcinoma and uterine cancer. In addition,
miRNA-122 was reported to be significantly downregulated in
cholangiocarcinoma and HCC (Table
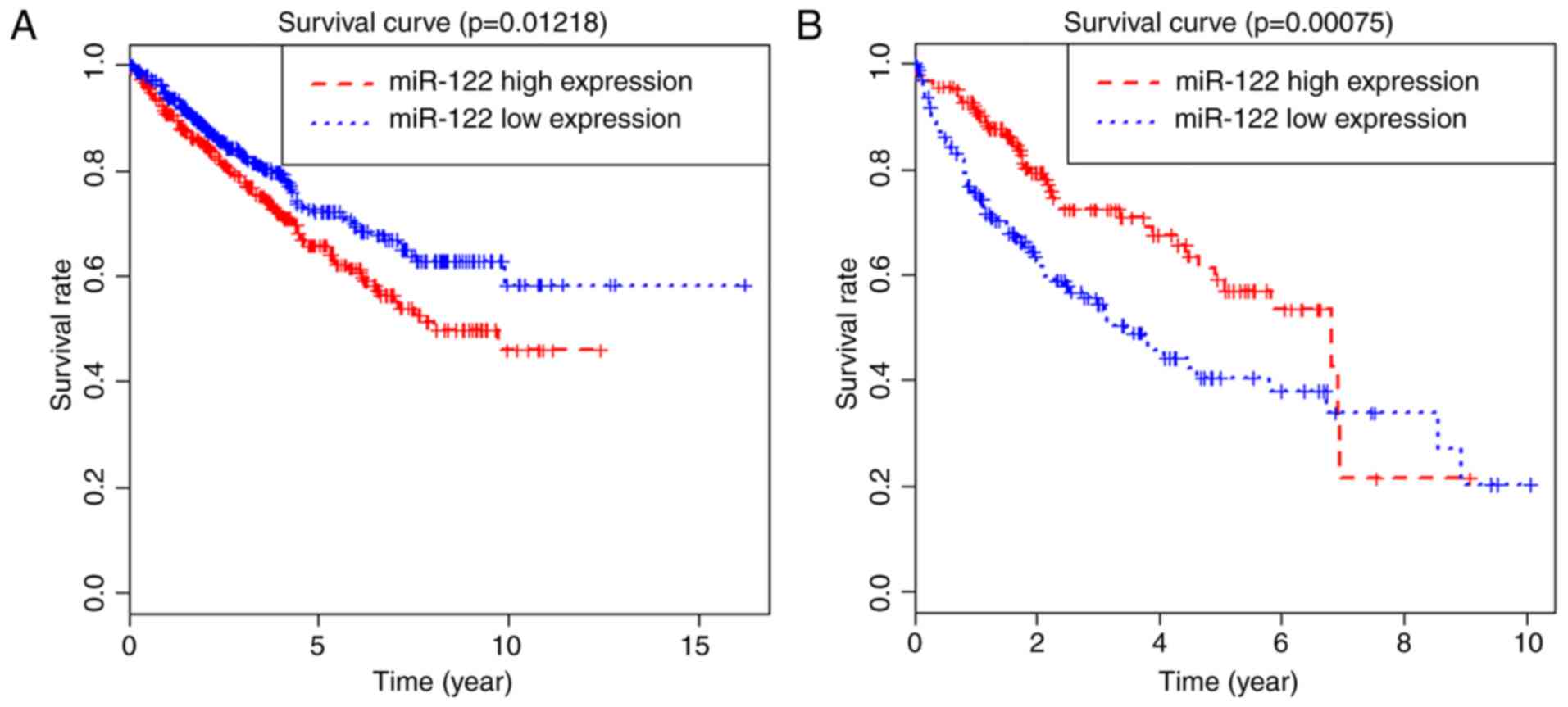
IV). Simultaneously, survival analysis was performed in the
types of cancer with differentially expressed miRNA-122. The
results of survival analysis demonstrated that high expression
levels of miRNA-122 were associated with poor prognosis in kidney
cancer, but good in patients with HCC (P=0.012 and P<0.001).
However, there was no significant effects on the prognoses of other
types of cancer (Fig. 2).
 | Table IV.Differential expression analysis of
microRNA-122 in 17 types of cancer from The Cancer Genome Atlas
database. |
Table IV.
Differential expression analysis of
microRNA-122 in 17 types of cancer from The Cancer Genome Atlas
database.
| Cancer type | Abbreviation | Cancer/normal,
n | False discovery
rate | LogFC |
|---|
| Colorectal
carcinoma | CRC | 619/11 |
4.62×10−2a | 8.18 |
| Adrenal gland
carcinoma | AGC | 264/3 | 1.00 | 7.02 |
|
Cholangiocarcinoma | CHOL | 36/9 |
5.01×10−8a | −4.16 |
| Bladder urothelial
carcinoma | BLCA | 418/19 |
3.59×10−1 | 2.04 |
| Brain glioma | BG | 530/5 | 1.00 | 0.45 |
| Breast cancer | BC | 1,103/104 |
1.80×10−13a | 7.34 |
| Cervical
cancer | CC | 309/3 | 1.00 | 6.52 |
| Esophageal
carcinoma | ESCA | 187/13 |
2.53×10−1 | 3.00 |
| Head and neck
squamous cell carcinoma | HNSC | 525/44 |
7.73×10−2 | −0.81 |
| Kidney cancer | KC | 1,074/147 |
9.40×10−36a | 5.43 |
| Hepatocellular
carcinoma | HCC | 375/50 |
1.14×10−3a | −0.61 |
| Lung cancer | LC | 999/91 |
9.40×10−11a | 5.75 |
| Pancreatic
adenocarcinoma | PAAD | 179/4 | 1.00 | 0.35 |
| Prostate
adenocarcinoma | PRAD | 499/52 |
1.19×10−6a | 3.84 |
| Stomach
adenocarcinoma | STAD | 446/45 |
9.86×10−4a | 3.44 |
| Thyroid
carcinoma | THCA | 514/59 |
7.77×10−3a | 1.64 |
| Uterine cancer | UC | 603/33 |
3.28×10−5a | 6.90 |
Literature summary of the expression
and prognosis of miRNA-122 via pan-cancer analysis
In addition to experimental validation and public
database mining, the collective findings of the literature were
also valuable. Information from available articles on the PubMed
database on differential expression levels and prognostic value of
miRNA-122 in a variety of cancer types was summarized. The results
are presented in Table V. Literature
data of the differential expression and survival analysis of
miRNA-122 between HCC and adjacent normal tissues were consistent
with the results of TCGA analysis. Additionally, the results in
colorectal cancer, renal carcinoma, cholangiocarcinoma, prostate
cancer and thyroid carcinoma were the same. However, there were
inconsistent conclusions of particular types of cancer, including
breast and gastric cancer. The inconsistencies need to be further
studied.
 | Table V.Differential expression and prognosis
evaluation of miRNA-122 in various types of cancer from the
reported literature. |
Table V.
Differential expression and prognosis
evaluation of miRNA-122 in various types of cancer from the
reported literature.
| Tumor type | Sample type | Differential
expression (cancer vs. normal) | Overall
survival | (Refs.) |
|---|
| Hepatocellular
carcinoma | Tissue | Downregulated | Poor | (52) |
| Hepatocellular
carcinoma | Plasma/serum | Upregulated | Poor | (57,58) |
| Hepatocellular
carcinoma | Plasma/serum |
| Better | (59,60) |
| Hepatocellular
carcinoma | Plasma/serum | Downregulated |
| (27) |
| Hepatocellular
carcinoma | Plasma/serum | No
significance |
| (49,63) |
| Glioma | Tissue | Downregulated |
| (32,64) |
| Glioma | Plasma | Downregulated | Poor | (54) |
| Lymphoma | Plasma | Downregulated |
| (65) |
| Renal cell
carcinoma | tissue | Upregulated |
| (33,66–71) |
| Gastric cancer | Tissue | Downregulated |
|
|
| Gastric cancer | Plasma | Downregulated | Poor | (55) |
| Pancreatic
cancer | Tissue | Downregulated |
| (35,72) |
| Pancreatic
cancer | Whole blood | Upregulated |
| (73) |
| Colorectal
carcinoma | Tissue | Upregulated |
| (53) |
| Colorectal
carcinoma | Plasma | Upregulated |
| (74) |
|
Cholangiocarcinoma | Tissue | Downregulated |
| (75) |
| Gallbladder
cancer | Tissue | Downregulated |
| (76) |
| Prostate
cancer | Tissue | Upregulated |
| (77) |
| Retinoblastoma | Tissue | Downregulated |
| (78) |
| Osteosarcoma | Tissue | Downregulated |
| (79) |
| Breast cancer | Tissue | Downregulated |
| (80–82) |
| Papillary thyroid
carcinoma | Tissue | Upregulated |
| (83) |
| Pituitary
carcinomas | Tissue | Upregulated |
| (84) |
| Acute myeloid
leukemia | Tissue | Downregulated | Poor | (56) |
| Bladder cancer | Tissue | Downregulated |
| (85) |
Target gene prediction of miRNA-122
and functional enrichment analysis
Due to the limitations of using a single prediction
software of miRNA target genes, four commonly used miRNA target
gene prediction software were used to simultaneously analyze. The
intersection of two or more of the employed software was selected.
Finally, 353 miRNA-122 target genes were included for functional
enrichment analysis (Fig. 3). MEF2C,
MASP1, SLC52A2, NONO, BRPF1, USP53, VPS4A, CCAR1, H1F0 and KIF5B
were the top 10 of the 353 target genes, and the others are not
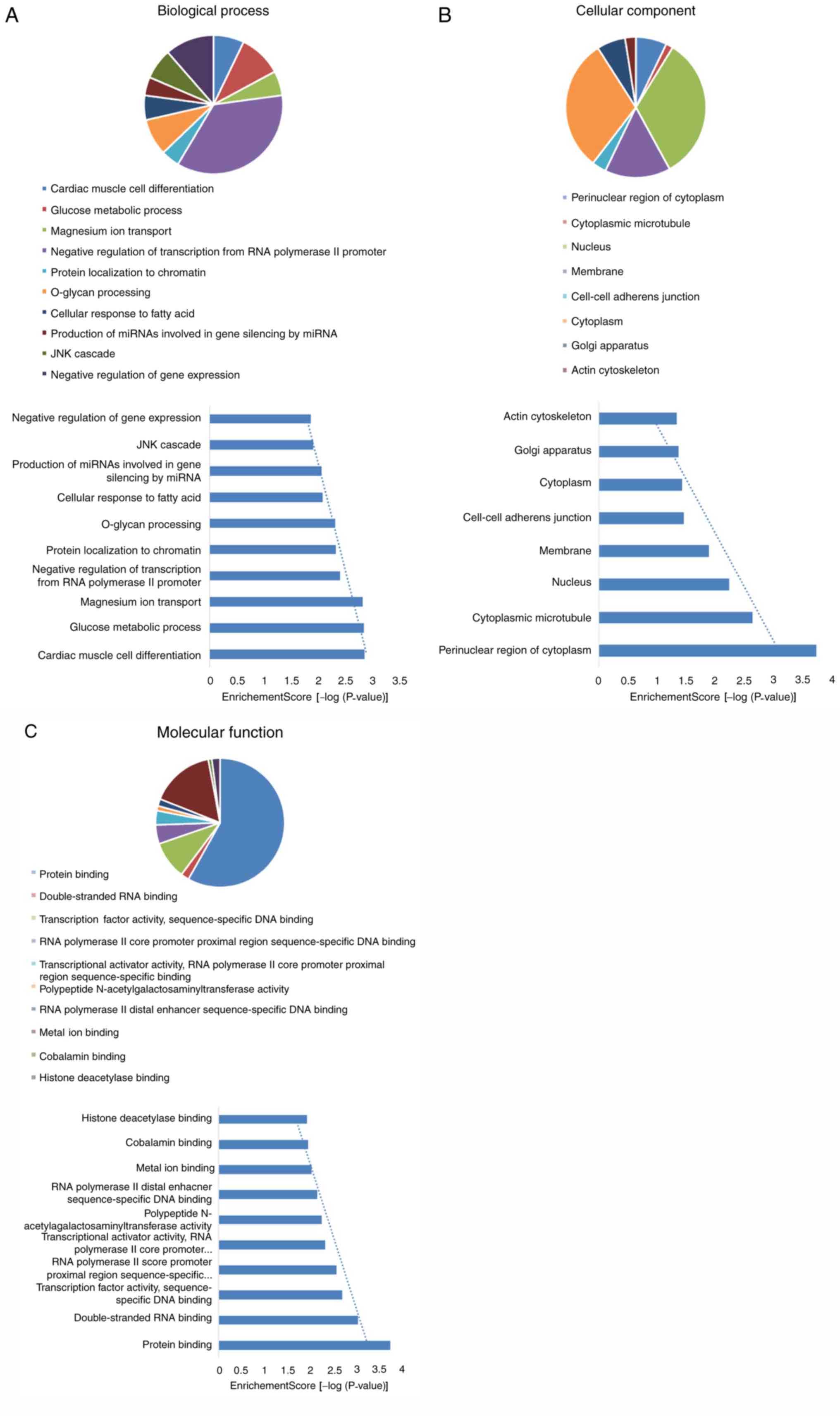
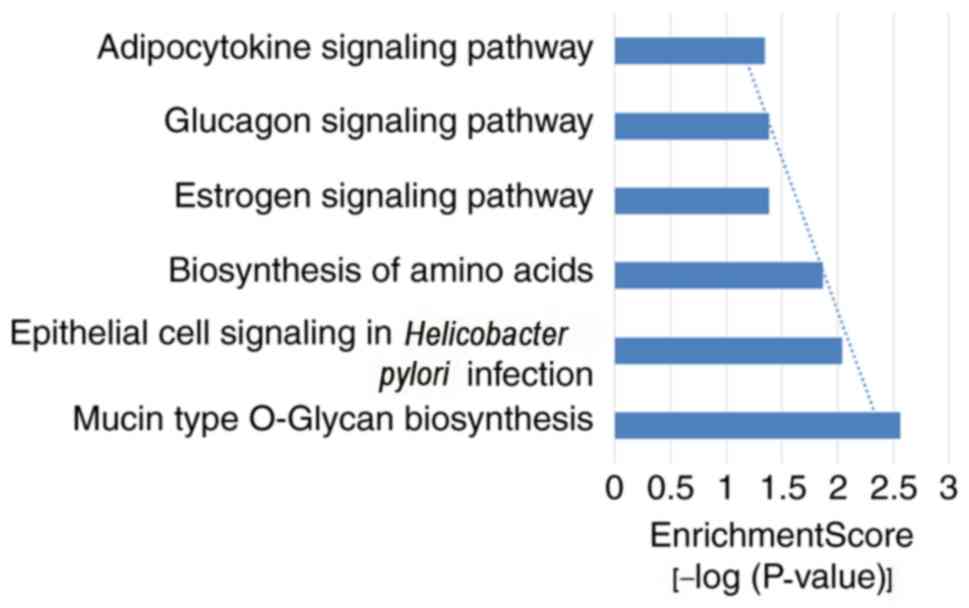
illustrated. In the present study, GO and KEGG pathway analysis
were carried on the target genes of miRNA-122. Following the
enrichment analysis by DAVID software, 68 notable GO items were
identified, of which 38 items for ‘biological process’ (BP), 8
items for ‘cellular component’ (CC) and 22 items for ‘molecular
function’ (MF). In addition, six important KEGG signaling pathways
were also identified. The top 10 items from different categories
are presented in Figs. 4 and
5. The results indicated that
cardiac muscle cell differentiation and glucose metabolic process
items in BP, the perinuclear region of cytoplasm and cytoplasmic
microtubule items in CC, protein binding and double-stranded RNA
binding items in MF, mucin type O-glycan biosynthesis and
epithelial cell signaling in Helicobacter pylori infection
items of the KEGG signaling pathways were the most closely
associated with the target genes of miRNA-122.
Discussion
Although research regarding the expression levels
and associated functions of miRNA-122 in HCC and other liver
diseases have been widely carried out (15,28), the
reports are diffuse and contradictory. For example, miRNA-122 acted
as a cancer suppressor gene and was downregulated in HCC tissues
and cells compared with the adjacent normal tissues and normal cell
lines (15,29), which was in line with the results of
TCGA analysis. Conversely, the serum expression levels of miRNA-122
increased significantly in HCC patients, hepatitis C patients and
other liver injury diseases compared with the healthy controls
(30,31). From this perspective, miRNA-122 may
be considered an onco-miRNA. In addition, the expression and
function of miRNA-122 in other tumors have also been widely
reported, including glioma (32),
renal cell carcinoma (33), gastric
(34) and pancreatic cancer
(35) and others. In addition to
cancer and liver injury diseases, miRNA-122 also served important
roles in cardiovascular diseases (36–38),
obesity (39–41), diabetes (42), immunological diseases (43–45) and
other non-cancer diseases (46).
The present study analyzed the expression levels of
miRNA-122 in tissues and serum samples of various types of cancer
systematically by RT-qPCR analysis, TCGA and literature data. The
lower expression levels of miRNA-122 in HCC tissues, compared with
the adjacent normal tissues, had been validated by a number of
researchers over the years (47,48);
however, one study has reported conflicting conclusions (49). The expression of miRNA-122 was
multifarious in different types and degrees of liver cancer and
liver injury diseases. From a large classification perspective, the
expression levels of miRNA-122 were significantly increased in the
serum of HCC patients compared with in healthy controls (50,51). The
expression levels of miRNA-122 in the serum of HBV-positive
patients with HCC were frequently decreased compared with patients
with benign liver lesions (27),
which was consistent with the results of serological analysis in
the present study. Meanwhile, miRNA-122 and AFP analyses were
combined for diagnosis in the present study, which may have
improved the non-invasive diagnostic accuracy of HCC.
TCGA database is one of the most widely used public
data platforms for a variety of types of cancer. Following
screening, 17 common cancer types were included in the differential
expression analysis of miRNA-122. Finally, it was demonstrated that
miRNA-122 was differentially expressed in 10 types of tumor from
TCGA. However, by summarizing the reported literature, differential
expression of miRNA-122 in a number of types of cancer was also
demonstrated. Additionally, there are several consistent types of
cancer between the two summary methods, including HCC (52), renal carcinoma (33) and colorectal cancer (53); however, certain results were
inconsistent. The number of samples, fixed tumor type in TCGA, the
difference in detection methods and platforms, etiology or
pathological classification, and clinical features, may lead to
inconsistent results. In future studies, an in-depth exploration of
inconsistencies can also be conducted. In addition, the prognostic
value of miRNA-122 in a variety of cancer was also analyzed and
summarized. Through the analysis of TCGA data, it was demonstrated
that the expression levels of miRNA-122 could affect the overall
survival rate of HCC and renal carcinoma. In addition to the
association between miRNA-122 and the prognosis of HCC, it was also
demonstrated that the low expression levels of miRNA-122 predicted
the poor prognosis of glioma (54),
gastric cancer (55) and leukemia
(56) by literature retrieval.
However, there were disagreements regarding the association between
the expression levels of miRNA-122 and the prognosis of HCC in the
serum or plasma. Specific studies revealed that high expression of
miRNA-122 indicated poor prognosis (57,58), but
certain findings suggested that the high expression of miRNA-122
predicted improved prognosis (59,60);
however, further investigation is required. Although there are a
number of reports on miRNA-122, most of them are limited to a
certain cancer, especially liver cancer. In the present study, a
more comprehensive pan-cancer analysis of miRNA-122 was performed
in combination with the current popular TCGA high-throughput
sequencing database and associated literature. The present study is
more general, gives a summary and can provide a macro guidance for
the diagnosis, prognosis and treatment of multiple cancers in
clinical practice.
In this study, the target genes of miRNA-122 were
predicted using numerous platforms and the GO and KEGG enrichment
analyses were performed on the reported target genes. The results
of GO enrichment analysis demonstrated that the target genes of
miRNA-122 may contribute to the composition of the nucleus and
cytoplasm, and regulate cardiac muscle cell differentiation in a
variety of biological processes, including cardiac muscle cell
differentiation and glucose metabolic processes through protein
binding. So far, numerous articles have reported that miRNA-122 was
closely associated with heart-related diseases, obesity and
diabetes (37,61,62,64). For
example, the expression of miR-122 was involved in hypoxia-induced
cardiomyocyte injury (37). In
experiments with miniature pigs, researchers demonstrated that
increases in weight and cholesterol levels could lead to a decline
in the expression levels of miRNA-122, which supported the
association of miRNA122 in obesity (61). KEGG pathway analysis revealed that
the target genes were mainly enriched in protein biosynthesis,
estrogen and glucagon associated signaling pathways. Munagala et
al (62) reported that miRNA-122
was one of the significantly modulated miRNAs in the process of
estrogen-mediated mammary carcinogenesis. miRNA-122 participated in
the regulation of insulin resistance, diabetes and metabolic
syndrome, which were all likely to be associated with glucagon
related signaling pathways (42).
In conclusion, the clinical significance of
miRNA-122 was systematically analyzed from multiple perspectives in
this study. The diagnostic value of miRNA-122 between the HCC and
healthy control via serology was further demonstrated, and the
differential diagnostic value in patients with HCC compared with in
benign lesions was also analyzed. In addition, it was demonstrated
that miRNA-122 exhibited significant differential expression and
prognostic ability in various types of cancer via TCGA database and
literature review. Finally, a number of important biological
functions and regulatory mechanisms have been reported through the
enrichment analysis of the miRNA-122 target genes. These analyses
demonstrated that miRNA-122 may be an indispensable biomarker for
the diagnosis, prognostic evaluation and targeted therapy in a
variety of cancer types.
Acknowledgements
The authors would like to thank Mrs Yujie Huang
(Department of Clinical Laboratory, First Affiliated Hospital of
Guangxi Medical University, Guangxi, China) for her work in
language editing.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the TCGA repository (https://cancergenome.nih.gov/).
Authors' contributions
MD contributed to the writing of the paper and
bioinformatics analysis; LL contributed to the sample collection
and experiment; XQ contributed to the overall design of this
paper.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of the First Affiliated Hospital of Guangxi Medical
University and written informed consent was obtained from all
individuals in this research project.
Patient consent for publication
All the patients have written informed consent for
the publication of any associated data or accompanying images.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang
B, Li BS and Yang SM: The emergence of long non-coding RNAs in the
tumorigenesis of hepatocellular carcinoma. Cancer Lett.
360:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273 e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu CJ and Kao JH: Global perspective on
the natural history of chronic hepatitis B: role of hepatitis B
virus genotypes A to J. Semin Liver Dis. 33:97–102. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fukaya T and Tomari Y: MicroRNAs mediate
gene silencing via multiple different pathways in drosophila.
Molecular cell. 48:825–836. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takasaki S: Roles of microRNAs in cancers
and development. Methods Mol Biol. 1218:375–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang LF, Jiang S and Liu MF: MicroRNA
regulation and analytical methods in cancer cell metabolism. Cell
Mol Life Sci. 74:2929–2941. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Castro D, Moreira M, Gouveia AM, Pozza DH
and De Mello RA: MicroRNAs in lung cancer. Oncotarget.
8:81679–81685. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ferracin M, Lupini L, Mangolini A and
Negrini M: Circulating non-coding RNA as biomarkers in colorectal
cancer. Adv Exp Med Biol. 937:171–181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Endzeliņš E, Melne V, Kalnina Z,
Lietuvietis V, Riekstiņa U, Llorente A and Linē A: Diagnostic,
prognostic and predictive value of cell-free miRNAs in prostate
cancer: A systematic review. Mol Cancer. 15:412016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bandiera S, Pfeffer S, Baumert TF and
Zeisel MB: miR-122-a key factor and therapeutic target in liver
disease. J Hepatol. 62:448–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wen J and Friedman JR: miR-122 regulates
hepatic lipid metabolism and tumor suppression. J Clin Invest.
122:2773–2776. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Y, He Y, Ding J, Wu K, Hu B, Liu Y, Wu
Y, Guo B, Shen Y, Landi D, et al: An insertion/deletion
polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′
untranslated region confers risk for hepatocellular carcinoma.
Carcinogenesis. 30:2064–2069. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma J, Li T, Han X and Yuan H: Knockdown of
LncRNA ANRIL suppresses cell proliferation, metastasis, and
invasion via regulating miR-122-5p expression in hepatocellular
carcinoma. J Cancer Res Clin Oncol. 144:205–214. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Caviglia GP, Abate ML, Gaia S, Petrini E,
Bosco C, Olivero A, Rosso C, Ciancio A, Pellicano R, Saracco GM, et
al: Risk of hepatocellular carcinoma in HBV cirrhotic patients
assessed by the combination of miR-122, AFP and PIVKA-II.
Panminerva Med. 59:283–289. 2017.PubMed/NCBI
|
|
17
|
Ding Y, Yan JL, Fang AN, Zhou WF and Huang
L: Circulating miRNAs as novel diagnostic biomarkers in
hepatocellular carcinoma detection: A meta-analysis based on 24
articles. Oncotarget. 8:66402–66413. 2017.PubMed/NCBI
|
|
18
|
Naderi M, Pazouki A, Arefian E, Hashemi
SM, Jamshidi-Adegani F, Gholamalamdari O, Soudi S, Azadmanesh K,
Mirab Samiee S, Merat S, et al: Two triacylglycerol pathway genes,
CTDNEP1 and LPIN1, are down-regulated by hsa-miR-122-5p in
hepatocytes. Arch Iran Med. 20:165–171. 2017.PubMed/NCBI
|
|
19
|
Schisterman EF, Perkins NJ, Liu A and
Bondell H: Optimal cut-point and its corresponding Youden Index to
discriminate individuals using pooled blood samples. Epidemiology.
16:73–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Dai M, Zhu H, Li J, Huang Z, Liu
X, Huang Y, Chen J and Dai S: Evaluation on the diagnostic and
prognostic values of long non-coding RNA BLACAT1 in common types of
human cancer. Mol Cancer. 16:1602017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goldvaser H, Gutkin A, Beery E, Edel Y,
Nordenberg J, Wolach O, Rabizadeh E, Uziel O and Lahav M:
Characterisation of blood-derived exosomal hTERT mRNA secretion in
cancer patients: A potential pan-cancer marker. Br J Cancer.
117:353–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:(Database Issue). D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:(Database Issue). D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
26
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiao DD, Yang J, Lei XF, Mi GL, Li SL, Li
K, Xu CQ and Yang HL: Expression of microRNA-122 and microRNA-22 in
HBV-related liver cancer and the correlation with clinical
features. Eur Rev Med Pharmacol Sci. 21:742–747. 2017.PubMed/NCBI
|
|
28
|
Wang Y, Zhu P, Qiu J, Wang J, Zhu H, Zhu
Y, Zhang L, Zhu J, Liu X and Dong C: Identification and
characterization of interferon signaling-related microRNAs in
occult hepatitis B virus infection. Clin Epigenetics. 9:1012017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dai R, Peng F, Xiao X, Gong X, Jiang Y,
Zhang M, Tian Y, Xu Y, Ma J, Li M, et al: Hepatitis B virus X
protein-induced upregulation of CAT-1 stimulates proliferation and
inhibits apoptosis in hepatocellular carcinoma cells. Oncotarget.
8:60962–60974. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ali HEA, Abdel Hameed R, Effat H, Ahmed
EK, Atef AA, Sharawi SK, Ali M, Abd Elmageed ZY and Abdel Wahab AH:
Circulating microRNAs panel as a diagnostic tool for discrimination
of HCV-associated hepatocellular carcinoma. Clin Res Hepatol
Gastroenterol. 41:e51–e62. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Murray DD, Suzuki K, Law M, Trebicka J,
Neuhaus Nordwall J, Johnson M, Vjecha MJ, Kelleher AD and Emery S:
Circulating miR-122 and miR-200a as biomarkers for fatal liver
disease in ART-treated, HIV-1-infected individuals. Sci Rep.
7:109342017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J,
Chen J, Liu L, Cai H, Li Z, et al: Knockdown of SOX2OT inhibits the
malignant biological behaviors of glioblastoma stem cells via
up-regulating the expression of miR-194-5p and miR-122. Mol Cancer.
16:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang
Y, Bao X, Du Q, Luo G, Liu K, et al: miR-122 promotes metastasis of
clear-cell renal cell carcinoma by downregulating Dicer. Int J
Cancer. 142:547–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rao M, Zhu Y, Zhou Y, Cong X and Feng L:
MicroRNA-122 inhibits proliferation and invasion in gastric cancer
by targeting CREB1. Am J Cancer Res. 7:323–333. 2017.PubMed/NCBI
|
|
35
|
Calatayud D, Dehlendorff C, Boisen MK,
Hasselby JP, Schultz NA, Werner J, Immervoll H, Molven A, Hansen CP
and Johansen JS: Tissue MicroRNA profiles as diagnostic and
prognostic biomarkers in patients with resectable pancreatic ductal
adenocarcinoma and periampullary cancers. Biomark Res. 5:82017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li XD, Yang YJ, Wang LY, Qiao SB, Lu XF,
Wu YJ, Xu B, Li HF and Gu DF: Elevated plasma miRNA-122, −140-3p,
−720, −2861, and −3149 during early period of acute coronary
syndrome are derived from peripheral blood mononuclear cells. PLoS
One. 12:e01842562017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Z, Li H, Chen S, Li Y, Cui Z and Ma
J: Knockdown of MicroRNA-122 protects H9c2 cardiomyocytes from
hypoxia-induced apoptosis and promotes autophagy. Med Sci Monit.
23:4284–4290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Šatrauskienė A, Navickas R, Laucevičius A
and Huber HJ: Identifying differential miR and gene consensus
patterns in peripheral blood of patients with cardiovascular
diseases from literature data. BMC Cardiovasc Disord. 17:1732017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Blum A, Yehuda H, Geron N and Meerson A:
Elevated levels of miR-122 in serum may contribute to improved
endothelial function and lower oncologic risk following bariatric
surgery. Isr Med Assoc J. 19:620–624. 2017.PubMed/NCBI
|
|
40
|
Jones A, Danielson KM, Benton MC, Ziegler
O, Shah R, Stubbs RS, Das S and Macartney-Coxson D: miRNA
signatures of insulin resistance in obesity. Obesity (Silver
Spring). 25:1734–1744. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao H, Shen J, Daniel-MacDougall C, Wu X
and Chow WH: Plasma MicroRNA signature predicting weight gain among
Mexican-American women. Obesity (Silver Spring). 25:958–964. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Willeit P, Skroblin P, Moschen AR, Yin X,
Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramírez CM,
Goedeke L, et al: Circulating MicroRNA-122 is associated with the
risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes.
66:347–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bijkerk R, Florijn BW, Khairoun M, Duijs
JMGJ, Ocak G, de Vries APJ, Schaapherder AF, Mallat MJK, de Fijter
JW, Rabelink TJ, et al: Acute rejection after kidney
transplantation associates with circulating MicroRNAs and vascular
injury. Transplant Direct. 3:e1742017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang T, Li F, Geng W, Ruan Q and Shi W:
MicroRNA-122 ameliorates corneal allograft rejection through the
downregulation of its target CPEB1. Cell Death Discov. 3:170212017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Selmaj I, Cichalewska M, Namiecinska M,
Galazka G, Horzelski W, Selmaj KW and Mycko MP: Global exosome
transcriptome profiling reveals biomarkers for multiple sclerosis.
Ann Neurol. 81:703–717. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Trzybulska D, Bobjer J, Giwercman A and
Tsatsanis C: Serum microRNAs in male subfertility-biomarkers and a
potential pathogenetic link to metabolic syndrome. J Assist Reprod
Genet. 34:1277–1282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Coulouarn C, Factor VM, Andersen JB,
Durkin ME and Thorgeirsson SS: Loss of miR-122 expression in liver
cancer correlates with suppression of the hepatic phenotype and
gain of metastatic properties. Oncogene. 28:3526–3536. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jin Y, Wang J, Han J, Luo D and Sun Z:
MiR-122 inhibits epithelial-mesenchymal transition in
hepatocellular carcinoma by targeting Snail1 and Snail2 and
suppressing WNT/β-cadherin signaling pathway. Exp Cell Res.
360:210–217. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Miquelestorena-Standley E, Tallet A,
Collin C, Piver E, De Muret A, Salamé E, Bourlier P, Kervarrec T,
Guyétant S and Pagès JC: Interest of variations in microRNA-152 and
−122 in a series of hepatocellular carcinomas related to hepatitis
C virus infection. Hepatol Res. 48:566–573. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH,
Wang JH, Huang CM, Lee CM, Lin CY, Yen YH and Chiu YC: Circulating
microRNAs as biomarkers for diagnosis of early hepatocellular
carcinoma associated with hepatitis B virus. Int J Cancer.
138:714–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
El-Garem H, Ammer A, Shehab H, Shaker O,
Anwer M, El-Akel W and Omar H: Circulating microRNA, miR-122 and
miR-221 signature in Egyptian patients with chronic hepatitis C
related hepatocellular carcinoma. World J Hepatol. 6:818–824. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu Q, Zhang M, Tu J, Pang L, Cai W and Liu
X: MicroRNA-122 affects cell aggressiveness and apoptosis by
targeting PKM2 in human hepatocellular carcinoma. Oncol Rep.
34:2054–2064. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kanaan Z, Rai SN, Eichenberger MR, Barnes
C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE,
et al: Differential microRNA expression tracks neoplastic
progression in inflammatory bowel disease-associated colorectal
cancer. Hum Mutat. 33:551–560. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tang Y, Zhao S, Wang J, Li D, Ren Q and
Tang Y: Plasma miR-122 as a potential diagnostic and prognostic
indicator in human glioma. Neurol Sci. 38:1087–1092. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang
Q, Chen L, Pang X, Leng W and Bi F: Plasma miR-122 and miR-192 as
potential novel biomarkers for the early detection of distant
metastasis of gastric cancer. Oncol Rep. 31:1863–1870. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yang J, Yuan Y, Yang X, Hong Z and Yang L:
Decreased expression of microRNA-122 is associated with an
unfavorable prognosis in childhood acute myeloid leukemia and
function analysis indicates a therapeutic potential. Pathol Res
Pract. 213:1166–1172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cho HJ, Kim JK, Nam JS, Wang HJ, Lee JH,
Kim BW, Kim SS, Noh CK, Shin SJ, Lee KM, et al: High circulating
microRNA-122 expression is a poor prognostic marker in patients
with hepatitis B virus-related hepatocellular carcinoma who undergo
radiofrequency ablation. Clin Biochem. 48:1073–1078. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu M, Liu J, Wang L, Wu H, Zhou C, Zhu H,
Xu N and Xie Y: Association of serum microRNA expression in
hepatocellular carcinomas treated with transarterial
chemoembolization and patient survival. PLoS One. 9:e1093472014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xu Y, Bu X, Dai C and Shang C: High serum
microRNA-122 level is independently associated with higher overall
survival rate in hepatocellular carcinoma patients. Tumour Biol.
36:4773–4776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Köberle V, Kronenberger B, Pleli T, Trojan
J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem
S, Piiper A and Waidmann O: Serum microRNA-1 and microRNA-122 are
prognostic markers in patients with hepatocellular carcinoma. Eur J
Cancer. 49:3442–3449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cirera S, Birck M, Busk PK and Fredholm M:
Expression profiles of miRNA-122 and its target CAT1 in minipigs
(Sus scrofa) fed a high-cholesterol diet. Comp Med. 60:136–141.
2010.PubMed/NCBI
|
|
62
|
Munagala R, Aqil F, Vadhanam MV and Gupta
RC: MicroRNA ‘signature’ during estrogen-mediated mammary
carcinogenesis and its reversal by ellagic acid intervention.
Cancer Lett. 339:175–184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
El-Abd NE, Fawzy NA, El-Sheikh SM and
Soliman ME: Circulating miRNA-122, miRNA-199a, and miRNA-16 as
biomarkers for early detection of hepatocellular carcinoma in
egyptian patients with chronic hepatitis C virus infection. Mol
Diagn Ther. 19:213–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang G, Zhao Y and Zheng Y:
MiR-122/Wnt/β-catenin regulatory circuitry sustains glioma
progression. Tumour Biol. 35:8565–8572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Khare D, Goldschmidt N, Bardugo A,
Gur-Wahnon D, Ben-Dov IZ and Avni B: Plasma microRNA profiling:
Exploring better biomarkers for lymphoma surveillance. PLoS One.
12:e01877222017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Conceição AL, Da Silva CT, Badial RM,
Valsechi MC, Stuqui B, Gonçalves JD, Jasiulionis MG, De Freitas
Calmon M and Rahal P: Downregulation of OCLN and GAS1 in clear cell
renal cell carcinoma. Oncol Rep. 37:1487–1496. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Nientiedt M, Deng M, Schmidt D, Perner S,
Müller SC and Ellinger J: Identification of aberrant tRNA-halves
expression patterns in clear cell renal cell carcinoma. Sci Rep.
6:371582016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jingushi K, Kashiwagi Y, Ueda Y, Kitae K,
Hase H, Nakata W, Fujita K, Uemura M, Nonomura N and Tsujikawa K:
High miR-122 expression promotes malignant phenotypes in ccRCC by
targeting occludin. Int J Oncol. 51:289–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Munari E, Marchionni L, Chitre A, Hayashi
M, Martignoni G, Brunelli M, Gobbo S, Argani P, Allaf M, Hoque MO
and Netto GJ: Clear cell papillary renal cell carcinoma: micro-RNA
expression profiling and comparison with clear cell renal cell
carcinoma and papillary renal cell carcinoma. Hum Pathol.
45:1130–1138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang Z, Qin C, Zhang J, Han Z, Tao J, Cao
Q, Zhou W, Xu Z, Zhao C, Tan R and Gu M: MiR-122 promotes renal
cancer cell proliferation by targeting Sprouty2. Tumour Biol.
39:10104283176911842017.PubMed/NCBI
|
|
71
|
Wotschofsky Z, Busch J, Jung M,
Kempkensteffen C, Weikert S, Schaser KD, Melcher I, Kilic E, Miller
K, Kristiansen G, et al: Diagnostic and prognostic potential of
differentially expressed miRNAs between metastatic and
non-metastatic renal cell carcinoma at the time of nephrectomy.
Clin Chim Acta. 416:5–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Papaconstantinou IG, Manta A, Gazouli M,
Lyberopoulou A, Lykoudis PM, Polymeneas G and Voros D: Expression
of microRNAs in patients with pancreatic cancer and its prognostic
significance. Pancreas. 42:67–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Schultz NA, Dehlendorff C, Jensen BV,
Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE,
Yilmaz M, Holländer NH, et al: MicroRNA biomarkers in whole blood
for detection of pancreatic cancer. JAMA. 311:392–404. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Carter JV, Roberts HL, Pan J, Rice JD,
Burton JF, Galbraith NJ, Eichenberger MR, Jorden J, Deveaux P,
Farmer R, et al: A highly predictive model for diagnosis of
colorectal neoplasms using plasma MicroRNA: Improving specificity
and sensitivity. Ann Surg. 264:575–584. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Liu N, Jiang F, He TL, Zhang JK, Zhao J,
Wang C, Jiang GX, Cao LP, Kang PC, Zhong XY, et al: The roles of
MicroRNA-122 overexpression in inhibiting proliferation and
invasion and stimulating apoptosis of human cholangiocarcinoma
cells. Sci Rep. 5:165662015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lu W, Zhang Y, Zhou L, Wang X, Mu J, Jiang
L, Hu Y, Dong P and Liu Y: miR-122 inhibits cancer cell malignancy
by targeting PKM2 in gallbladder carcinoma. Tumour Biol. Nov
6–2015.(Epub ahead of print).
|
|
77
|
Walter BA, Valera VA, Pinto PA and Merino
MJ: Comprehensive microRNA profiling of prostate cancer. J Cancer.
4:350–357. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Venkatesan N, Deepa PR, Khetan V and
Krishnakumar S: Computational and in vitro investigation of
miRNA-Gene regulations in retinoblastoma pathogenesis: miRNA mimics
strategy. Bioinform Biol Insights. 9:89–101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xiao F, Chen J, Lian C, Han P and Zhang C:
Tumor necrosis factor-related apoptosis-inducing ligand induces
cytotoxicity specific to osteosarcoma by microRNA response
elements. Mol Med Rep. 11:739–745. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ergün S, Ulasli M, Igci YZ, Igci M,
Kırkbes S, Borazan E, Balik A, Yumrutaş Ö, Camci C, Cakmak EA, et
al: The association of the expression of miR-122-5p and its target
ADAM10 with human breast cancer. Mol Biol Rep. 42:497–505. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yan Y, Zhang F, Fan Q, Li X and Zhou K:
Breast cancer-specific TRAIL expression mediated by miRNA response
elements of let-7 and miR-122. Neoplasma. 61:672–679. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang B, Wang H and Yang Z: MiR-122
inhibits cell proliferation and tumorigenesis of breast cancer by
targeting IGF1R. PLoS One. 7:e470532012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Peng Y, Li C, Luo DC, Ding JW, Zhang W and
Pan G: Expression profile and clinical significance of microRNAs in
papillary thyroid carcinoma. Molecules. 19:11586–11599. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Stilling G, Sun Z, Zhang S, Jin L, Righi
A, Kovācs G, Korbonits M, Scheithauer BW, Kovacs K and Lloyd RV:
MicroRNA expression in ACTH-producing pituitary tumors:
up-regulation of microRNA-122 and −493 in pituitary carcinomas.
Endocrine. 38:67–75. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wang Y, Xing QF, Liu XQ, Guo ZJ, Li CY and
Sun G: MiR-122 targets VEGFC in bladder cancer to inhibit tumor
growth and angiogenesis. Am J Transl Res. 8:3056–3066.
2016.PubMed/NCBI
|