Introduction
Esophageal cancer is considered as the 7th most
common type of cancer worldwide. It is also the 6th leading cause
of cancer-associated mortality (1,2).
According to different histological types, esophageal cancer can be
further divided into squamous cell carcinoma and adenocarcinoma.
Squamous cell carcinoma is regarded as the most prevalent
esophageal cancer in the world, especially in some developing
countries (3). Although some
advances has been made in diagnosis and treatments, the incidence
of esophageal cancer has markedly increased in the past years
(3), and the 5-year survival rate of
patients with advanced esophageal cancer is less than 20% (4). Therefore, the development of more
effective therapeutic methods and novel prognostic molecular
markers are necessary to improve the patient survival rate.
Numerous microRNAs (miRNAs or miRs) have been
demonstrated to act as oncogenes or tumor suppressor genes. During
the process of cancer development and progression, miRNAs have been
revealed to play a significant role in regulation. In addition,
these miRNAs also can be used as novel molecular biomarkers for
cancer prognosis, even cancer targeted therapies (5–8). Among
these miRNAs, researchers have demonstrated that microRNA-193b
(miR-193b) functions as a tumor suppressor in multiple
malignancies, such as prostate cancer (9), melanoma (10), and breast cancer (11). In addition, the level of miR-193b was
revealed to be different between chemosensitive and chemoresistant
cell lines, and the chemoresistant cell lines had lower expression
than chemosensitive cell lines (12). The overexpression of miR-193b was
revealed to significantly impede the ability of esophageal cancer
cells to recover following 5-fluorouracil treatment, and markedly
induce autophagic flux and non-apoptotic cell death, indicating the
important role of miR-193b in esophageal cancer chemotherapy
(12,13).
Molecular alterations associated with esophageal
cancer progression have been extensively investigated in recent
years (3,14). One of the most frequent molecular
alterations is KRAS, an oncogene. Activated KRAS could cause cell
growth and survival, which is important during cancer development
(15,16). Although KRAS mutations are regarded
as a key event in carcinogenesis, the targeting upstream signaling
which modulates KRAS activity is still a promising future approach
for the treatment of esophageal cancer.
In present study, KRAS was confirmed as the direct
target gene of miR-193b in human esophageal squamous cell
carcinoma. It was also revealed that miR-193b overexpression could
induce esophageal squamous cell carcinoma cell apoptosis and
suppress cancer cell proliferation, as well as migration/invasion,
indicating that miR-193b has the potential to become a novel
diagnosis marker and therapy target for human esophageal squamous
cell carcinoma.
Materials and methods
Esophageal squamous cell carcinoma
samples
In our study, 53 different patients (38 males and 15
females) donated their esophageal squamous cell carcinoma tissues
and paracancerous tissues for our research. All samples belonged to
primary tumors. The cancer tissues from 31 patients were
infiltrating esophageal squamous cell carcinoma and the cancer
tissues from 22 patients were superficial esophageal squamous cell
carcinoma. The number of patients in the different age-groups was
as follows: n(50–59)=9; n(60–69)=30; n(70=79)=14. The number of
patients in the different cancer stages wasas follows: n(Stage
I)=6; n(stage II)=16; n(stage III)=20; n(stage IV)=11.
Cell culture
Esophageal squamous cell carcinoma cell lines,
KYSE450 and TE1, and normal epithelial cell line, Het-1A, were used
in the present study, and were purchased from the Cell Bank of the
Chinese Academy of Sciences (Shanghai, China). The cells were
cultured using Dulbecco's Modified Eagle's Medium (DMEM; Hyclone;
GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences), 0.1
g/ml streptomycin and 100 U/ml penicillin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) in a humidified 37°C incubator with 5%
CO2. The culture medium was changed every two days, and
the cells were passaged by 1:4 dilution every 5–6 days.
miR-193b preparation and
transfection
miR-193b (5′-AACUGGCCCUCAAAGUCCCGCU-3′) was
synthesized from Sengong Biotech, Shanghai, China. The unspecific
miRNA (5′-ACGUGACACGUUCGGAGAAUU-3′) was used as the negative
control (ctrl miRNA). The reverse complementary miRNA
(5′-AGCGGGACUUUGAGGCCAGUU-3′) was applied as a miR-193b inhibitor
in this study. Cell transfection was performed using Lipofectamine
3000 transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions.
Real-Time quantitative PCR
The real-time quantitative PCR procedure was the
same as previously described (17–19). In
the present study, KRAS was normalized to 18S rRNA and miR-193b was
normalized to U6. The primer sequences (5′-3′) were as follows:
KRAS forward, 5′-GCCTTGACGATACAGCTAAT-3′ and reverse,
5′-GCTGTGTCGAGAATATCCAA-3′; 18S rRNA forward,
5′-CCTGGATACCGCAGCTAGGA-3′ and reverse,
5′-GCGGCGCAATACGAATGCCCC-3′; miR-193b forward,
5′-ACACTCCAGCTGGGAACTGGCCCTCAAAGTC-3′ and reverse,
5′-AGCCTCTGCGCACGTGTTC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Dual luciferase assay
The potential target genes of miR-193b were analyzed
using TargetScan (http://www.targetscan.org/vert_72/). The 3′UTR
sequence of the human KRAS gene was amplified using PCR and further
cloned into a psiCHECK-based luciferase plasmid (Addgene, Inc.,
Cambridge, MA, USA). The establishment of mutated psiCHECK-KRAS
3′UTR, as well as the procedure of the dual luciferase assay was
same as previously described (20,21). In
brief, the WT psiCHECK-KRAS 3′UTR was used as the template for PCR,
and the primers which matched the 3′UTR part and contained the
mutated sequence were synthesized by IDT (Beijing, China). The
extension of the PCR template could generate nicked circular DNA
molecules, followed by DpnI endonuclease digestion,
competent cell transformation and mini-plasmid preparation. Herein,
3–4 bases could be mutated using the above method to induce
mutation in psiCHECK-KRAS 3′UTR at once. Therefore, to obtain the
whole mutated sequence (11-base mutation), the mutation induction
was repeated for 3 times with different primers. The sequence of
primers (5′-3′) were as follows: M1-F,
GCTTGTGACATTAAAAGATTAAAACGGCCAGTTATAGCTTATTAGGTGTTGA and M1-R,
TCAACACCTAATAAGCTATAACTGGCCGTTTTAATCTTTTAATGTCACAAGC; M2-F,
GCTTGTGACATTAAAAGATTAAAACCCGGAGTTATAGCTTATTAGGTGTTGA and M2-R,
TCAACACCTAATAAGCTATAACTCCGGGTTTTAATCTTTTAATGTCACAAGC; M3-F,
GCTTGTGACATTAAAAGATTAAAACCCGGTCATATAGCTTATTAGGTGTTGA and M3-R,
TCAACACCTAATAAGCTATATGACCGGGTTTTAATCTTTTAATGTCACAAGC.
The dual luciferase assay was performed using a Dual
Luciferase Assay Kit according to the manufacturer's instructions
(Promega Corporation, Madison, WI, USA). In the present study, both
Firefly and Renilla luciferase values were detected. In
addition, for the evaluation of relative luciferase activity, the
firefly value was used in Renilla value normalization.
Colony-forming ability assay and
various other assays
To evaluate the colony-forming ability of ctrl miRNA
group, miR-193b group and inhibitor group, 100 cancer cells were
seeded into 12-well plates and incubated for 7 days at 37°C in an
incubator with 5% CO2. Then the cancer cells were fixed
with 75% ethanol. The plate was further stained using crystal
violet for 20 min. Finally, an Epson Perfection V600, Epson, Japan
scanner was used to scan the plate and the results were further
analyzed using BioSpot® software 5.0, Cellular
Technology Limited (CTL), USA.
Cell proliferation, cell cycle analysis, cell
apoptosis, and cell migration/invasion assays were performed as
previously described (22).
Statistical analysis
In the present study, the results were expressed as
the means ± SEM, and analysis was performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA. Unpaired Student's t-tests
were used to analyze the means of two groups. One-way ANOVA with
Bonferroni's correction was used to analyze the means of three or
more groups. P<0.05 was considered to indicate a statistically
significant difference. In Fig. 1A and
B, the level of paracancerous tissue group was regarded as ‘1’.
In Fig. 2A and B, the level of the
control group (Ctrl miRNA) was regarded as ‘1’. In Fig. 2C, the level of the control group
[(KRAS-3′-UTR(WT) and KRAS-3′-UTR(Mu)] was regarded as ‘1’.
Results
KRAS is the direct target of miR-193b
in esophageal cancer cells
In the present study, esophageal squamous cell
carcinoma tissues and paracancerous tissues from 53 different
patients were harvested and qPCR was used to evaluate the
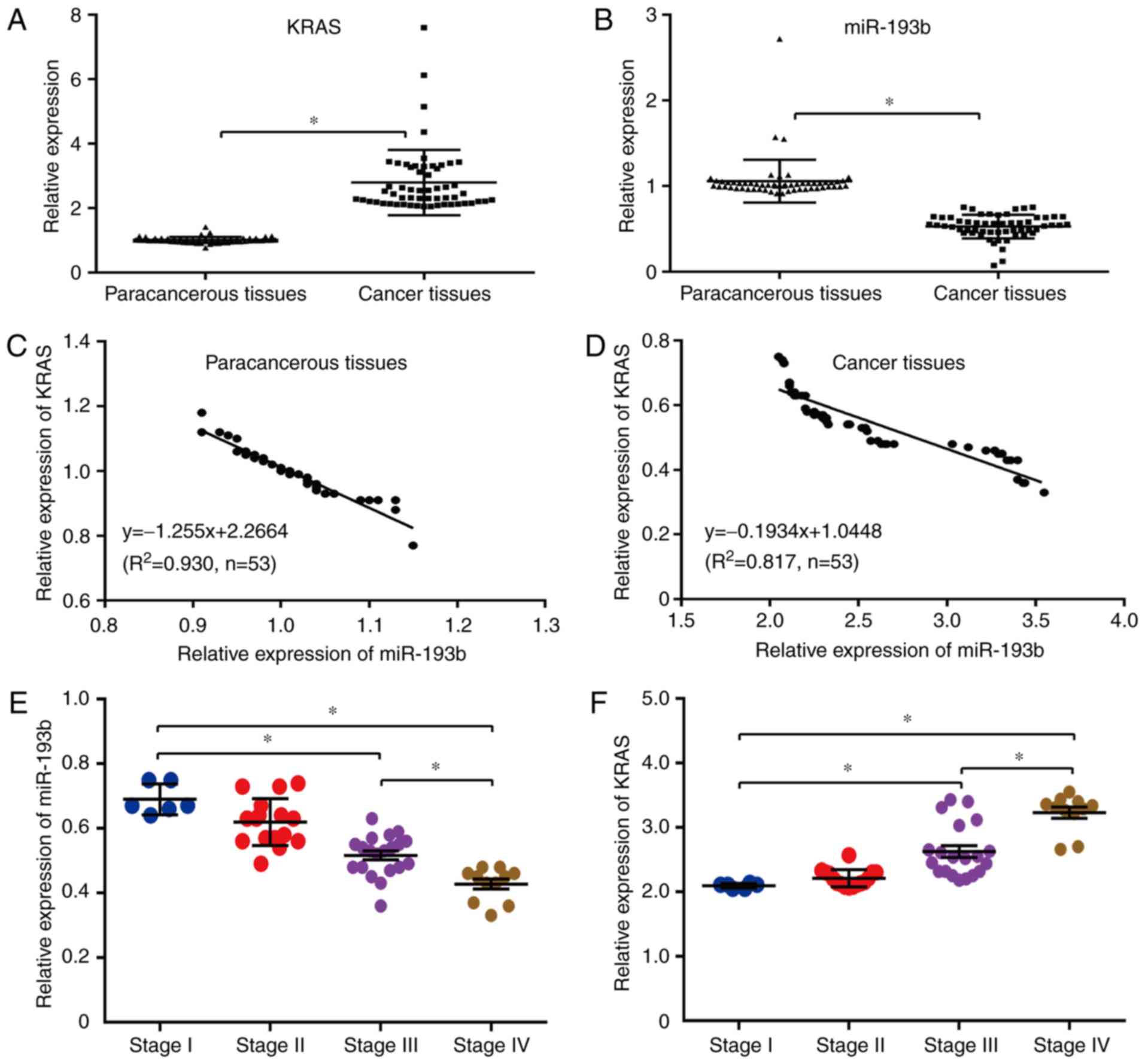
relationship between KRAS and miR-193b. The results revealed that
the mRNA level of miR-193b was significantly higher in human
paracancerous tissues than human esophageal cancer tissues
(P<0.05; Fig. 1B), while the
miRNA level of KRAS revealed the opposite tendency in human
esophageal cancer tissues and paracancerous tissues (P<0.05;
Fig. 1B), indicating that there may
be a negative regulatory relationship between miR-193b and KRAS in
the esophageal cancer tissues of patients. In addition, the
correlation between KRAS and miR-193b expression was also confirmed
through linear regression analysis, and the results indicated that
the increased miR-193b expression was significantly correlated with
decreased KRAS expression in both paracancerous tissues
(y=−1.255×+2.2664, R2=0.930; Fig. 1C) and cancer tissues
(y=−0.1934×+1.0448, R2=0.817; Fig. 1D). In addition, the relationship
between miR-193b/KRAS expression and stage of cancers was further
analyzed herein. The results revealed no obvious difference between
stages I and II, while increased expression of KRAS and decreased
expression of miR-193b could be observed in stages III and IV
compared to stage I. Therefore, the expression level of
miR-193b/KRAS was stage-dependent in human esophageal cancers, and
may potentially be novel diagnosis and prognostic molecular markers
(Fig. 1E and F).
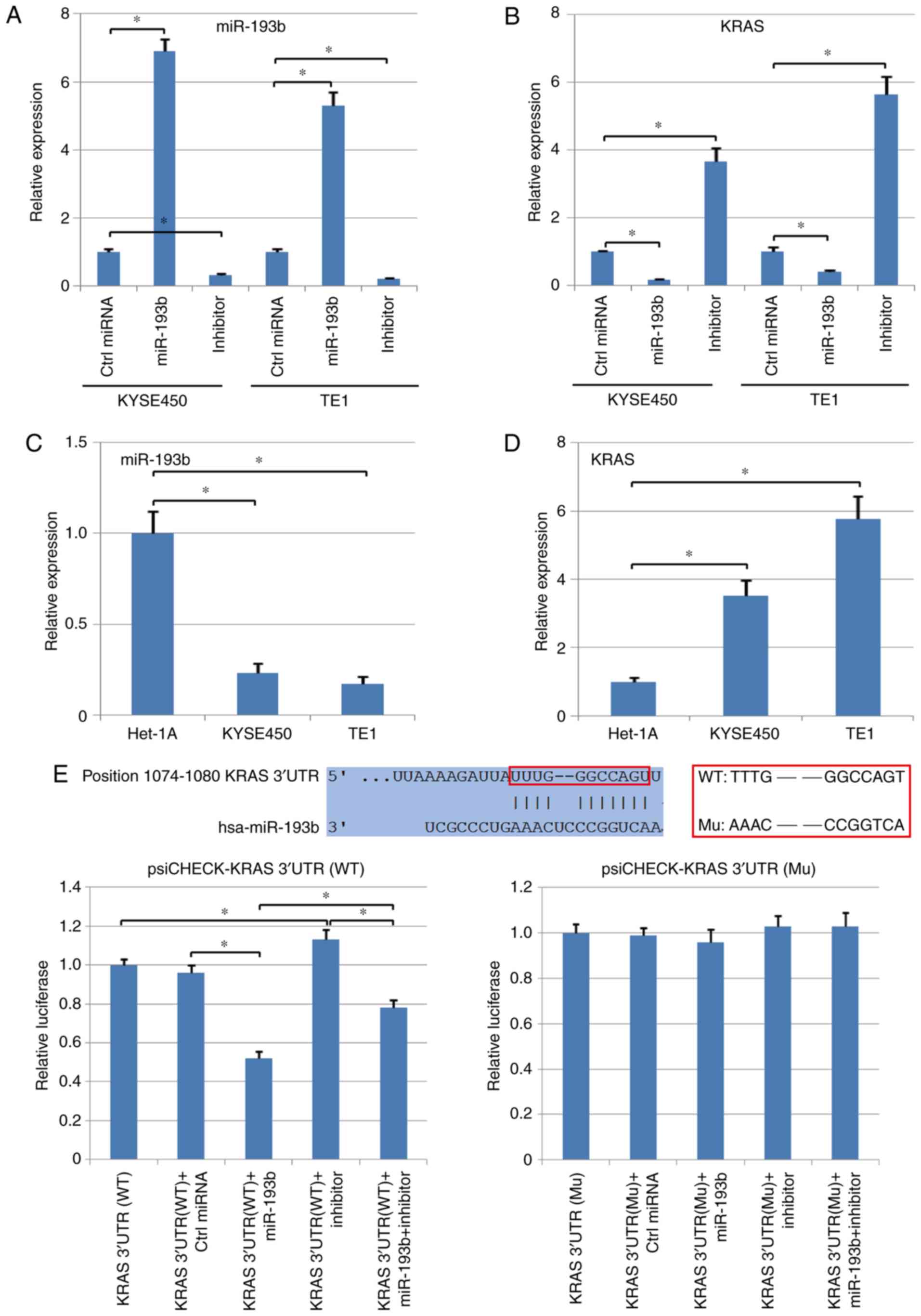
In addition, two different esophageal squamous cell
carcinoma cell lines, KYSE450 and TE1, were used to analyze the
negative regulatory relationship between miR-193b and KRAS in
esophageal cancer, and the level of KRAS and miR-193b in Het-1A,
normal epithelial cell line, was also detected using qPCR. It was
determined that the mRNA level of KRAS was inhibited by the
upregulation of miR-193b in human esophageal squamous cell
carcinoma cells. In addition, the cancer cells transfected with the
miR-193b inhibitor revealed significantly higher levels of KRAS
compared with the control group (P<0.05; Fig. 2A and B). Compared with normal
epithelial cells, Het-1A, both KYSE450 and TE1 exhibited a
significantly higher expression of KRAS and a lower expression of
miR-193b (P<0.05, Fig. 2C and
D).
The potential target genes of miR-193b were analyzed
using TargetScan, indicating that miR-193b could target the 3′UTR
of KRAS and regulate this gene directly (the binding relationship
was revealed in Fig. 2E). Therefore,
both wild-type (WT) and mutant-type (Mu) 3′UTR of KRAS were cloned
into the psi-CHECK vector, followed by TE1 cell transfection and
dual-luciferase assay. The results revealed that miR-193b
significantly suppressed the luciferase activity in the WT
KRAS-3′-UTR group, but did not reveal a significant effect in the
Mu KRAS-3′-UTR group (P<0.05; Fig.
2E). In addition, the inhibitory effect of miR-193b was
reversed by the transfection with the miR-193b inhibitor in the WT
KRAS-3′-UTR group, but no significant difference was observed in
the Mu KRAS-3′-UTR group (P<0.05; Fig. 2E). Collectively, these assays
revealed the direct targeted regulation between KRAS and
miR-193b.
miR-193b inhibits the growth and
proliferation abilities of esophageal cancer cell lines
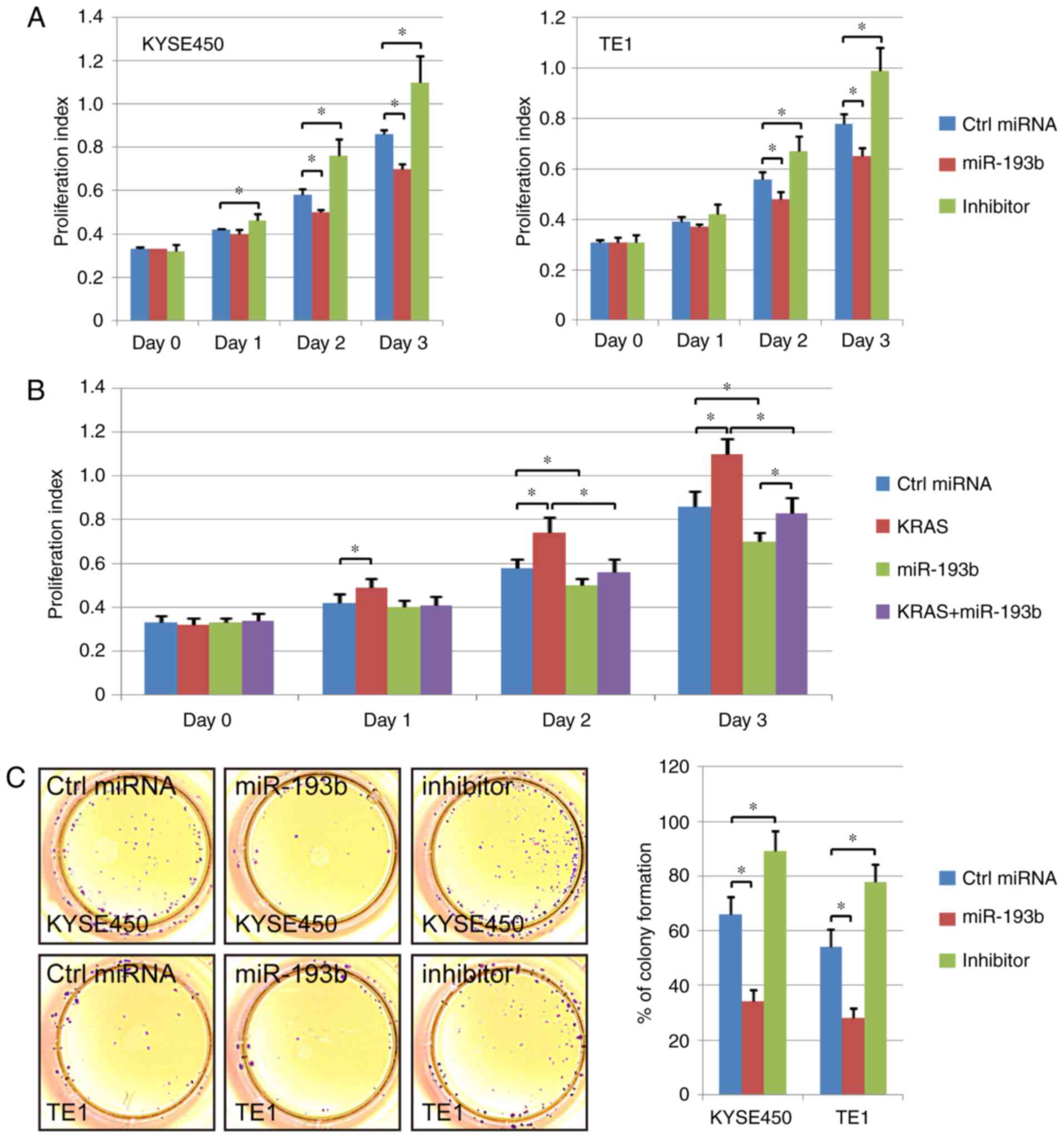
The difference in cell proliferation ability among
the control miRNA transfection group, the miR-193b overexpression
group and the miR-193b inhibitor group was analyzed in the present
study. The CCK-8 results revealed that esophageal cancer cell
proliferation ability was enhanced via miR-193b inhibitor
transfection. In addition, the overexpression of miR-193b
significantly suppressed the cell proliferation ability compared to
the control cells (Fig. 3A). A
rescue experiment was also performed, and KRAS overexpression was
revealed to significantly increase the proliferation ability of
esophageal cancer cells. miR-193b overexpression inhibited the
promoting effect of KRAS overexpression, indicating the effect of
miR-193b on cell viability via regulation of KRAS (Fig. 3B).
Furthermore, the proliferation ability of different
groups was further detected via colony formation assay. The results
revealed that the colony formation ability was increased in the
miR-193b-inhibitor transfection group. miR-193b overexpression
significantly decreased the number of colonies in esophageal
squamous cell carcinoma cells, which was consistent with the CCK-8
results (Fig. 3C).
In addition, the regulation of miR-193b and KRAS on
the cell cycle was assessed by flow cytometric assay. The results
revealed that miR-193b overexpression increased the percentage of
cells at the G0/G1 phase and decreased the percentage of S-phase
and G2/M-phase cells of KYSE450 and TE1. The opposite phenomenon
was revealed in the miR-193b inhibitor group (P<0.05, Fig. 3D). Collectively, these results
indicated that miR-193b inhibited the proliferation ability of
esophageal cancer cell lines.
Effect of miR-193b on esophageal
cancer cell apoptosis
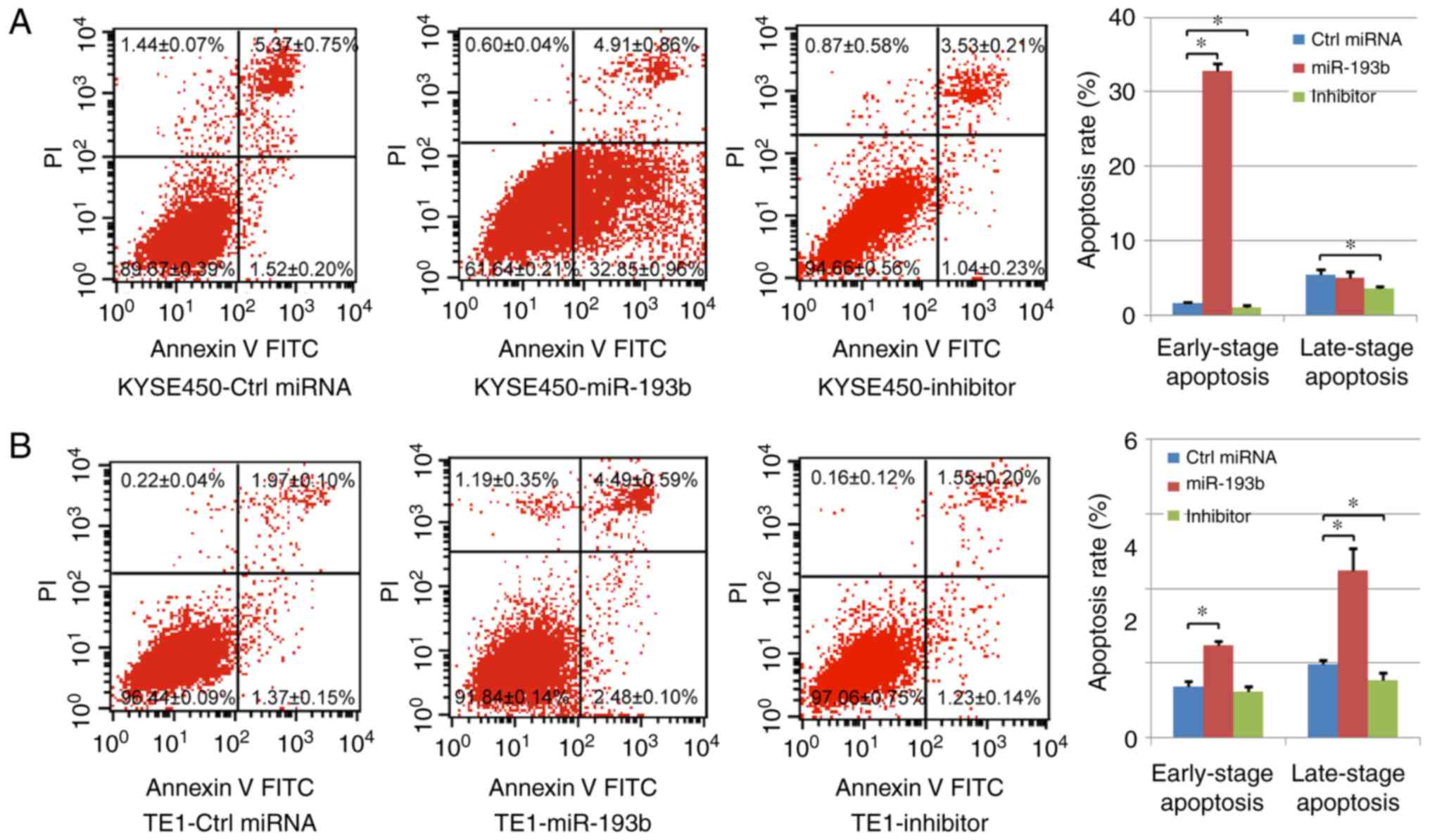
In our study, cell apoptosis was assessed using
Annexin V/PI staining. Both early-stage apoptotic cells (Annexin
V-positive and PI-negative) and late-stage apoptotic cells (Annexin
V-positive and PI-positive) were analyzed. In the KYSE450 cells, it
was revealed that the percentage of early-stage apoptotic cells was
significantly increased in the miR-193b overexpression group, while
transfection with the miR-193b inhibitor significantly decreased
the level of both early-stage and late-stage apoptotic cells
compared with the control group. In the TE1 cells, both the
percentages of early-stage and late-stage apoptotic cells were
significantly increased with the overexpression of miR-193b,
however, miR-193b-inhibitor transfection only reduced the
percentage of late-stage apoptotic cells (Fig. 4A and B), indicating that miR-193b
regulated cell apoptosis of esophageal cancer cells.
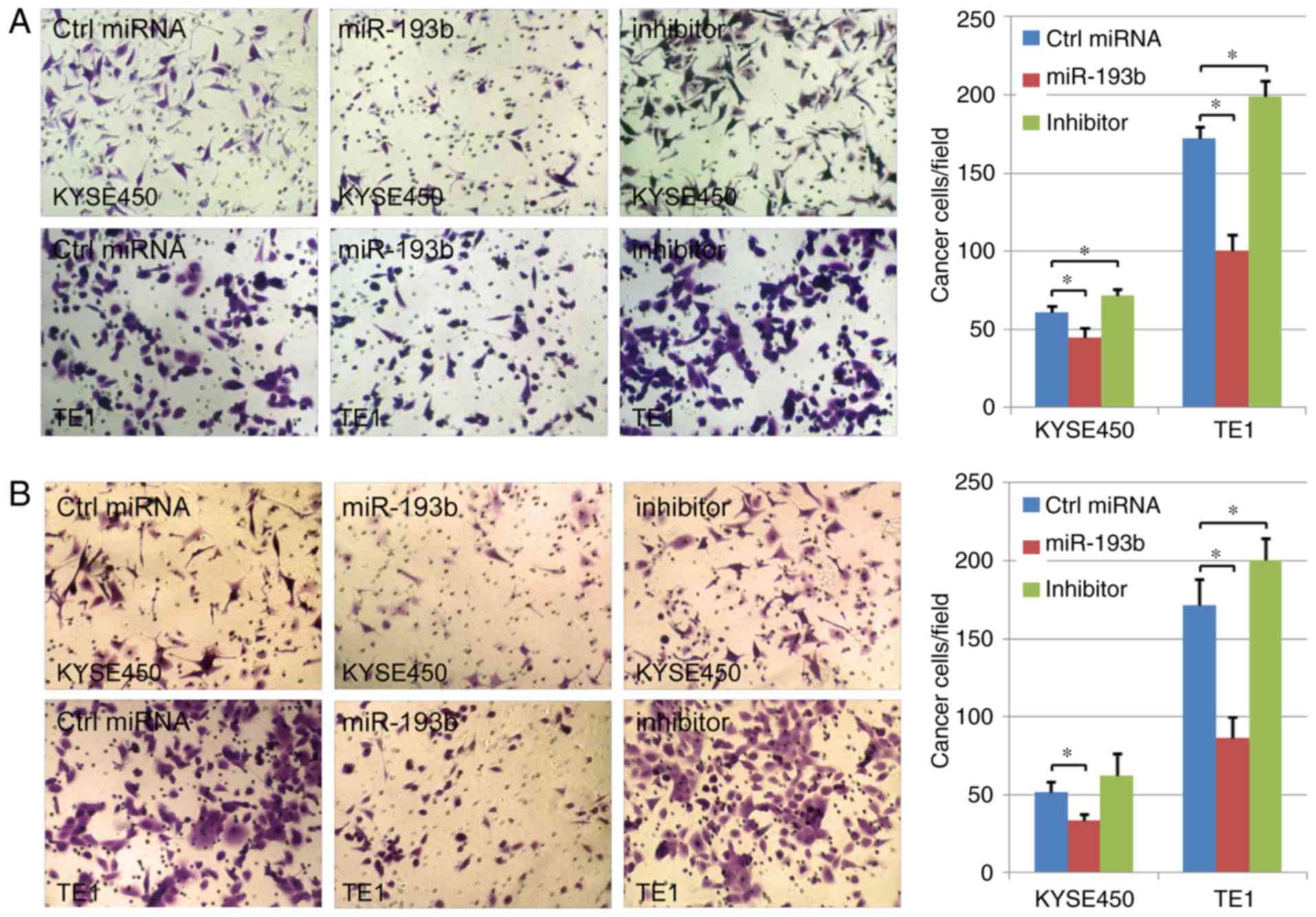
miR-193b inhibits esophageal cancer
cell migration and invasion
The effect of miR-193b on cell migration and
invasion abilities was further analyzed in the present study. In
TE1 cancer cells, the results indicated that both cell migration
and invasion could significantly be enhanced via miR-193b-inhibitor
transfection. Compared with the control group, the migration and
invasion abilities of human esophageal cancer cells were suppressed
in the miR-193b overexpression group, indicating the key role of
miR-193b in the migration/invasion of human esophageal cancer cells
(P<0.05; Fig. 5A and B). A
similar tendency was revealed in the results of KYSE450 cells,
however, in this cell line, miR-193b-inhibitor transfection did not
significantly affect cell invasion ability.
Discussion
The key role of miR-193b in the progression and
development of various cancers has been revealed in studies
(10,23–25). In
recent years, researchers revealed that miR-193b could promote
autophagy as well as non-apoptotic cell death in esophageal cancer
cells (13). In addition, although
critical mRNA targets of miR-193b were unknown, both target
prediction and siRNA data analysis indicated that miR-193b may
mediate these effects through stathmin 1 regulation (13). Researchers also revealed that the
silencing of stathmin 1 could at least partially reverse the
enhanced sensitivity to 5-fluorouracil, similar to the results
obtained with overexpression of miR-193b, and miR-193b was revealed
to inhibit tumor growth and metastasis, consequently through
regulation of the expression of stathmin 1 in other cancer cells
(26). Therefore, these data
revealed the important role of miR-193b in the regulation of
esophageal cancer progression. In addition, a previous study
revealed the target relationship between miR-193b and KRAS in
pancreatic cancer (24). However,
whether miR-193b can negatively regulate the expression of KRAS,
and whether it is related to clinical and pathological features in
human esophageal cancer, has not been investigated intensively. In
the present study, it was demonstrated that miR-193b targeted the
3′UTR of KRAS, and regulated the expression of KRAS negatively in
both human esophageal squamous cell carcinoma tissues and cells.
The expression level of miR-193b/KRAS was stage-dependent in human
esophageal cancer. miR-193b overexpression promoted cell apoptosis
and significantly inhibited the proliferation and
migration/invasion abilities of esophageal squamous cell carcinoma
cells, indicating the key role of miR-193b in the development and
progression of human esophageal cancer.
However, the detailed mechanism concerning the
regulatory function of miR-193b in human esophageal squamous cell
carcinoma still remains to be fully elucidated. Why the development
of esophageal squamous cell carcinoma is accompanied with
downregulation of miR-193b still requires further investigation.
Some studies indicated that the level of hypermethylation in the
promoter of some miRNAs could be enhanced in some cancer cells,
resulting in the downregulation of those miRNAs (27). Therefore, hypermethylation in the
promoter of miRNAs, may play an important role in the regulation of
miRNAs and in the development and progression of cancers.
Nevertheless, whether such a hypermethylation mechanism is still
applicable for the regulation of miR-193b still requires further
investigation. Recently, research revealed that certain
microenvironments induce the downregulation of miR-193b. In a
previous study, the direct interaction between cancer cells and
mesothelial cells, which covered the surface of the omentum, could
decrease the expression level of miR-193b via DNA methyltransferase
1, providing another possible mechanism for the regulation of
miR-193b in cancer progression (28).
Moreover, researchers have revealed other genes
which could be regulated by miR-193b directly. For example, it was
revealed that miR-193b was significantly downregulated in human
Ewing sarcoma, and ErbB4 was further identified as a target of
miR-193b. The overexpression of miR-193b significantly inhibited
the expression of ErbB4 and the restoration of ErbB4 expression
could reverse the inhibitory function of miR-193b on cancer cell
proliferation as well as metastasis (29). Therefore, miR-193b can regulate the
viability of cancer cells through different pathways. Thus, it may
be useful to analyze different signaling pathways associated with
miR-193b. A better understanding of the regulatory mechanism of
miR-193b in different cancers and which gene plays the most
important role during the process of miR-193b regulation, can help
us develop more reliable and effective methods to treat patients
with cancer.
In present study, our results revealed that miR-193b
overexpression could decrease the viability of human esophageal
cancer cell lines, and enhance cell apoptosis. However, slight
differences were observed between the different cell lines. For
example, miR-193b overexpression led to an increase in the
percentage of both early-stage apoptotic cells and late-stage
apoptotic cells in TE1 cells, while, overexpression of miR-193b
only significantly increased the percentage of early-stage
apoptotic cells in KYSE450 cells, and no obvious change was
observed in late-stage apoptotic cells. It is possible that cancer
cells with a high expression level of miR-193b or a low expression
level of KRAS exhibit some resistance to miR-193b overexpression.
Nevertheless, the application of more cell lines is required to
confirm such a hypothesis and detailed mechanism.
In conclusion, the present study revealed that KRAS
can be targeted by miR-193b in human esophageal squamous cell
carcinoma, and miR-193b negatively regulated the expression of KRAS
via binding to the 3′UTR of KRAS directly. miR-193b overexpression
increased the level of apoptosis and inhibited cell proliferation
ability as well as cell migration/invasion abilities in human
esophageal squamous cell carcinoma cell lines via suppression of
the expression of KRAS. Collectively, our results indicated that
miR-193b holds promise as a novel diagnosis marker in human
esophageal squamous cell carcinoma, as well as a target for gene
therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81570507, 81670474,
81302160).
Availability of data and materials
The analyzed datasets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
PL, SG and SZhang conceived and designed the
experiments. MK, SG, and YL performed the experiments. MK and SZhu
analyzed the data. MK and SZhu contributed the
reagents/materials/analysis tools. MK and SZhang wrote the paper.
PL and SZhang supervised the experiments. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Committee on the
Ethics of Animal Experiments and Human Subject Research of Capital
Medical University (Beijing, China). All of the volunteers who
donated cancer tissues and paracancerous tissues had provided
written informed consent. The Ethics Committees of Capital Medical
University had reviewed and approved this consent procedure.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A Review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wen SW, Zhang YF, Li Y, Liu ZX, Lv HL, Li
ZH, Xu YZ, Zhu YG and Tian ZQ: Characterization and effects of
miR-21 expression in esophageal cancer. Genet Mol Res.
14:8810–8818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gabra MM and Salmena L: microRNAs and
acute myeloid leukemia chemoresistance: A mechanistic overview.
Front Oncol. 7:2552017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng L, Zhan B, Luo P and Wang B:
miRNA375 regulates the cell survival and apoptosis of human
nonsmall cell carcinoma by targeting HER2. Mol Med Rep.
15:1387–1392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Poltronieri P: Editorial: Overview on
microRNAs in cancer development and virus infection. Microrna.
5:80–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tutar L, Tutar E and Tutar Y: MicroRNAs
and cancer; an overview. Curr Pharm Biotechnol. 15:430–437. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rauhala HE, Jalava SE, Isotalo J, Bracken
H, Lehmusvaara S, Tammela TL, Oja H and Visakorpi T: miR-193b is an
epigenetically regulated putative tumor suppressor in prostate
cancer. Int J Cancer. 127:1363–1372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen J, Feilotter HE, Paré GC, Zhang X,
Pemberton JG, Garady C, Lai D, Yang X and Tron VA: MicroRNA-193b
represses cell proliferation and regulates cyclin D1 in melanoma.
Am J Pathol. 176:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li XF, Yan PJ and Shao ZM: Downregulation
of miR-193b contributes to enhance urokinase-type plasminogen
activator (uPA) expression and tumor progression and invasion in
human breast cancer. Oncogene. 28:3937–3948. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hummel R, Sie C, Watson DI, Wang T, Ansar
A, Michael MZ, Van der Hoek M, Haier J and Hussey DJ: MicroRNA
signatures in chemotherapy resistant esophageal cancer cell lines.
World J Gastroenterol. 20:14904–14912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nyhan MJ, O'Donovan TR, Boersma AW, Wiemer
EA and McKenna SL: MiR-193b promotes autophagy and non-apoptotic
cell death in oesophageal cancer cells. BMC Cancer. 16:1012016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gupta B and Kumar N: Worldwide incidence,
mortality and time trends for cancer of the oesophagus. Eur J
Cancer Prev. 26:107–118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Petty RD, Dahle-Smith A, Stevenson DAJ,
Osborne A, Massie D, Clark C, Murray GI, Dutton SJ, Roberts C,
Chong IY, et al: Gefitinib and EGFR gene copy number aberrations in
esophageal cancer. J Clin Oncol. 35:2279–2287. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang R, Li Y, Chen Y, Liu X, Wang Z, Sun
H, Zheng Y, Ding Z, Lan L, Li M, et al: Clinical implications of
the concentration and EGFR/KRAS mutations of plasma cell free DNA
of patients with lung cancer and esophageal cancer. Zhonghua Yi Xue
Za Zhi. 95:3839–3842. 2015.(In Chinese). PubMed/NCBI
|
|
17
|
Liu P, Feng Y, Dong D, Liu X, Chen Y, Wang
Y and Zhou Y: Enhanced renoprotective efect of IGF-1 modifed human
umbilical cord-derived mesenchymal stem cells on gentamicin-induced
acute kidney injury. Sci Rep. 6:202872016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu P, Cai J, Dong D, Chen Y, Liu X, Wang
Y and Zhou Y: Effects of SOX2 on proliferation, migration and
adhesion of human dental pulp stem cells. PLoS One.
10:e01413462015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu P, Chen S, Li X, Qin L, Huang K, Wang
L, Huang W, Li S, Jia B, Zhong M, et al: Low immunogenicity of
neural progenitor cells differentiated from induced pluripotent
stem cells derived from less immunogenic somatic cells. PLoS One.
8:e696172013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Wu T, Song J, Chen X, Zhang Y and
Wan Y: A mutant screening method by critical annealing
temperature-PCR for site-directed mutagenesis. BMC Biotechnol.
13:212013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu P, Zhang Y, Chen S, Cai J and Pei D:
Application of iPS cells in dental bioengineering and beyond. Stem
Cell Rev. 10:663–670. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao Q, Pei Y, Zhang X and Xie B:
microRNA-96 acts as a tumor suppressor gene in human osteosarcoma
via target regulation of EZRIN. Life Sci. 203:1–11. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen K, Liu MX, Mak CS, Yung MM, Leung TH,
Xu D, Ngu SF, Chan KK, Yang H, Ngan HY and Chan DW:
Methylation-associated silencing of miR-193a-3p promotes ovarian
cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways.
Theranostics. 8:423–436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin X, Sun Y, Yang H, Li J, Yu S, Chang X,
Lu Z and Chen J: Deregulation of the MiR-193b-KRAS axis contributes
to impaired cell growth in pancreatic cancer. PLoS One.
10:e01255152015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gastaldi C, Bertero T, Xu N, Lebrigand K,
Fourre S, Popa A, Cardot-Leccia N, Meneguzzi G, Sonkoly E, et al:
miR-193b/365a cluster controls progression of epidermal squamous
cell carcinoma. Carcinogenesis. 35:1110–1120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Kong F, Wu K, Song K, He J and Sun
W: miR-193b directly targets STMN1 and uPA genes and suppresses
tumor growth and metastasis in pancreatic cancer. Mol Med Rep.
10:2613–2620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lü L, Liu T, Gao J, Zeng H, Chen J, Gu X
and Mei Z: Aberrant methylation of microRNA-193b in human Barrett's
esophagus and esophageal adenocarcinoma. Mol Med Rep. 14:283–288.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mitra AK, Chiang CY, Tiwari P, Tomar S,
Watters KM, Peter ME and Lengyel E: Microenvironment-induced
downregulation of miR-193b drives ovarian cancer metastasis.
Oncogene. 34:5923–5932. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moore C, Parrish JK and Jedlicka P:
MiR-193b, downregulated in Ewing Sarcoma, targets the ErbB4
oncogene to inhibit anchorage-independent growth. PLoS One.
12:e01780282017. View Article : Google Scholar : PubMed/NCBI
|