Introduction
Lung adenocarcinoma (LAC) is a serious disease, and
accounts for ~40% of all lung cancer cases (1,2).
Researches revealed that LAC is a leading cause of
cancer-associated mortalities, particularly in developing countries
(1,3). In China, there are >1 million
patients with LAC and >200,000 mortalities associated with lung
cancer annually (4). Notably, LAC is
consistently identified among non-smokers (5). Despite recent advances in diagnosis,
surgery, chemotherapy, targeted therapy, radiation therapy,
cellular immunotherapy and radiofrequency ablation, the overall
survival rate of patients with LAC remains low at an advanced
stage, with a 5-year survival rate of only ~18% (6–8).
Additionally, these patients also suffer complications, including
postoperative metastasis and side effects from drug treatments
(9). Thus, the identification of
novel diagnostic methods for patients with LAC will help monitor
tumor progression and guide clinical treatment, which may assist in
the development of gene target-based therapy.
microRNAs (miRNAs) are small non-coding RNAs ~20–22
nt in length that serve vital roles in diseases, which threaten
human health by coding specific mRNAs (10,11). In
the past two decades, increasing evidence indicated that miRNAs are
involved in the pathogenesis of LAC (12,13).
Berrout et al (14)
demonstrated that miRNA-142-3p functions as a regulatory oncogenic
driver by binding transient receptor potential cation channel
subfamily A member 1-fibroblast growth factor receptor 2 in LAC.
Yan et al (15) reported that
miR-503 modulated epithelial-mesenchymal transition in
silica-induced pulmonary fibrosis by targeting phosphoinositide
3-kinase p85. Additionally, the study of Pan et al (16) indicated that miR-944 served a tumor
suppressive role via the metastasis associated in colon cancer 1
(MACC1)/Met/AKT signaling pathway in gastric cancer (16). The aforementioned studies
demonstrated that miRNAs serve crucial roles in LAC and other
diseases. However, the role of miR-944 in LAC requires further
investigation.
Signal transducer and activator of transcription
(STAT)-1, a member of the STAT super family, has a number of
biological functions, including acting as a tumor suppressor and
preventing tumor development, and also exhibits a role in
immunotherapy (17–19). A previous study reported that STAT1
served vital roles in the miR-15A and miR-16-1 signaling pathways
in the regulation of colorectal tumors (20). Additionally, Zhang et al
(21) reported that miR-181a/STAT1
inhibited colorectal cancer cell proliferation by regulating the
phosphatase and tensin homolog/AKT signaling pathway (22). However, to the best of our knowledge,
at present there is has been no report investigating whether
miR-944 has a role in LAC. Collectively, the present study aimed to
investigate the effects of miR-944 on cell proliferation and
apoptosis in LAC.
Materials and methods
Tissue collection
A total of 25 LAC tissues from 13 males and 12
females, with a median age of 57.6 years, were obtained from
patients who underwent surgery at the Third Hospital of Qiqihar
Medical College (Qiqihar, China), between September 2014 and
September 2016. The present study was approved by the Research
Ethics Committee of Third Hospital of Qiqihar Medical College, and
written informed consent was obtained from all patients. All the
specimens, including cancer tissues, were diagnosed with LAC
(stages I, II, and III) (23). The
patients received no local or systemic treatments prior to surgery
(Table I). All collected tissues
were placed in liquid nitrogen immediately and stored at −80°C
until required.
 | Table I.Patient clinical information. |
Table I.
Patient clinical information.
| Variables | Patients
(n=25) |
|---|
| Age (years) | 57.6 (range 30–80
years) |
| Sex |
|
|
Male | 13 |
|
Female | 12 |
| Tumor size |
|
| ≤5
cm | 16 |
| >5
cm | 9 |
| TNM stage |
|
|
I/II | 15 |
|
III/IV | 10 |
| Lymph node
metastasis |
|
|
Yes | 18 |
| No | 7 |
Cell culture
LAC cells (A549, H1299, SK-Lu-1, and PC-9) were
obtained from the Chinese Academy of Science Shanghai Cell Bank
(Shanghai, China) and cultured in Dulbecco's modified Eagle's
medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% heat-inactivated fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml
penicillin, and 100 g/ml streptomycin (Beyotime Institute of
Biotechnology, Shanghai, China). Human bronchial epithelial cells
(16HBE), obtained from the Henan Xingfa Bio-Technology Co., Ltd.
(Henan, China), were maintained in RPMI-1640 (HyClone; GE
Healthcare Life Sciences) supplemented with 10% FBS, 100 µg/ml
penicillin, and 100 µg/ml streptomycin. These cells were placed in
a humidified atmosphere containing 5% CO2 at 37°C.
Gene silencing of STAT1
STAT1-small interfering (si)RNA was purchased
from Shanghai GeneChem Co., Ltd. (Shanghai, China). The sequence
was as follows:
5′-CCGGCTGGAAGATTTACAAGATGAACTCGAGTTCATCTTGTAAATCTTCCAGTTTTTG-3′. A
control siRNA
(5′-CCGGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTG-3′)
was used as a negative control. Cells were transfected with 80 pmol
siRNA plasmid for 48 h using Lipofectamine™ 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h following
transfection A549 cells were harvested for subsequent
experimentation.
Colony formation assay
A total of 8×102 A549 or H1229 cells were
plated in triplicate into 60 mm dishes and cultured for 14 days in
a humidified atmosphere containing 5% CO2 at 37°C.
Following 14 days, colonies were stained with 0.1% crystal violet
in 20% methanol for 15 min. Colonies consisting of >50 cells
were counted as a single colony.
Dual luciferase reporter assay
Wild-type (WT) miR-944 (miR-944-WT), mutant miR-944
(miR-944-Mut), STAT1-WT, and STAT1-Mut were cloned into separate
pMIR-REPORT Luciferase vectors (Ambion; Thermo Fisher Scientific,
Inc.). A total of 8×103 A549 cells/well were seeded in
6-well plates and transfected with the specific vectors using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h as described previously. Luciferase
activity was assessed using the Dual Luciferase-reporter 1000 assay
system (Promega Corporation, Madison, WI, USA). Renilla
activity was used for normalization.
Kaplan-Meier method
All clinical data and the Tier 3 RNASeqV2 mRNA
expression data were downloaded from https://tcga-data.nci.nih.gov/tcga/. Patients with a
follow-up time or time to mortality >0 days were included in the
analysis. For each gene, all samples were divided to two groups
based on the median expression values, with samples with a median
value placed in the high expression group. Kaplan-Meier analysis
was then performed to examine the significance between the two
groups. Cox proportional hazards regression was also performed with
the coxph function from the R survival library (version 2.43–3;
http://cran.r-project.org/web/packages/survival/index.html).
Hazard ratios with 95% confidence intervals were obtained.
Cell Counting Kit (CCK)-8 assay
CCK-8 assays (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) were used to determine cell viability following
transfection with 50 pmol miR-944 mimic and miR-negative control
(NC) in A549 and H1299 cells, according to the manufacturer's
protocol and the aforementioned protocol. LAC cells were seeded
into 96-well plates at a density of 2×103 cells/well and
cultured for 48 h in a humidified atmosphere containing 5%
CO2 at 37°C. The sequences of miR-944 mimic and NC were
as follows: miR-944 NC, 5′-ACUUCAGUGGAUGUUUGCAGC-3′; and miR-944
mimic, 5′-GAGUAGGCUAAUGUUAUAAA-3′.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from A549 cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was
synthesized using High-Capacity cDNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers
were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
qPCR was performed using Power SYBR Green PCR master mix (Thermo
Fisher Scientific, Inc.) for 35 cycles at 95°C for 30 sec, 60°C for
30 sec and 72°C for 35 sec. Gene expression levels were normalized
with β-actin and analyzed using the 2−ΔΔCq method
(20). The primer sequences were as
follows: STAT1 forward, 5′-AGCCAGTGCAAATCACGATG-3′ and reverse,
5′-CGTCAGCAAAGCCCATTTGA-3′; β-actin forward,
5′-TGTCACCAACTGGGACGATA-3′ and reverse,
5′-GGGGTGTTGAAGGTCTCAAA-3′.
Western blot analysis
All proteins were obtained from LAC cells and
patients. A549 cells and patient tissues were lysed with
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology). The proteins were then quantified using a
bicinchoninic acid assay (Beyotime Institute of Biotechnology). The
proteins (60–80 µg) were separated by 10% SDS-PAGE and transferred
to a nitrocellulose membrane. Following blocking with 5% non-fat
milk for 2 h at room temperature, the blots were probed with
primary antibodies against STAT1 (catalog no. ab30645; 1:500;
Abcam, Cambridge, MA, USA) and β-actin (catalog no. 4970; 1:500;
Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C
overnight. Following threes washes with PBS and Tween-20 for 15
min, the membranes were incubated with rabbit (catalog no.
926-32211-00; 1:10,000) or mouse (catalog no. 926-32211-01;
1:10,000) secondary antibodies (LI-COR Biosciences, Lincoln, NE,
USA) at room temperature in the dark for 1 h. The blots were then
visualized using an Infrared Imaging System (LI-COR Biosciences)
and the band density was quantified using Odyssey 3.0 software
(LI-COR Biosciences) (22). Using
β-actin as an internal control, the blots were subjected to
densitometry.
Tumor xenograft nude mouse model
Transfected A549 cells were subcutaneously injected
into the back of total of 30 male BALB/c nude mice, aged 6 weeks
old with a median weight of ~20 g (Vital River Laboratory Animal
Technology, Beijing, China). The mice were kept at 21–24°C in a
light/dark cycle with food and water available ad libitum.
Tumor volumes were measured once each week at equal intervals. A
tumor growth curve was constructed to determine the effects of the
miR-944-mimic and miR-NC on tumor growth. At 21 days following
injection, the mice were sacrificed with carbon dioxide at a
displacement rate of 20%/min and imaged, and the tumors were
dissected. The tumors in each group were harvested and weighed.
Total proteins and RNAs were extracted for western blotting and
RT-qPCR. All animal experiments were approved by the Third Hospital
of Qiqihar Medical College's Animal Care and Use Committee and
conducted according to the National Institutes of Health guidelines
(23).
Statistical analysis
Statistical analysis was conducted using SPSS
software (version 13.0; SPSS Inc., Chicago, IL, USA). All data were
expressed as means ± standard deviation. Statistical analysis was
performed using Student's non-paired t-test or one-way analysis of
variance followed by Tukey's post-test. miR-944 targets were
predicted using Targetscan software 7.2 (24). P<0.05 was considered to indicate a
statistically significant difference.
Results
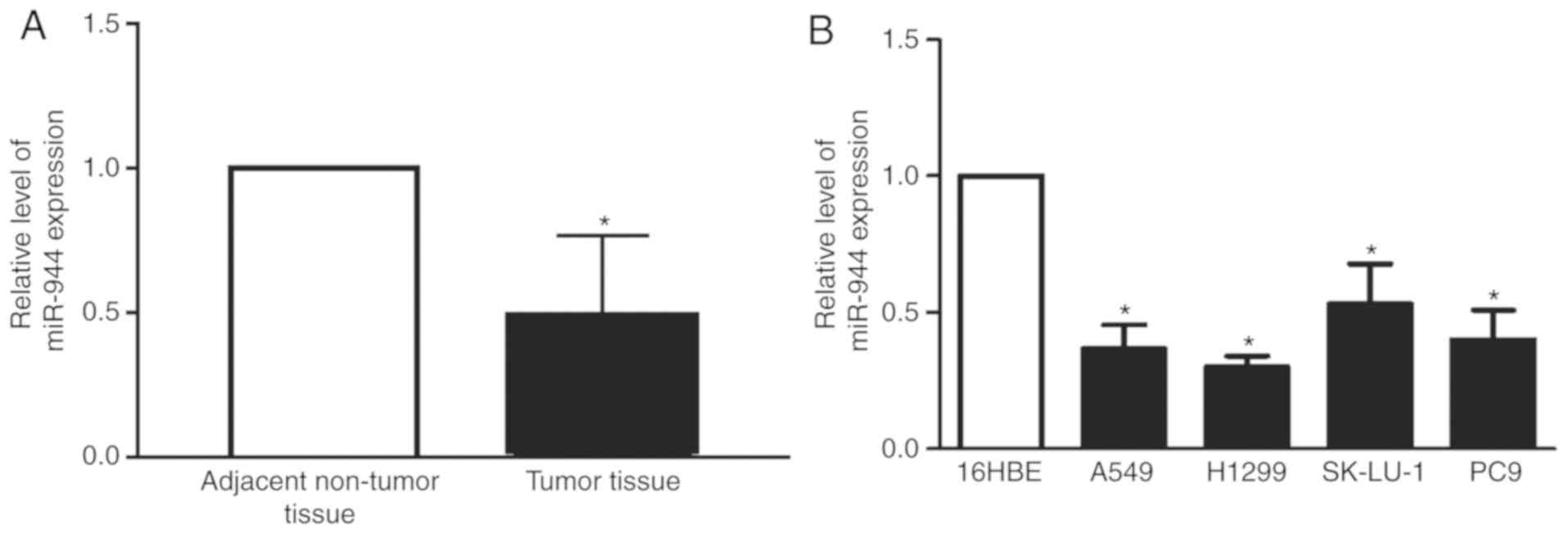
miR-944 is underexpressed in LAC
tissues and cell lines
To determine the biological function of miR-944 in
patients with LAC and cell lines, the present study initially
evaluated the expression levels of miR-944 in 25 pairs of LAC
tissues. The expression of miR-944 was significantly reduced in the
LAC group compared with the normal group (Fig. 1A). Additionally, the expression of
miR-944 in the four lung cancer cell lines, A549, H1299, SK-Lu-1
and PC-9, was significantly decreased, compared with normal 16HBE
lung cells (Fig. 1B). However, the
expression of miR-944 was reduced in A549 and H1299 cell lines,
compared with SK-Lu-1 and PC9 cell lines. Therefore, for subsequent
experiments the A549 and H1299 cell lines were selected. This data
demonstrated that miR-944 functions as a tumor suppressor gene in
LAC tissues and cell lines.
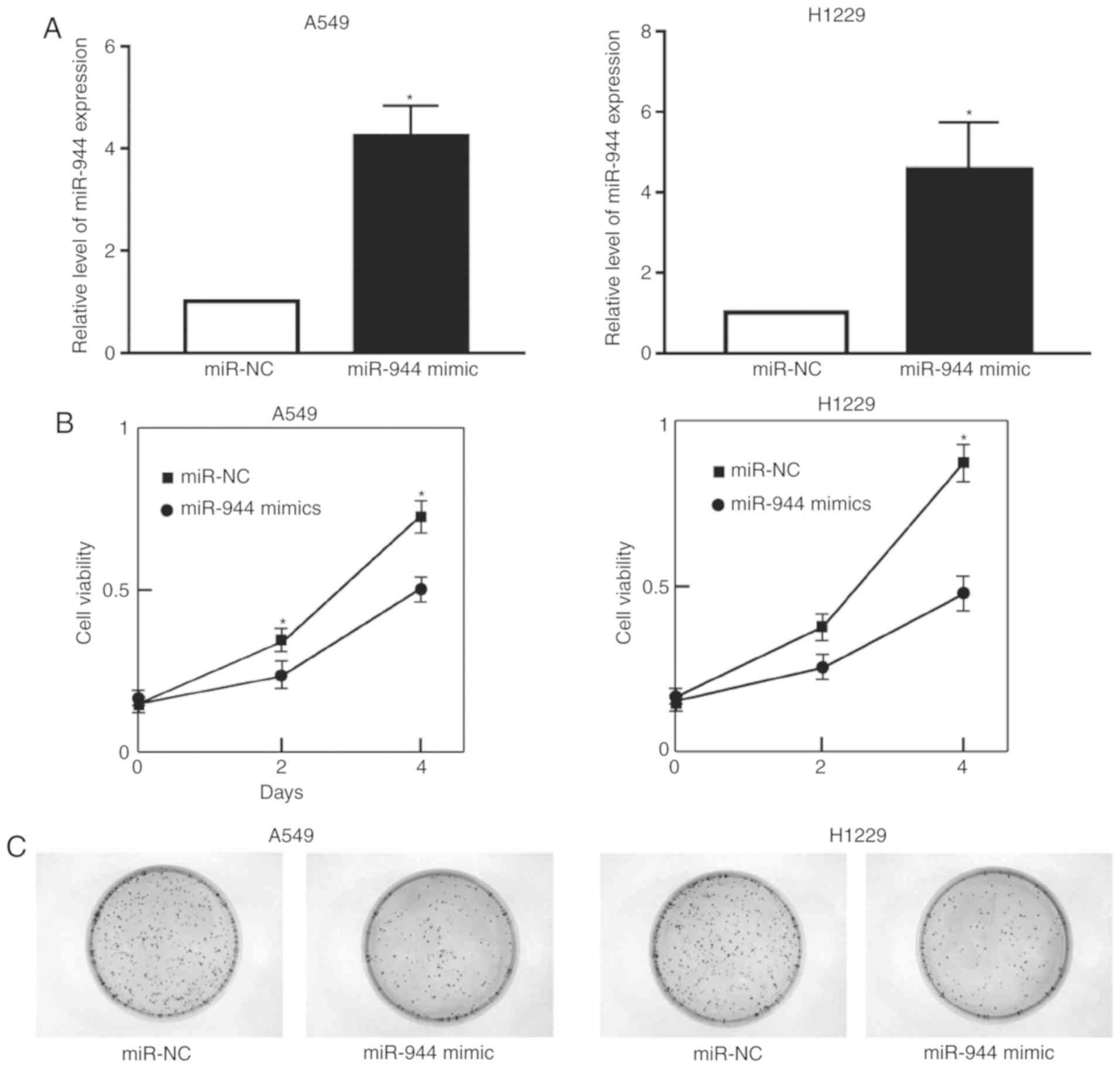
miR-944 inhibits cell
proliferation
To improve the understanding of the effect of
miR-944 on LAC cell proliferation, A549 and H1299 cells were
treated with the miR-944 mimic or miR-NC. The expression of miR-944
was significantly increased following treatment with the miR-944
mimic in A549 and H1299 cells (Fig.
2A). Cell growth was also observed to be attenuated in
miRNA-944 mimic cells, compared with miR-NC cells, as illustrated
by the CCK-8 and colony formation assays (Fig. 2B and C). Collectively, these results
indicate that miRNA-944 had a negative effect on the proliferation
of LAC cells.
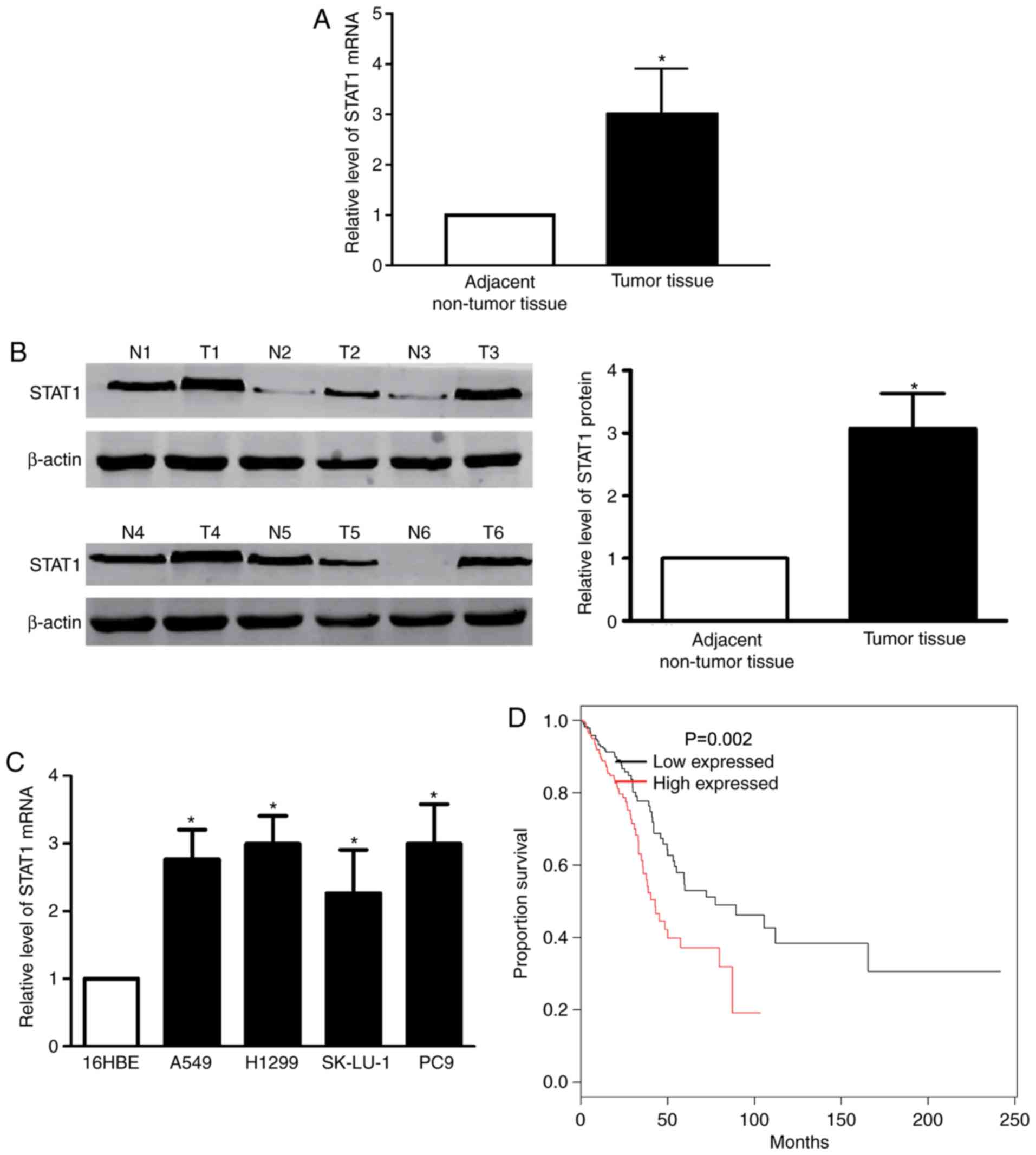
STAT1 is upregulated in LAC tissue and
cells
The RT-qPCR demonstrated that the STAT1 mRNA level
was significantly upregulated in LAC samples, compared with paired
non-malignant samples (Fig. 3A).
Western blotting confirmed the results of the STAT1 mRNA level
(Fig. 3B). Subsequently, the present
study determined the expression of STAT1 in the LAC cell lines
A549, H1299, SK-Lu-1 and PC-9. The data revealed that significantly
increased levels of STAT1 mRNA expression were identified in LAC
cell lines, compared with 16HBE cells (Fig. 3C). Using the Kaplan-Meier method, the
overall survival times in patients with high-STAT1 expression were
revealed to be significantly reduced, compared with patients with
negative and low-STAT1 expression (Fig.
3D).
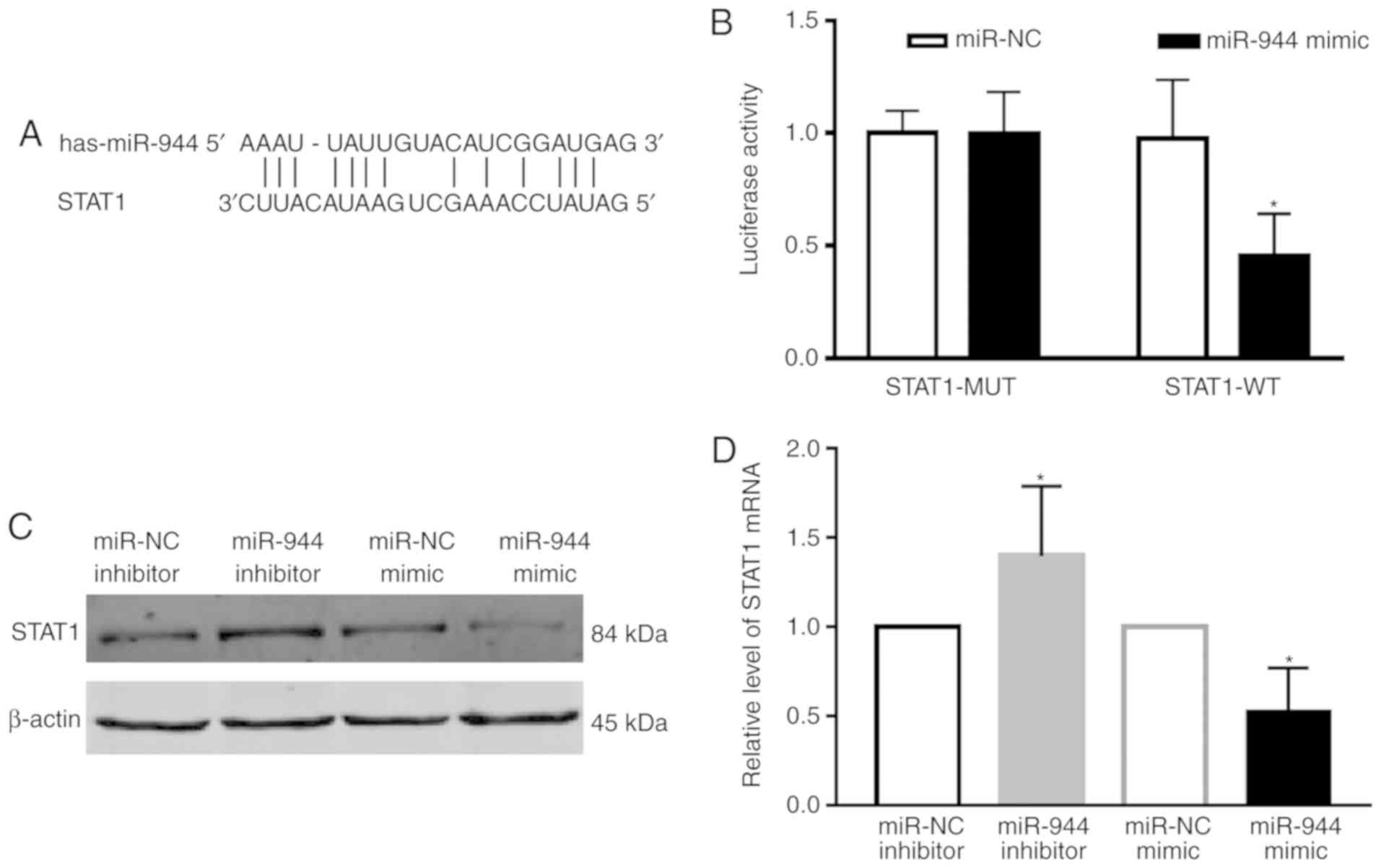
miR-944 directly downregulates the
expression of STAT1
To understand how miR-944 functions in LAC, the
Microrna search program (www.targetscn.org) was used to predict targets of
miR-944, which revealed that STAT1 is considered to be a putative
target of miR-944 (Fig. 4A). The
luciferase reporter assay also demonstrated that STAT1 was a direct
target gene of miR-944 (Fig. 4B).
The protein level of STAT1 following transfection with the miR-944
and miR-NC mimics or miR-944 inhibitor and miR-NC inhibitor was
then determined in A549 cells. The results demonstrated that the
protein level of STAT1 was downregulated by treatment with the
miR-944 mimic, and upregulated following treatment with the miR-944
inhibitor (Fig. 4C). These results
were also confirmed at the mRNA level (Fig. 4D). Collectively, these results
indicate that STAT1 was a direct target of miR-944 in LAC A549
cells.
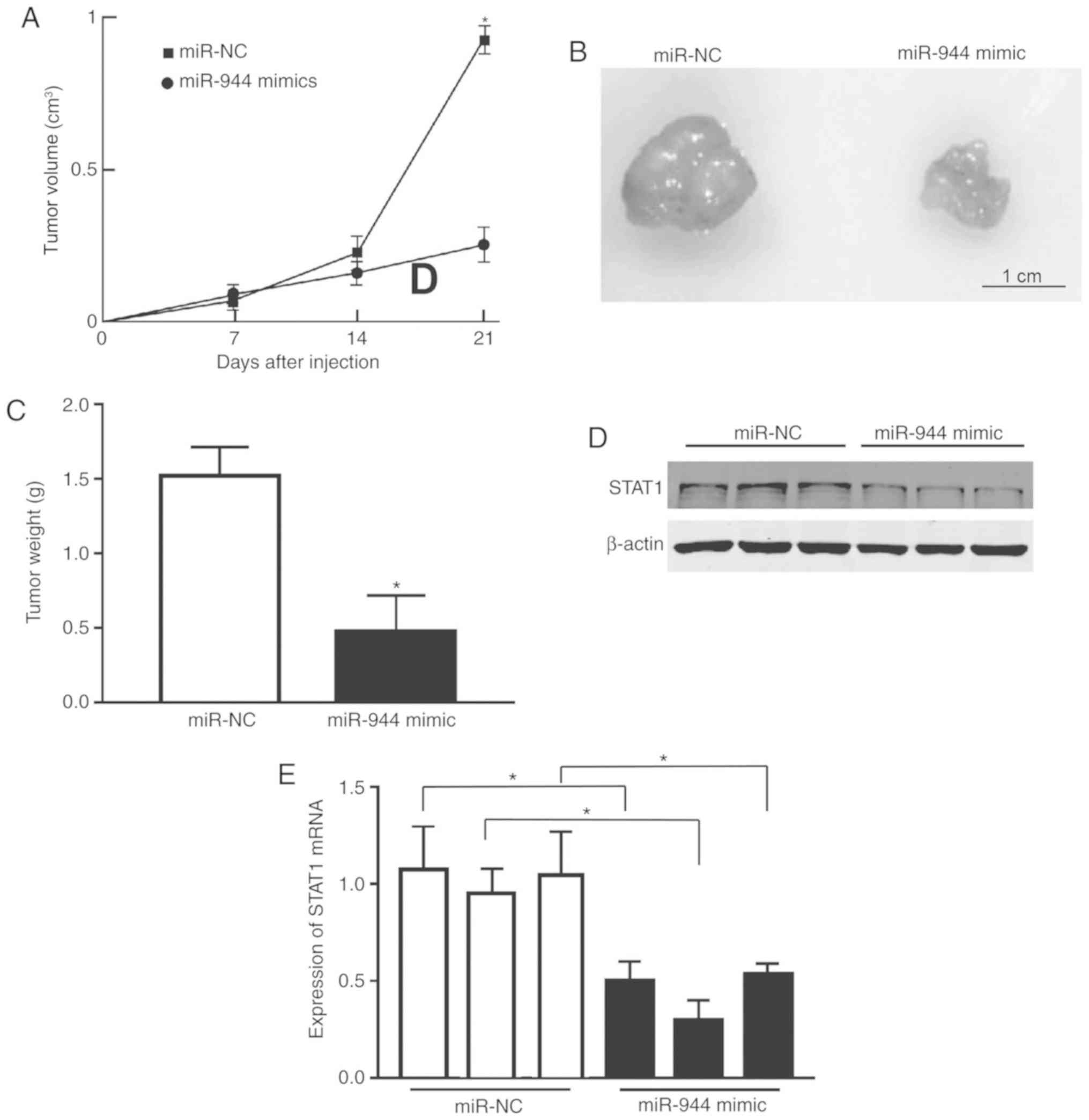
miR-944 inhibits tumor growth in
vivo
To examine the role of miR-944 in tumor
proliferation in vivo, a nude mouse xenograft model was
used. A549 cells were transfected with miR-NC or miR-944 mimics and
injected into mice. Tumor size was measured once each week, and the
growth curve as a function of the average tumor size was plotted
following the injection of cells. The mice were sacrificed after 2
weeks and their bodies and xenografts were weighed. As expected,
there was a significant decrease in tumor size and weight in the
miR-944-overexpressing groups compared with the NC group (Fig. 5A-C). Consistent with the in
vitro studies, the protein level of STAT1 in tumor tissues from
the miR-944 mimic group was markedly reduced, compared with the
miR-NC group, as illustrated by the immunoblotting assay (Fig. 5D). The RT-qPCR data on STAT1 mRNA
measurement also exhibited the same pattern (Fig. 5E). Collectively, these observations
indicate that miR-944 was a tumor suppressor in LAC.
Discussion
Over the past two decades, a number of miRNAs,
including LAC, have been demonstrated to exhibit functions in
numerous diseases, including circulatory system diseases,
cerebrovascular diseases and cancer (25–27).
Recently, increasing evidence has implicated STAT1 in a number of
processes associated with cancer, including cell cycle regulation,
motility, differentiation and proliferation (18,20,28). The
present study identified that miR-944 is downregulated in LAC
tissues and cells, indicating a potential role for miR-944 in LAC.
STAT1 is a direct target gene of miR-944, and overexpression of
miR-944 significantly inhibited LAC cell proliferation, survival
and tumor growth.
miRNAs are small non-coding RNAs that regulate 30%
of gene expressions (29,30). miRNAs act as master regulators of
gene expression in numerous important biological pathways,
including cell cycle, apoptosis and proliferation (31), particularly in cancer. miRNAs can act
as oncogenes or tumor suppressors and regulate their target genes,
which are dysregulated in a numbr of cancer types, including
prostate cancer, colon cancer and gastrointestinal cancer (32–34).
Recent studies have demonstrated that miR-944 acted as a tumor
suppressor in numerous cancer types, for example Wen et al
(35) reported that miR-944 inhibits
cell migration and invasion by targeting MACC1 in colorectal
cancer. Additionally, He et al (36) demonstrated that miR-944 acts as a
prognostic marker and promotes tumor progression in endometrial
cancer. However, there is currently no data regarding the role of
miR-944 in LAC proliferation. In the present study, miR-944
expression was identified to be downregulated in LAC tissue
samples, compared with normal tissues. These results indicate that
miR-944 may exhibit an anticancer effect of LAC. Additionally, the
overexpression of miR-944 significantly inhibits LAC cell
proliferation and tumor growth, which demonstrated that miR-1994
has a notable anti-proliferation effect in vivo and
vitro.
STAT1 belongs to the STAT super family and has
numerous functions, including reducing apoptosis, attenuating
inflammation and modulating oxidative stress (37,38).
Carbotti et al (39) reported
that interleukin-27 triggered STAT1 phosphorylation in small cell
lung cancer cells. Furthermore, Zhang et al (40) discussed the role of STAT1 in cancer.
Recently, evidence indicated that STAT1 can be regulated by miRNAs.
For example, Xi et al (41)
reported that miR-21 depletion in macrophages promotes tumoricidal
polarization and enhances PD-1 immunotherapy by STAT1 (42). Additionally, Li et al
(43) indicated that miR-194
promotes osteoblast differentiation via downregulating STAT1. In
the present study, STAT1 was identified as a direct target gene of
miR-944. To further investigate the role of STAT1 in LAC, the
expression of STAT1 in patients with LAC and LAC cell lines was
examined. The results demonstrated that the expression of STAT1 is
upregulated in LAC tissues. The study by Gujam et al
(44) demonstrated that STAT1 and
STAT3 regulate tumor microenvironment and survival in patients with
invasive ductal breast cancer. In the present study, the
overexpression of STAT1 was observed to decrease the 5-year
survival rate of patients with LAC, these data demonstrated that
STAT1 functions as an oncogene in LAC. Additionally, downregulated
miR-944 can upregulate STAT1 protein and mRNA expression levels,
furthermore, overexpression of miR-944 can increase the expression
of STAT1. These results demonstrated that the STAT1 oncogene was
validated experimentally as the novel target of miR-944.
Collectively, the present study demonstrated that miR-944
significantly suppressed LAC growth through inhibition of STAT1
translation.
In summary, the present study demonstrated that
miR-944/STAT1 is a novel constituent of LAC tumorigenesis and
proliferation, and miR-944 regulates cell growth in LAC by
targeting STAT1. This data contributes towards the improved
understanding of LAC and indicates that miR-944 may be used as a
clinical agent in the treatment of LAC. Although miR-944/STAT1 is
not the only signaling pathway to regulate LAC cell proliferation
and migration, it may provide the foundation for the development of
novel methods for the diagnosis and therapy of LAC.
Acknowledgements
Not applicable.
Funding
This work was supported by the Qiqihar Science and
Technology Bureau project (grant no. SFZD-2017001).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request,
while preserving the necessary confidentiality and anonymity.
Authors' contributions
BM conceived and designed the study and critically
revised the manuscript. JCA and HBS designed and performed the
experiments, analyzed the data and wrote the manuscript. WBH and KZ
were involved in drafting the manuscript and revising it critically
for important intellectual content, gave advice on the experiments,
designed and performed the experiments, analyzed the data, and
contributed with reagents and technical assistance. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Third Hospital of Qiqihar Medical College
(Qiqihar, China), and written informed consent was obtained from
all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang TW, Lin KF, Lee CH, Chang H, Lee SC
and Shieh YS: The role of thyroid transcription factor-1 and tumor
differentiation in resected lung adenocarcinoma. Sci Rep.
7:142222017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vieira A and Ugalde Figueroa P: Anatomic
bisegmentectomy for synchronous lung adenocarcinoma. J Vis Surg.
3:642017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang T, Chen L, Zhang S, Xu Y, Fan Y and
Zhang L: Effects of high-intensity focused ultrasound on
cisplatin-resistant human lung adenocarcinoma in vitro and in vivo.
Acta Biochim Biophys Sin (Shanghai). 49:1092–1098. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu H, Meng S, Xu Q, Wang X, Wang J, Gong
R, Song Y, Duan Y and Zhang Y: Gene expression profiling of lung
adenocarcinoma in Xuanwei, China. Eur J Cancer Prev. 25:508–517.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ni J, Zhou LL, Ding L, Zhao X, Cao H, Fan
F, Li H, Lou R, Du Y, Dong S, et al: PPARγ agonist efatutazone and
gefitinib synergistically inhibit the proliferation of
EGFR-TKI-resistant lung adenocarcinoma cells via the PPARγ/PTEN/Akt
pathway. Exp Cell Res. 361:246–256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bernhardt D, Adeberg S, Bozorgmehr F,
Opfermann N, Hörner-Rieber J, König L, Kappes J, Thomas M,
Unterberg A, Herth F, et al: Outcome and prognostic factors in
single brain metastases from small-cell lung cancer. Strahlenther
Onkol. 194:98–106. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim CS and Jeter MD: Radiation therapy,
early stage non-small cell lung cancer. StatPearls. Treasure.
(Island (FL)). 2018.
|
|
8
|
Li-Ming X, Zhao LJ, Simone CB II, Cheng C,
Kang M, Wang X, Gong LL, Pang QS, Wang J, Yuan ZY and Wang P:
Receipt of thoracic radiation therapy and radiotherapy dose are
correlated with outcomes in a retrospective study of three hundred
and six patients with extensive stage small-cell lung cancer.
Radiother Oncol. 125:331–337. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peng Y, Ren W, Wang H, Li M, Feng Z and
Peng Z: Surgical treatment is an effective approach for patients
with synchronous multiple primary lung cancers. J Cancer Res Ther.
13:702–706. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nhat Tran, Alipourfard B, Abhyankar V,
Nguyen K, Weidanz J and Gao J: Improved microRNA biomarkers for
pathological stages in lung adenocarcinoma via clustering of
dysregulated microRNA-target associations. Conf Proc IEEE Eng Med
Biol Soc. 2017:2708–2711. 2017.PubMed/NCBI
|
|
11
|
Chang SM and Hu WW: Long non-coding RNA
MALAT1 promotes oral squamous cell carcinoma development via
microRNA-125b/STAT3 axis. J Cell Physiol. 233:3384–3396. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bian T, Jiang D, Liu J, Yuan X, Feng J, Li
Q, Zhang Q, Li X, Liu Y and Zhang J: miR-1236-3p suppresses the
migration and invasion by targeting KLF8 in lung adenocarcinoma
A549 cells. Biochem Biophys Res Commun. 492:461–467. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sui J, Yang RS, Xu SY, Zhang YQ, Li CY,
Yang S, Yin LH, Pu YP and Liang GY: Comprehensive analysis of
aberrantly expressed microRNA profiles reveals potential biomarkers
of human lung adenocarcinoma progression. Oncol Rep. 38:2453–2463.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Berrout J, Kyriakopoulou E, Moparthi L,
Hogea AS, Berrout L, Ivan C, Lorger M, Boyle J, Peers C, Muench S,
et al: TRPA1-FGFR2 binding event is a regulatory oncogenic driver
modulated by miRNA-142-3p. Nat Commun. 8:9472017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan W, Wu Q, Yao W, Li Y, Liu Y, Yuan J,
Han R, Yang J, Ji X and Ni C: MiR-503 modulates
epithelial-mesenchymal transition in silica-induced pulmonary
fibrosis by targeting PI3K p85 and is sponged by lncRNA MALAT1. Sci
Rep. 7:113132017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan T, Chen W, Yuan X, Shen J, Qin C and
Wang L: miR-944 inhibits metastasis of gastric cancer by preventing
the epithelial-mesenchymal transition via MACC1/Met/AKT signaling.
FEBS Open Bio. 7:905–914. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang J, Zheng C and Shao J, Chen L, Liu X
and Shao J: Overexpression of eEF1A1 regulates G1-phase progression
to promote HCC proliferation through the STAT1-cyclin D1 pathway.
Biochem Biophys Res Commun. 494:542–549. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hiller J, Hagl B, Effner R, Puel A,
Schaller M, Mascher B, Eyerich S, Eyerich K, Jansson AF, Ring J, et
al: STAT1 gain-of-function and dominant negative STAT3 mutations
impair IL-17 and IL-22 immunity associated with CMC. J Invest
Dermatol. 138:711–714. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Collins-McMillen D, Stevenson EV, Kim JH,
Lee BJ, Cieply SJ, Nogalski MT, Chan GC, Frost RW III, Spohn CR and
Yurochko AD: HCMV utilizes a non-traditional STAT1 activation
cascade via signaling through EGFR and integrins to efficiently
promote the motility, differentiation, and polarization of infected
monocytes. J Virol pii. JVI.00622-17. 2017. View Article : Google Scholar
|
|
20
|
Liu R, Lu Z, Gu J, Liu J, Huang E, Liu X,
Wang L, Yang J, Deng Y, Qian J, et al: MicroRNAs 15A and 16-1
activate signaling pathways that mediate chemotaxis of immune
regulatory B cells to colorectal tumors. Gastroenterology.
154:637–651.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Li X, Tan F, Yu N and Pei H:
STAT1 inhibits MiR-181a expression to suppress colorectal cancer
cell proliferation through PTEN/Akt. J Cell Biochem. 118:3435–3443.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang M, Du Y, Lu R, Shu Y, Zhao W, Li Z,
Zhang Y, Liu R, Yang T, Luo S, et al: Cholesterol retards
senescence in bone marrow mesenchymal stem cells by modulating
autophagy and ROS/p53/p21Cip1/Waf1 pathway. Oxid Med
Cell Longev. 2016:75243082016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Daugherty A, Hegele RA, Mackman N, Rader
DJ, Schmidt AM and Weber C: Complying with the national institutes
of health guidelines and principles for rigor and reproducibility:
Refutations. Arterioscler Thromb Vasc Biol. 36:1303–1304. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li M, Liang Z, He S, Zeng Y, Jing Y, Fang
W, Wu K, Wang G, Ning X, Wang L, et al: Genome-wide identification
of leaf abscission associated microRNAs in sugarcane (Saccharum
officinarum L.). BMC Genomics. 18:7542017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao S, Gao X, Zang S, Li Y, Feng X and
Yuan X: MicroRNA-383-5p acts as a prognostic marker and inhibitor
of cell proliferation in lung adenocarcinoma by cancerous inhibitor
of protein phosphatase 2A. Oncol Lett. 14:3573–3579. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhuang L, Shou T, Li K, Gao CL, Duan LC,
Fang LZ, Zhang QY, Chen ZN, Zhang C, Yang ST and Li GF:
MicroRNA-30e-5p promotes cell growth by targeting PTPN13 and
indicates poor survival and recurrence in lung adenocarcinoma. J
Cell Mol Med. 21:2852–2862. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Chen Y, Yun H, Liu Z, Su M and
Lai R: STAT1β enhances STAT1 function by protecting STAT1α from
degradation in esophageal squamous cell carcinoma. Cell Death Dis.
8:e30772017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu Y, Wang L, Gu J, Qu K and Wang Y:
Identification of microRNA differentially expressed in three
subtypes of non-small cell lung cancer and in silico functional
analysis. Oncotarget. 8:74554–74566. 2017.PubMed/NCBI
|
|
30
|
Li W, Yang W, Liu Y, Chen S, Chin S, Qi X,
Zhao Y, Liu H, Wang J, Mei X, et al: MicroRNA-378 enhances
inhibitory effect of curcumin on glioblastoma. Oncotarget.
8:73938–73946. 2017.PubMed/NCBI
|
|
31
|
Zhang QL, Zhu QH, Zhang F, Xu B, Wang XQ
and Chen JY: Transcriptome-wide analysis of immune-responsive
microRNAs against poly (I:C) challenge in Branchiostoma belcheri by
deep sequencing and bioinformatics. Oncotarget. 8:73590–73602.
2017.PubMed/NCBI
|
|
32
|
Yang Y, Ji C, Guo S, Su X, Zhao X, Zhang
S, Liu G, Qiu X, Zhang Q, Guo H and Chen H: The miR-486-5p plays a
causative role in prostate cancer through negative regulation of
multiple tumor suppressor pathways. Oncotarget. 8:72835–72846.
2017.PubMed/NCBI
|
|
33
|
Yan S, Dang G, Zhang X, Jin C, Qin L, Wang
Y, Shi M, Huang H and Duan Q: Downregulation of circulating
exosomal miR-638 predicts poor prognosis in colon cancer patients.
Oncotarget. 8:72220–72226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fawzy MS, Toraih EA, Ibrahiem A,
Abdeldayem H, Mohamed AO and Abdel-Daim MM: Evaluation of
miRNA-196a2 and apoptosis-related target genes: ANXA1, DFFA and
PDCD4 expression in gastrointestinal cancer patients: A pilot
study. PLoS One. 12:e01873102017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wen L, Li Y, Jiang Z, Zhang Y, Yang B and
Han F: miR-944 inhibits cell migration and invasion by targeting
MACC1 in colorectal cancer. Oncol Rep. 37:3415–3422. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He Z, Xu H, Meng Y and Kuang Y: miR-944
acts as a prognostic marker and promotes the tumor progression in
endometrial cancer. Biomed Pharmacother. 88:902–910. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu Y, Zhang J, Sun X, Su Q and You C:
Down-regulation of miR-29b in carcinoma associated fibroblasts
promotes cell growth and metastasis of breast cancer. Oncotarget.
8:39559–39570. 2017.PubMed/NCBI
|
|
38
|
Borsini A, Cattaneo A, Malpighi C, Thuret
S, Harrison NA; MRC ImmunoPsychiatry Consortium, ; Zunszain PA and
Pariante CM: Interferon-alpha reduces human hippocampal
neurogenesis and increases apoptosis via activation of distinct
STAT1-dependent mechanisms. Int J Neuropsychopharmacol. 21:187–200.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Carbotti G, Nikpoor AR, Vacca P, Gangemi
R, Giordano C, Campelli F, Ferrini S and Fabbi M: IL-27 mediates
HLA class I up-regulation, which can be inhibited by the IL-6
pathway, in HLA-deficient small cell lung cancer cells. J Exp Clin
Cancer Res. 36:1402017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y and Liu Z: STAT1 in cancer: Friend
or foe? Discov Med. 24:19–29. 2017.PubMed/NCBI
|
|
41
|
Xi J, Huang Q, Wang L, Ma X, Deng Q, Kumar
M, Zhou Z, Li L, Zeng Z, Young KH, et al: miR-21 depletion in
macrophages promotes tumoricidal polarization and enhances PD-1
immunotherapy. Oncogene. 37:3151–3165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li J, He X, Wei W and Zhou X: MicroRNA-194
promotes osteoblast differentiation via downregulating STAT1.
Biochem Biophys Res Commun. 460:482–488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y,
Pan X, Zhang X, Zhou L, Yu D, et al: FLI1 exonic circular RNAs as a
novel oncogenic driver to promote tumor metastasis in small cell
lung cancer. Clin Cancer Res. Nov 14–2018.(Epub ahead of
print).
|
|
44
|
Gujam FJ, McMillan DC and Edwards J: The
relationship between total and phosphorylated STAT1 and STAT3
tumour cell expression, components of tumour microenvironment and
survival in patients with invasive ductal breast cancer.
Oncotarget. 7:77607–77621. 2016. View Article : Google Scholar : PubMed/NCBI
|