Introduction
Cutaneous melanoma has a high incidence and it is
responsible for most skin cancer deaths in humans, the main risk
factor being exposure to ultraviolet radiation. According to World
Health Organization, 132,000 melanoma skin cancers occur globally
each year. Cutaneous melanoma is the most aggressive type of skin
cancer, with a high resistance to classical therapies as
chemotherapy and radiotherapy (1).
Melanoma is usually highly immunogenic and spontaneous remissions
have been observed (2,3).
The immune system plays a major role in regulating
tumor cell proliferation by initiating defence responses against
tumor aggression. In recent years, there has been increasing
interest in understanding the role of the immune system in tumor
development and progression (4–6). In
melanoma, skin's immune system and tumor cells are interconnected
from the very beginning of the tumorigenesis process, including
initiation, progression, tumor invasion and metastasis. The
cellular components of the skin immune system, in particular
regulatory T cells, NK and dendritic cells, are the main components
of the immunosuppressive network. The failure of antitumor immune
response stems from alterations of local immune suppressor cells
and factors. In this complex microenvironment, interactions of
melanocytes with these factors can lead to malignant transformation
(7).
Recent studies reflect the concern to identify
immune markers by minimally invasive methods to monitor and guide
the treatment in skin melanoma. NK and dendritic cells, important
components of innate immune surveillance, have not been extensively
studied in peripheral blood (PB) in cutaneous melanoma; however,
recent data indicate a significant alteration of NK cells: A
decrease in their activity, a reduction in the percentage of IFN-γ
secreting NK cells and a predominance of the CD16dim/neg
subpopulation (8).
There is strong evidence that an effective innate
immune response plays an important role in tumor growth and
progression. NK cells are innate effector cells that substantially
contribute to antitumor immune responses, low activity of PB NK
cells is associated with an increased risk of cancer (9). Monitoring NK cell functions is
important in diagnosis, prognosis, or follow-up during therapy in
many diseases, including cancer (10). NK cells have the ability to induce
direct cytotoxicity of target cells, without prior sensitization.
Target recognition and effector function by NK cells are controlled
by both activating and inhibitory receptors signals.
NK cells are a heterogeneous population divided into
different subsets that can be defined both functionally and by a
combination of surface markers (11–13).
Based on the CD56 expression, two human NK subsets have been
identified, CD56dim and CD56bright.
CD56dim cellular subset has cytotoxic function and is
found mostly in PB, while CD56bright subset has a lower
cytolytic activity and is found mainly in lymphoid organs. Mouse NK
cells can be subdivided into 4 differentiation stages based on
surface density expression of CD27 and CD11b (14). The maturation of NK cells appears to
be a continuous process that starts with a double negative stage,
CD27−CD11b− cells (the most immature stage)
and ends with CD27−CD11b+ phenotype, most
mature cells (15,16). In healthy mice the majority of
CD11b+ NK cells are found in peripheral organs such as
the spleen, blood, liver and lung (17).
The purpose of this study, was to characterize
T-CD4+ and T-CD8+ lymphocytes, B lymphocytes
and NK cells in both PB and secondary lymphoid organ like the
spleen from melanoma-bearing mice (MbM). The investigation aimed
through these cellular populations to assess the immunosuppression
potential of the tumor in order to find the best peripheral immune
cell population that can be further developed as an indicator of
tumor evolution.
Materials and methods
Animal strain
C57BL/6 mice (males and females), 8–10 weeks old,
purchased from Jackson Laboratory (Bar Harbor, ME, USA), were
provided by the Animal Husbandry of Victor Babes National Institute
of Pathology. The animals were maintained in optimal conditions:
temperature 22±2°C, humidity 55±10%, artificial ventilation,
lighting 12/12, light/dark cycle. The mice received food (special
granulated fodder for mice) and water (filtered and sterilized)
ad libitum. They were accommodated in different special
cages with bedding according to sex, with an optimal density of
individuals per cage. All mice were kept under a rigorous cleaning
and hygiene program and were monitored daily. The experiments were
done in accordance with recognized principles of Laboratory Animal
Care in the framework of EU Directive 2010/63/EU for animal
experiments (18). The study was
approved by the Ethics Committee from ‘Victor Babeș’ Institute and
by the National Sanitary Veterinary and Food Safety Authority
through project authorization, no. 388/22.03.2018 (Bucharest,
Romania).
Murine experimental model of
melanoma
We developed the standard animal model for cutaneous
melanoma using C57BL/6 mice by inoculating subcutaneously in the
right flank (day 1) 0.5×106 B16-F10 (ATCC®
CRL-6475™; ATCC, Manassas, VA, USA) melanoma cells line/mouse.
Tumor-bearing mice were housed in separate boxes from healthy
animals, under the same feeding and microclimate conditions.
Two groups of C57BL/6 mice were considered (8–10
weeks old, 1:1 sex ratio): i) the control group (20 mice) - healthy
mice; ii) the melanoma group (15 mice) - inoculated with B16-F10
cells on day 1 and sacrificed on day 21 of the experiment.
Tumor growth monitoring
Two perpendicular diameters of the developed tumors
were measured weekly using a calliper. The tumor volume was
calculated by the following formula: Tumor volume = 4 π r3/3, where
r is the mean of the two perpendicular diameters (19).
Blood and tissues sampling
On day 21 of the experiment all animals were
anesthetized with ketamine/acepromazine cocktail (ketamine 100
mg/kg, ketamin 10%, Medistar Arzneimittelvertrieb Gmbh, Ascheberg,
Germany; acepromazine 5 mg/kg, Calmivet Solution Injectable
Acepromazine 5 mg, Vétoquinol SA, Lure, France) prior to blood
collection and spleen sampling, then sacrificed. Blood was
collected by retro-orbital veni-puncture in K2-EDTA coated tubes
(Microvette, Sarstedt AG & Co., Numbrecht, Germany).
Isolation of spleen cells
Spleens were harvested in RPMI-1640 media
supplemented with 5% fetal bovine serum (FBS) (Biochrom GmbH,
Berlin, Germany); spleen cell suspensions were prepared by
mechanical disruption and passed through a 70 µm cell strainer (BD
Falcon; BD Biosciences, San Jose, CA, USA). The cell suspensions
were centrifuged at 350 × g for 5 min (20°C) and resuspended in RBC
Lysis Buffer (BioLegend, San Diego, CA, USA). After 5-min
incubation on ice, the cell lysis was stopped by adding 10 ml Cell
Staining Buffer (BioLegend). Cell suspensions were centrifuged at
350 × g for 5 min (20°C) and the cell pellet was resuspended twice
in Cell Staining Buffer. Viable cells were counted and resuspended
in Cell Staining Buffer at 1×106 cells/ml.
Flow cytometry analysis
Lymphocyte immunophenotyping and NK degranulation
assays were performed by flow cytometry, based on the expression of
surface or intracellular markers. Unlabeled cells were used as
controls; nonspecific fluorescence signals due to spectral overlap
were automatically compensated (UltraComp eBeads; Thermo Fischer
Scientific, Inc., San Diego, CA, USA). Data acquisition and
analysis were performed on a BD FACSCanto II cytometer with BD
FACSDiva v.6.1 software (BD Biosciences).
Immunophenotyping of lymphocytes
Blood samples and spleen cell suspensions were
incubated with TruStain fcX (anti-mouse CD16/32) antibody (clone
93; isotype Rat IgG2a, λ; diluted 2/100, cat. no. 101319;
BioLegend, San Diego, CA, USA) in order to block non-specific
antibody binding, then stained for 20 min at room temperature with
the following antibodies: Alexa Fluor 647 anti-mouse CD3ε (clone
145-2C11, isotype Armenian Hamster IgG, diluted 0.5/100, cat. no.
100322), PE/Cy7 anti-mouse CD4 (clone GK1.5, isotype Rat IgG2b, κ,
diluted 1.25/100, cat. no. 100422), Alexa Fluor 488 anti-mouse CD8a
(clone 53–6.7, isotype Rat IgG2a, κ, diluted 0.5/100, cat. no.
100723), PerCP/Cy5.5 anti-mouse CD19 (clone 6D5, isotype Rat IgG2a,
κ, diluted 1.25/100, cat. no. 115534) and PE anti-mouse NK1.1
(clone PK136, isotype mouse IgG2a, κ, diluted 1.25/100, cat. no.
108708) (all from BioLegend). Surface staining for blood samples
was followed by red blood cell lysis with RBC Lysis Buffer
(BioLegend) for 10 min at room temperature in the dark and
centrifugation at 350 × g for 5 min, 20°C. Cells were washed twice
with Cell Staining Buffer (BioLegend) and analysed by flow
cytometry.
NK cells phenotype
Blood samples and spleen cell suspensions were
labelled according to the above described protocol, with the
following monoclonal antibodies: BV 510 anti-mouse NK1.1 (clone
PK136, isotype mouse IgG2a, κ, diluted 5/100, cat. no. 108738),
FITC anti-mouse CD3ε (clone 145-2C11, isotype Armenian Hamster IgG,
diluted 1/100, cat. no. 152304), PE/Cy7 anti-mouse CD335 (NKp46)
(clone 29A1.4, isotype rat IgG2a, κ, diluted 5/100, cat. no.
137618), PE/Cy7 anti-mouse CD69 (clone H1.2F3, isotype Armenian
Hamster IgG, diluted 5/100, cat. no. 104511), APC/Cy7 anti-mouse
CD45R (B220) (clone RA3-6B2, isotype Rat IgG2a, κ, diluted
1.25/100, cat. no. 103223), PerCP/Cy5.5 anti-mouse CD11c (clone
N418, isotype Armenian Hamster IgG, diluted 5/100, cat. no.
117328), APC/Cy7 anti-mouse CD43 (clone 1B11, isotype Rat IgG2a, κ,
diluted 1.25/100, cat. no. 121219), PerCP/Cy5.5
anti-mouse/rat/human CD27 (clone LG.3A10, isotype Armenian Hamster
IgG, diluted 1.25/100, cat. no. 124213), APC/Cy7 anti-mouse CD25
(IL-2Rα) (clone PC61, isotype Rat IgG1, λ, diluted 5/100, cat. no.
102025), PerCP/Cy5.5 anti-mouse CD122 (IL-2R/IL-15Rβ) (clone TM-β1,
isotype Rat IgG2b, κ, diluted 2.5/100, cat. no. 123211), PE
anti-mouse CD132 (common γ chain) (clone TUGm2, isotype Rat IgG2b,
κ, diluted 5/100, cat. no. 132305) (all from BioLegend), eFluor 450
anti-mouse CD49b (DX5) (clone DX5, isotype Rat IgM, κ, diluted
5/100, cat. no. 48–5971-82), APC anti-mouse CD11b (clone M1/70,
isotype Rat IgG2b, κ, diluted 2.5/100, cat. no. 50–0112-80), PE
anti-mouse KLRG1 (clone 2F1, isotype Syrian Hamster IgG, diluted
1.25/100, cat. no. 12–5893-80) (all from eBioscience Inc., San
Diego, CA, USA).
NK antitumoral activity measured as
degranulation capacity
CD107a (LAMP-1) conjugated with APC/Cy7 was used to
measure NK cell degranulation after exposure to YAC-1 tumour target
cells. The cell suspension obtained from splenocytes was incubated
with or without target cells at an E:T ratio = 2/1 for 1 h. The
APC/Cy7 anti-mouse CD107a (clone 1D4B, isotype Rat IgG2a, κ,
diluted 5/100, cat. no. 121616; BioLegend) antibody and GolgiStop
(BD GolgiStop; BD Biosciences) reagent (diluted 1/151) were then
added and cell were incubated for 3 h. Next, cells were labelled
with anti-CD3 (FITC anti-mouse CD3ε, clone 145–2C11, isotype
Armenian Hamster IgG; BioLegend) and NK1.1 (BV 510 anti-mouse
NK1.1, clone PK136, isotype Mouse IgG2a, κ) (BioLegend) according
to the protocol described before, and analyzed by flow
cytometry.
Lactate dehydrogenase (LDH) release
assay
NK cell cytotoxicity was determined with CytoTox 96
Non-Radioactive Cytotoxicity assay (Promega Co., Madison, WI, USA)
using YAC-1 cells as target cell line (ATCC® TIB-160).
This colorimetric assay quantitatively measures LDH, a stable
cytosolic enzyme that is released upon cell lysis, in much the same
way that 51Cr is released in a radioactive assay.
Briefly, spleen cells (effector cell, E) and YAC-1 cells
(2×104 cells/well; targeted cell, T) were mixed in
different E:T ratios (20:1 and 10:1) in a 96-well and incubated at
37°C with 5% CO2 overnight according to the
manufacturer's instructions. The effector:target cellular ratio and
incubation time were prior tested to optimize the experimental
conditions. The NK cell cytotoxicity of effector cells was measured
at 490 nm (Absorbance Microplate Reader Sunrise; Tecan, Grödig,
Austria) and was calculated using the following equation:
(Experimental - Effector Spontaneous - Target Spontaneous)/(Target
Maximum - Target Spontaneous) × 100, where ‘Experimental’ is the
experimental LDH release of co-cultured effector and target cells,
‘Effector Spontaneous’ and ‘Target Spontaneous’ express the
spontaneous released LDH of the effector and target cells alone,
respectively, and ‘Target Maximum’ is the maximum LDH release of
target cells.
Statistical analysis
Data analysis was performed using BD FACSDiva (v.6.1
software; BD Biosciences), with CD3ε+ lymphocytes gated
as CD4+CD8a− and
CD4−CD8a+ populations, and CD3ε−
lymphocytes as CD19+NK1.1− and
CD19-NK1.1+ populations. Unlabeled cells were used as
controls; nonspecific fluorescence signals due to spectral overlap
were automatically compensated. Data was analysed using Microsoft
Excel (Microsoft, Redmond, CA, USA). Results are presented as mean
± SD of cells percentage. For statistical analysis, the Students
t-test (two-tailed, assuming equal variance) was used for
statistical evaluation of the differences between the experimental
groups. P-value <0.05 was considered to indicate a statistically
significant difference.
Results
Immunophenotyping of lymphocytes
In order to assess the immune cell status of MbM, T,
B and NK cell populations were analysed by flow cytometry. The
results were expressed as a percentage from the CD3+
lymphocytes, respectively CD3− lymphocytes (mean ± SD)
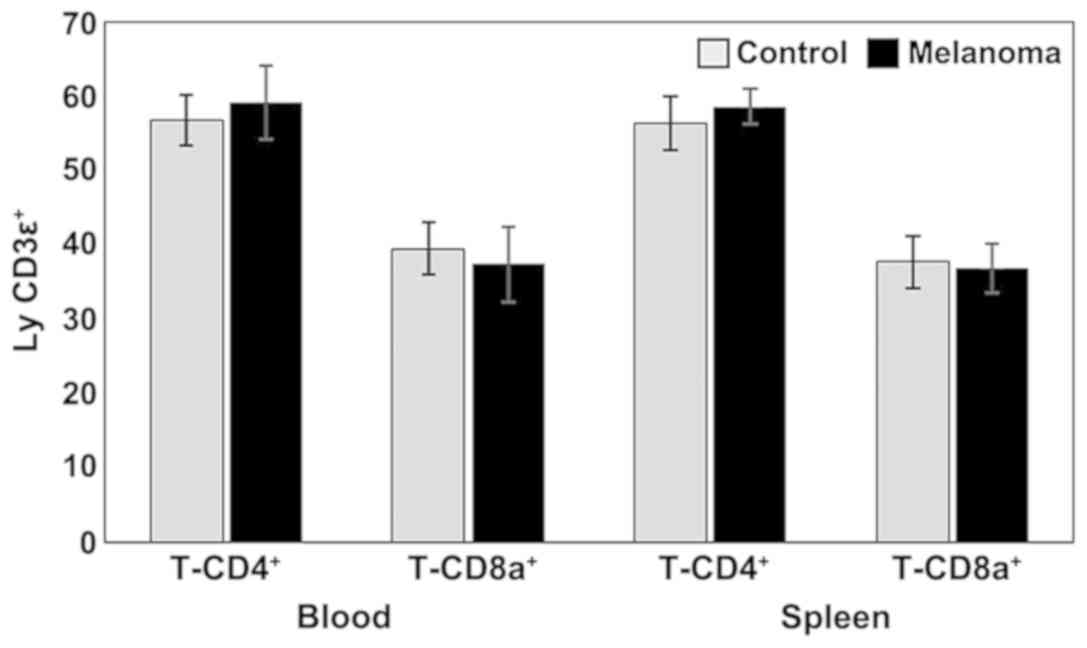
(Figs. 1 and 3).
Analysis of T cell subpopulations in melanoma mice
showed a slight increase in the percentage of T-CD4+
lymphocytes in both PB and spleen cell suspension. The percentages
of T-CD8+ lymphocytes were lower in MbM compared to the
control group (in both PB and spleen cell suspension) but with no
statistical significance (Fig. 1).
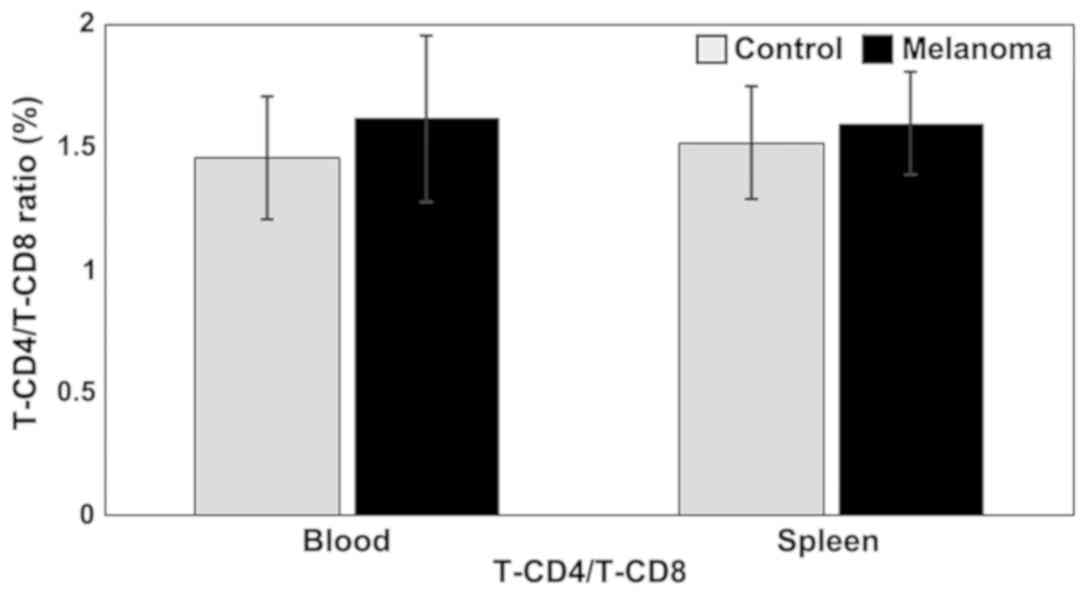
T-CD4/T-CD8 ratio is higher in MbM compared to control group, but
the differences between experimental groups are not statistically
significant (Fig. 2).
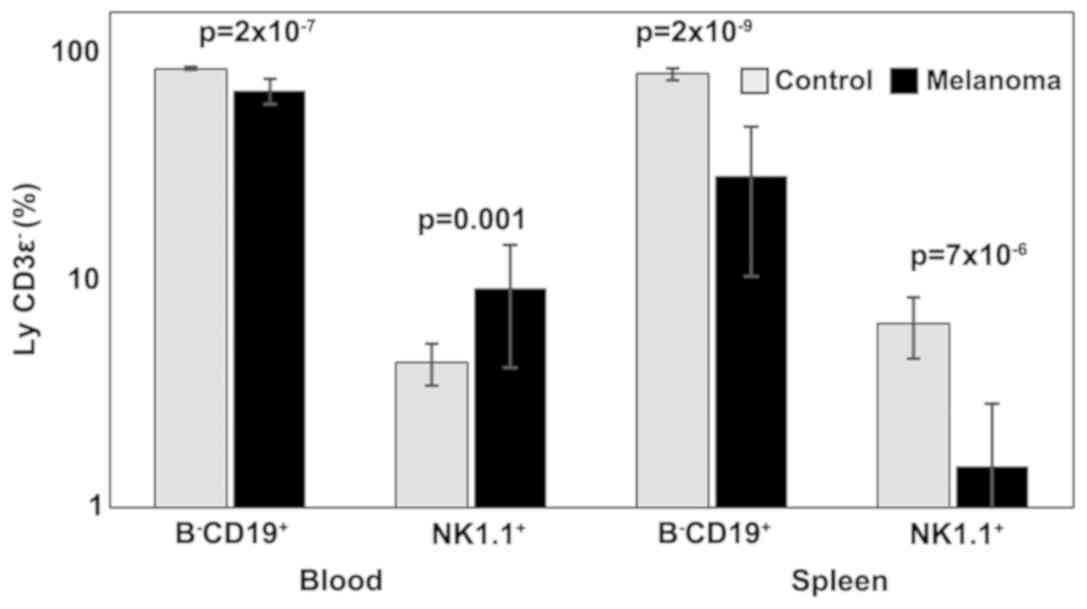
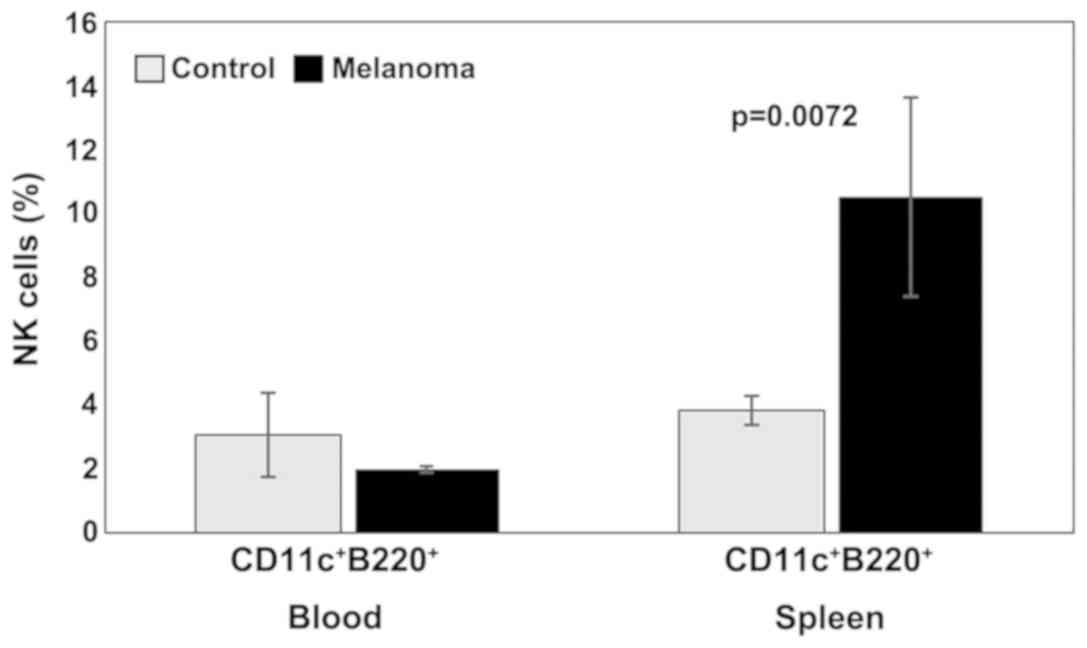
Analysis of B lymphocytes and NK cells revealed
statistically significant differences between the two experimental
groups in both PB and spleen cell suspension. B-CD19+
lymphocytes were significantly decreased in MbM (68±9,
p=2×10−7 in PB; 29±18, p=2×10−9 in spleen
cell suspension) compared to the control group (85±2 in PB; 81±5 in
spleen cell suspension). Percentages of NK1.1+ cells
increased significantly in melanoma-bearing mice in the PB (9±5,
p=0.001) and decreased in spleen cells suspension (2±1,
p=7×10−6) compared to the control group (4±1 in PB,
respectively 6±2 in spleen cell suspension) (Fig. 3).
NK cells phenotype
A large panel of surface markers was investigated in
PB and spleen cell suspension to assess NK cells phenotype, thus
lineage markers: CD161 (NK1.1), CD3ε; activation and maturation
markers: CD335 (NKp46), CD69, CD45R (B220), CD11c, CD49b (DX5),
CD11b, CD43, CD27, KLRG1; markers for cytokine receptors: CD25
(IL-2Rα), CD122 (IL-2R/IL-15Rβ), CD132 (common γ chain).
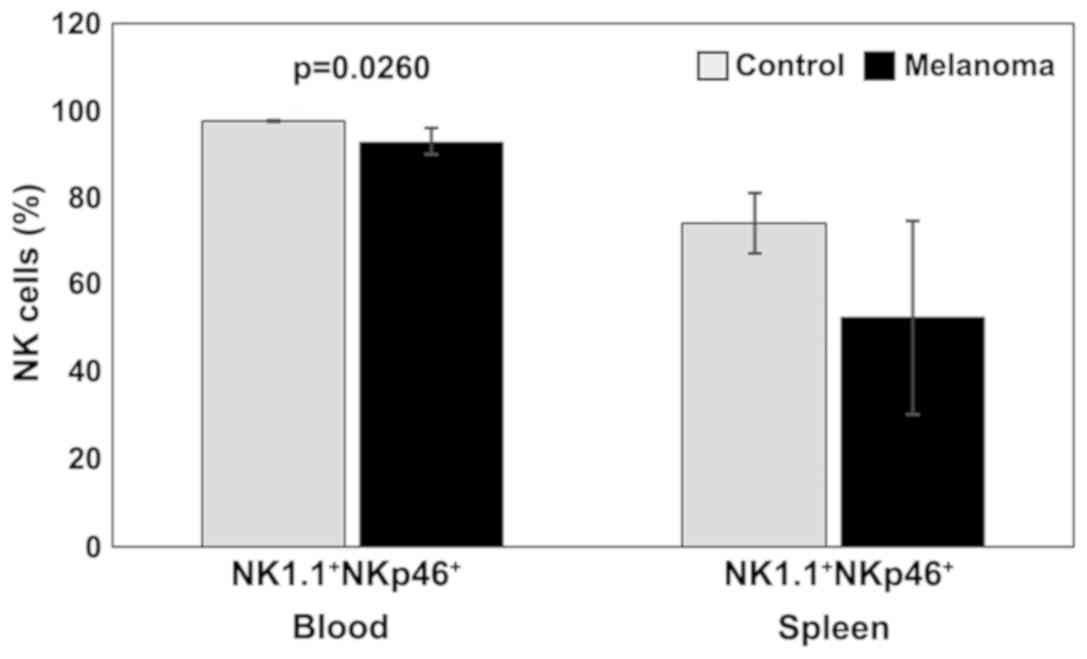
Our experimental data suggested a decrease in NKp46
(activating receptor) expression in both PB and spleen cell
suspension for MbM compared to the control group. Significant
differences between the two experimental groups were observed in PB
(p<0.05) (Fig. 4).
For activation and maturation markers of NK cells
(CD69, B220, CD11c), the main change observed in PB is the
significant decrease of CD11c in melanoma (p<0.05); the levels
of expression for CD69 and B220 are similar for the two
experimental groups (Fig. 5A). An
increased expression of these activation markers was observed, on
the other hand, in the spleen cell suspension. Significant
differences (p<0.05) were obtained between the experimental
groups for all three activation/maturation markers analyzed
(Fig. 5B).
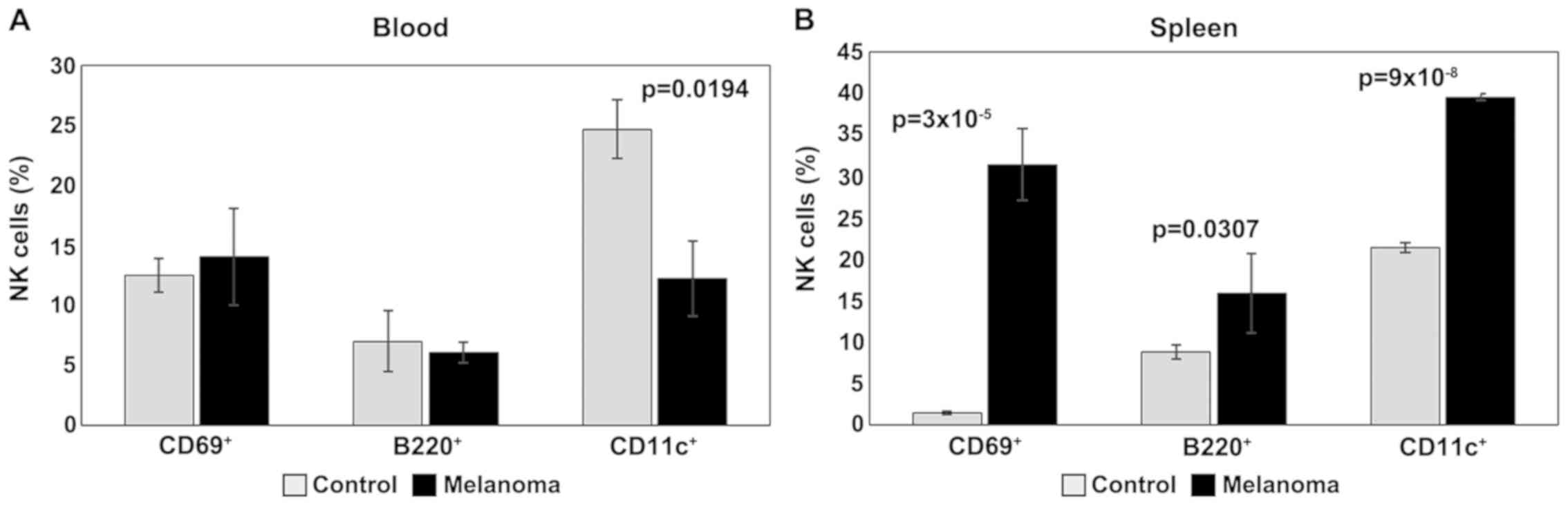 | Figure 5.Expression of CD69, B220 and CD11c
for NK1.1+ cells. (A) Expression of CD69, B220 and CD11c
for NK1.1+ cells in PB. CD69+,
B220+ and CD11c+ cells in MbM (n=5) (14±4,
6±0.9 and 12±3.1, p=0.0194) as compared to control group (n=5)
(12±1.4, 7±2.6 and 25±2.4) in PB. (B) Expression of CD69, B220 and
CD11c for NK1.1+ cells in spleen cell suspension.
CD69+, B220+ and CD11c+ cells in
MbM (n=5) (31±4.4, p=3×10−5; 16±4.8, p=0.0307; 40±0.4,
p=9×10−8) as compared to control group (n=5) (2±0.2,
9±0.8 and 21±0.6) in spleen cell suspension. The results are
presented as a percentage from NK1.1+ cells (mean ±
SD). |
Analysis of B220 and CD11c on NK1.1+
cells revealed that in control tumor-free mice, the
B220+CD11c+NK1.1+ subset in both
PB and spleen cell suspension was at ~3% of NK1.1+
cells. In tumor-bearing mice, a decrease in the percentage of
B220+CD11c+NK1.1+ cells in the PB
was observed, as well as a significant increase in the spleen
(p<0.01) (Fig. 6).
In PB, analysis of CD49b, CD11b, CD43 and KLRG1
expression revealed similar levels for the two experimental groups.
The expression of CD27 is downregulated- in MbM, and the
differences between the experimental groups are significant
(p=0.001) (Fig. 7A). Significantly
lower levels of NK cells expressing CD49b, CD11b, CD43 and KLRG1
were observed in spleen cell suspension in MbM compared to the
control group (p<0.05). NK cells expressing CD27 are slightly
increased in percentage in MbM, with no statistical significance
(Fig. 7B).
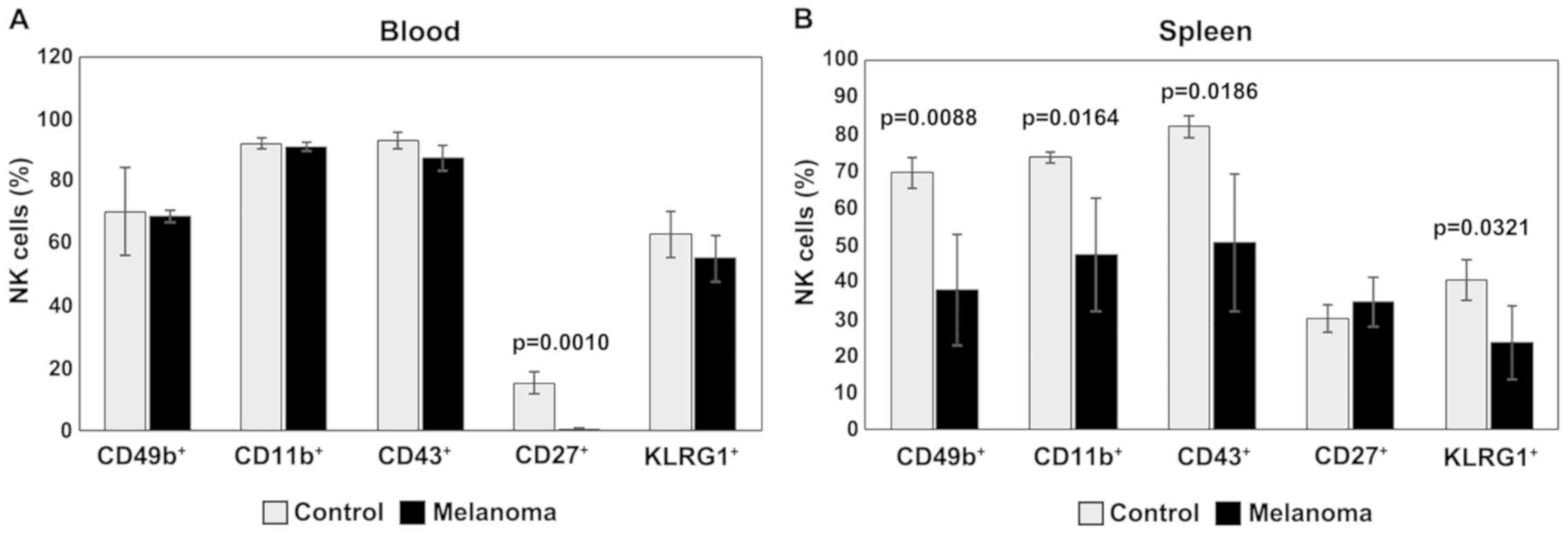 | Figure 7.Expression of CD49b, CD11b, CD43,
CD27 and KLRG1 for NK1.1+ cells. (A) Expression of
CD49b, CD11b, CD43, CD27 and KLRG1 for NK1.1+ cells in
PB. CD49b+, CD11b+, CD43+,
CD27+ and KLRG1+ cells in MbM (n=5) (69±2;
91±1.4; 88±4; 0.4±0.3, p=0.001 and 55±7.5) as compared to control
group (n=5) (70±14.1, 92±1.8, 93±2.6, 15±3.7 and 63±7.5) in PB. (B)
Expression of CD49b, CD11b, CD43, CD27 and KLRG1 for
NK1.1+ cells in spleen cell suspension.
CD49b+, CD11b+, CD43+,
CD27+ and KLRG1+ cells in MbM (n=5) (38±14.9,
p=0.0088; 47±15.4, p=0.0164; 51±18.6, p=0.0186; 35±6.6 and 23±10,
p=0.0321) as compared to control group (n=5) (69±4.2, 74±1.4, 82±3,
30±3.7 and 40±5.4) in spleen cell suspension. The results are
presented as a percentage from NK1.1+ cells (mean ±
SD). |
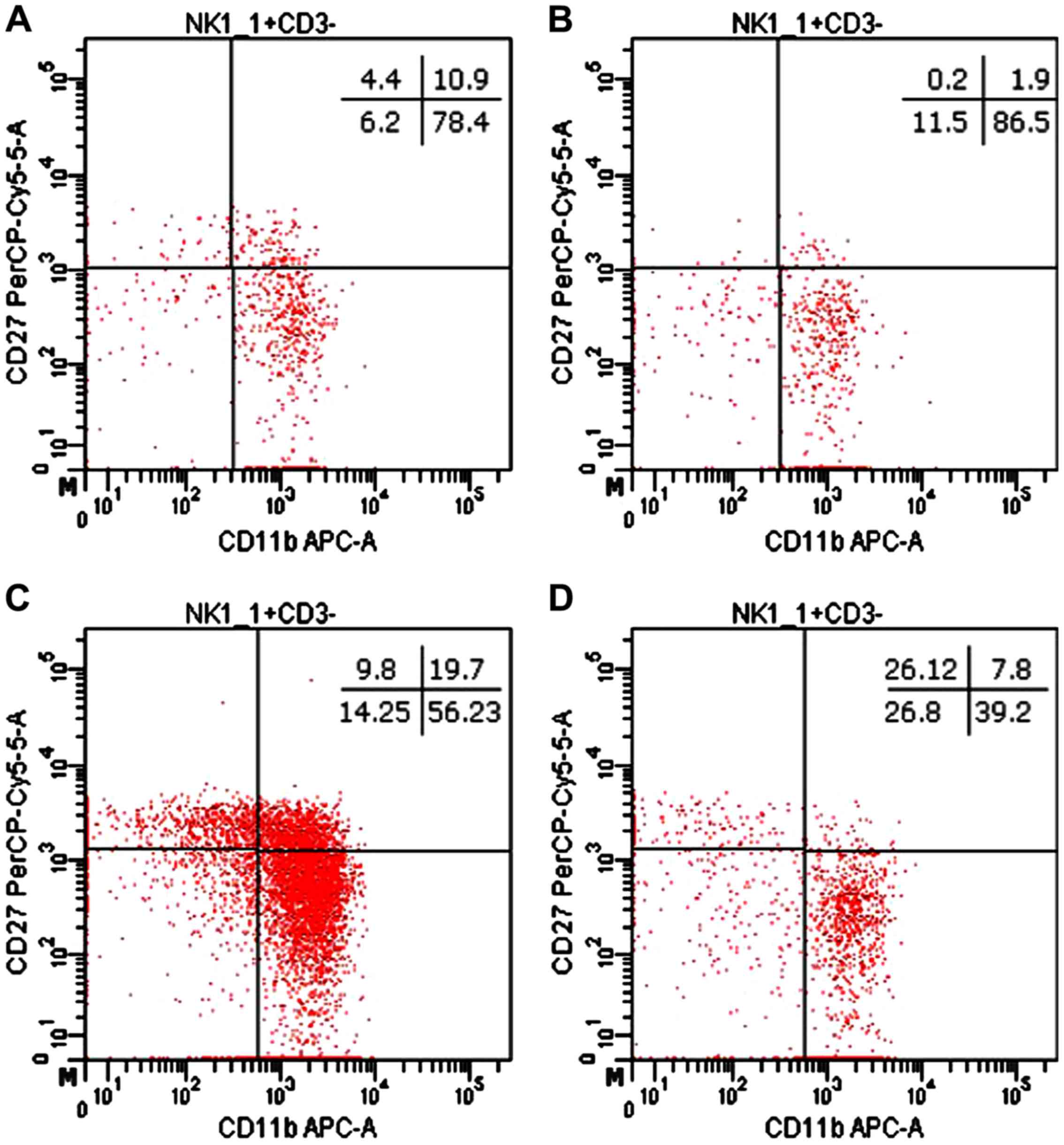
The distribution of CD27 and CD11b markers can be
used to divide murine NK cells into four subsets defining different
maturation stages. The CD27−CD11b− phenotype
defines immature NK cells, CD27+CD11b−
suggest an early maturation stage, while
CD27+CD11b+ and
CD27−CD11b+ are specific to mature NK cells.
In our experimental data, the distribution of mature NK cell
subsets in tumor-bearing mice was different from that of healthy
mice (Figs. 8 and 9). The analysis of PB NK cells revealed
significant differences between MbM and the control mice for
immature CD27−CD11b− NK cells (p<0.05),
early mature CD27+CD11b− (p<0.01) and
mature CD27+CD11b+ (p<0.01) NK cells. The
CD27−CD11b+ NK subset was increased in MbM
but without statistical significance (Fig. 9A). CD27−CD11b−,
CD27+CD11b− and
CD27+CD11b+ NK subsets in spleen cell
suspension revealed increased values for immature (p<0.05) and
early mature (p<0.01) NK cells, while the percentage of mature
NK cells is lower in MbM than in the control group (p<0.01).
CD27−CD11b+ NK subset was decreased in MbM,
but without statistical significance (Fig. 9B).
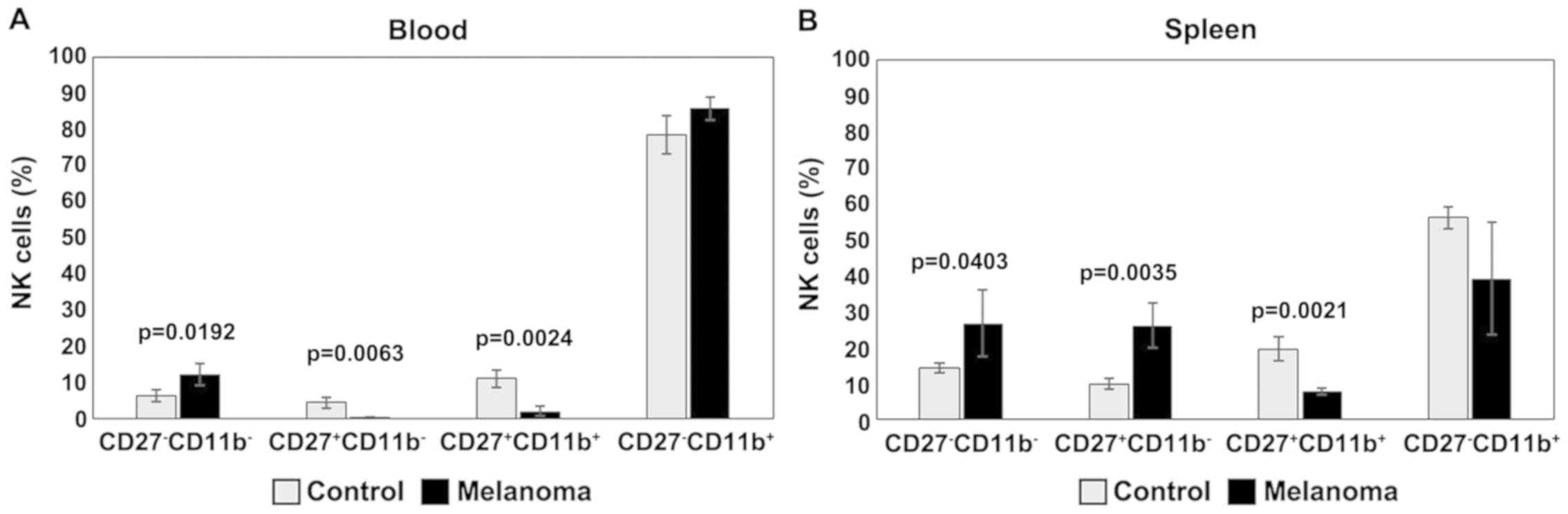 | Figure 9.Distribution of
CD27/CD11b/NK1.1+ subsets. (A) Distribution of CD27
CD11b NK1.1+ subsets in PB.
CD27−CD11b−,
CD27+CD11b−,
CD27+CD11b+ and
CD27−CD11b+ cells in MbM (n=5) (12±3,
p=0.0192; 0.2±0.2, p=0.0063; 2±1, p=0.0024 and 86±3) and the
control group (n=5) (6±1.6, 4±1.5, 11±2.4 and 78±5.3) in PB. (B)
Distribution of CD27CD11bNK1.1+ subsets in spleen cell
suspension. CD27−CD11b−,
CD27+CD11b−,
CD27+CD11b+ and
CD27−CD11b+ cells in MbM (n=5) (27±9.3,
p=0.0403; 26±6.2, p=0.0035; 8±1, p=0.0021 and 39±15.7) and the
control group (n=5) (14±1.4, 10±1.5, 20±3.4 and 56±3) in PB. The
results are presented as a percentage from NK1.1+ cells
(mean ± SD). |
Data obtained for the levels of markers for cytokine
receptors showed a decreased expression in the common γ receptor
subunit, both in blood and spleen cell suspension (p<0.001) in
MbM, and a decrease in receptor subunit β in spleen (p<0.01)
(Fig. 10A and B). Also, we noted a
decrease in IL-2α receptor subunit (CD25) expression in PB
(p<0.05) in MbM, and an increase in spleen cell suspension
(p<0.001).
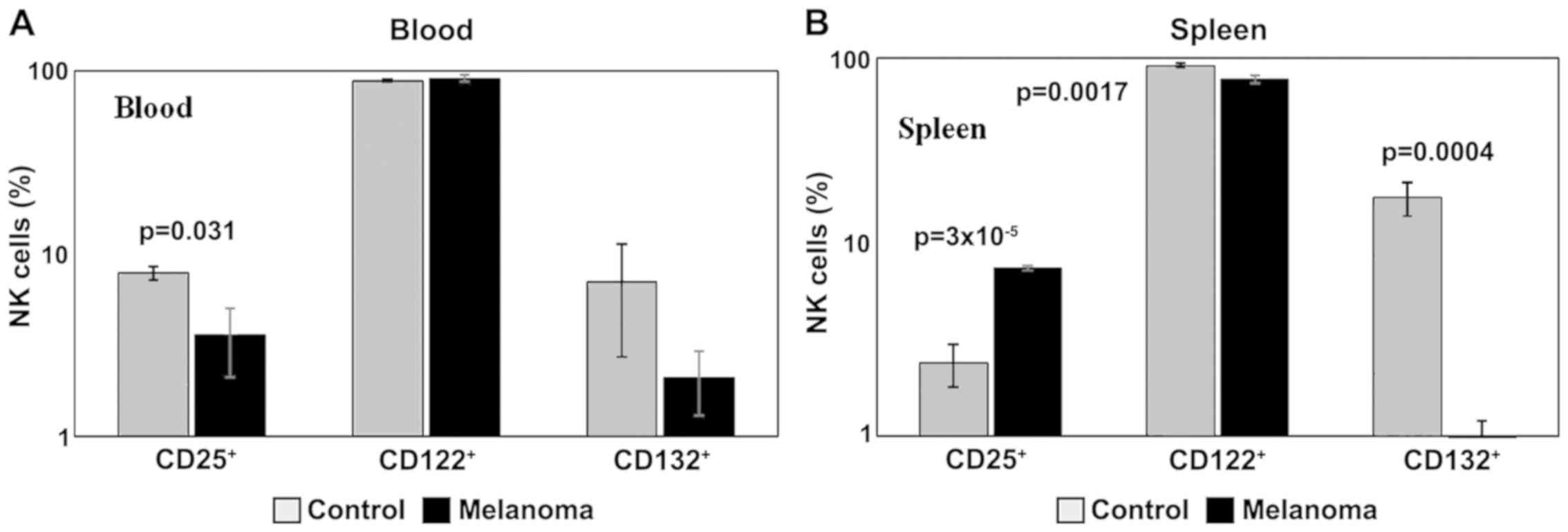 | Figure 10.Expression of CD25, CD122 and CD132
for NK1.1+ cells. (A) Expression of CD25, CD122 and
CD132 for NK1.1+ cells in PB. CD25+,
CD122+ and CD132+ cells in MbM (n=5) (4±1.5,
p=0.031; 91±4.1 and 2±0.8) and the control group (n=5) (8±0.6,
89±1.3 and 7±4.3) in PB. (B) Expression of CD25, CD122 and CD132
for NK1.1+ cells in spleen cell suspension.
CD25+, CD122+ and CD132+ cells in
MbM (n=5) (8±0.2, p=3×10−5; 77±3.6, p=0.0017 and 1±0.4,
p=0.0004) as compared to control group (n=5) (2±0.6, 91±2.6 and
18±3.6) in spleen cell suspension. The results are presented as a
percentage from NK1.1+ cells (mean ± SD). |
Functional evaluation of NK cells
Functional characterization of NK cells from cell
suspensions was performed by evaluating the degranulation of NK
cells (effector cells) after exposure to target tumor cells (YAC-1
cells) and the release of LDH to evaluate the cytotoxic effect that
NK cells exhibit on tumor cells. CD107a expression change is
reflecting NK cell degranulation and it was measured by flow
cytometry, assessing thus NK cytotoxicity.
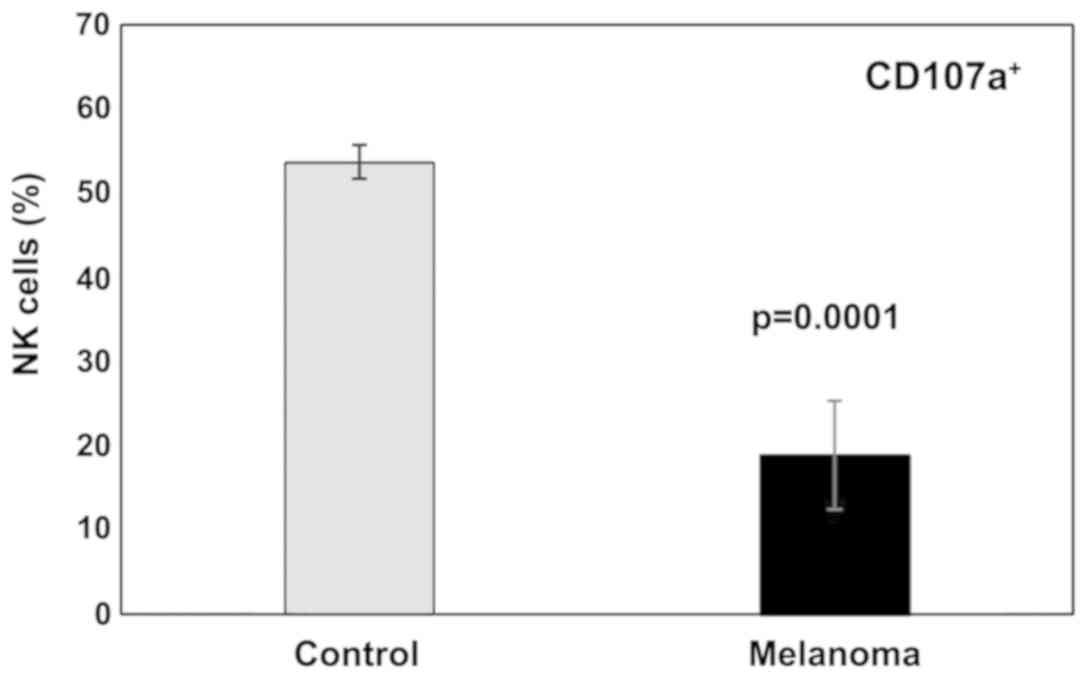
In this study a significant decrease in NK cells
degranulation capacity (the effector function) was detected in MbM
(Fig. 11). The differences between
the experimental groups were statistically significant
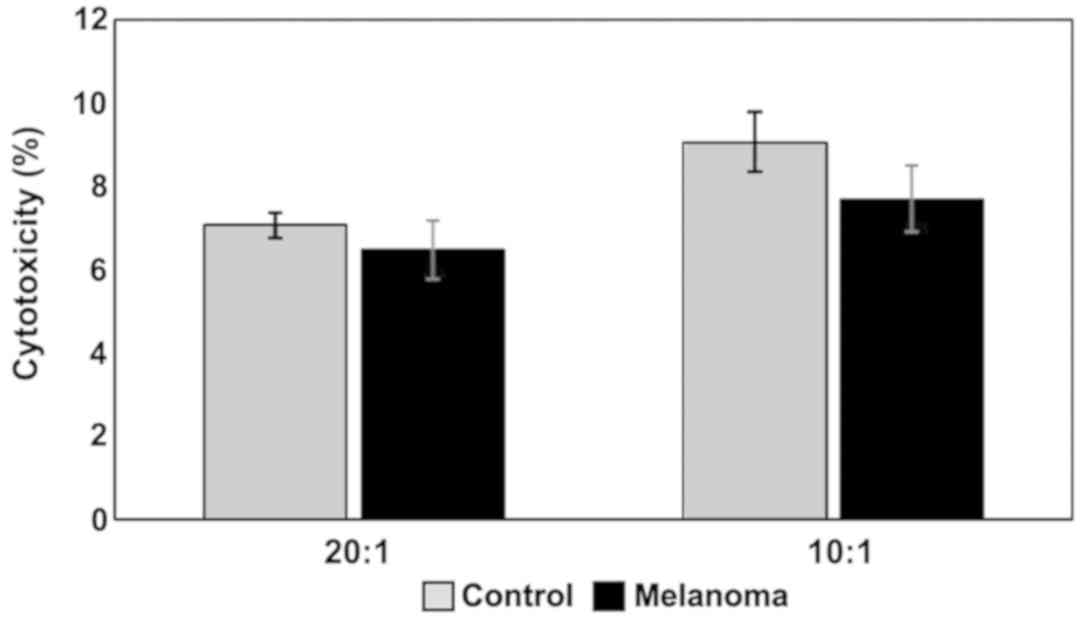
(p<0.001). Regarding cytotoxicity inflicted upon YAC-1 cells
(Fig. 12) the actual differences
between studied groups can be seen only in the 10:1 (effector
cell:target cell) ratio, with a decrease in melanoma-bearing mice
compared to controls.
NK cytotoxicity to tumor target cells was assessed
by measuring the release of LDH from dead cells in a culture
medium. MbM NK cells showed a lower cytotoxicity against target
cells (YAC-1) compared to the control group (Fig. 12). Even though the decrease in
cytotoxicity is not statistically significant, the data suggest a
decrease of the tumoricidal efficiency of NK cells.
Discussion
We investigated T-CD4+ and
T-CD8+ lymphocytes, B lymphocytes and NK cells in PB and
spleen cell suspension from MbM compared to healthy controls in
order to assess the potential for tumor growth-promoting
immunosuppression.
Analysis of T cell subpopulations in melanoma mice
showed a slight increase in the percentage of T-CD4+
lymphocytes and a decrease of T-CD8+ lymphocytes in MbM
compared to the control group, in both PB and spleen cell
suspension, but with no statistical significance. T-CD4/T-CD8 ratio
is higher in MbM compared to control group, but the differences
between experimental groups are not statistically significant.
Analysis of B lymphocytes and NK cells revealed statistically
significant differences between the two experimental groups in both
PB and spleen cell suspension. B-CD19+ lymphocytes were
significantly decreased in MbM (p=2×10−7 in PB;
p=2×10−9 in spleen cell suspension) compared to the
control group. Percentages of NK1.1+ cells increased
significantly in melanoma-bearing mice in the PB (p=0.001) and
decreased in spleen cell suspension (p=7×10−6).
Evaluation of lymphocyte populations from PB and spleen cell
suspension revealed an altered distribution in tumor-bearing mice
as compared to control mice. The main changes were observed in B
lymphocytes and NK cells, the data obtained for these populations
in MbM differed significantly from those obtained for the control
group, in both PB and spleen cell suspension.
Several surface markers were investigated in MbM
(both in PB and spleen cell suspension) to assess NK cell
phenotype, thus lineage markers: CD161 (NK1.1), CD3ε; activation
and maturation markers: CD335 (NKp46), CD69, CD45R (B220), CD11c,
CD49b (DX5), CD11b, CD43, CD27, KLRG1; markers for cytokine
receptors: CD25 (IL-2Rα), CD122 (IL-2R/IL-15Rβ), CD132 (common γ
chain).
NKp46 is one of the three members of the natural
cytotoxicity receptor group and an important regulator of NK cell
function. Signalling through the NKp46 receptor induces NK cell
activation processes resulting in an increased cytokine production
and release of cytolytic granules. NKp46 is involved in tumor cell
recognition (20,21) and is found on all mature NK cells. It
is upregulated in humans and mice during NK cell maturation. In
this study we observed a decrease in NKp46 expression in both PB
and spleen cell suspension for MbM compared to the control group.
Only in PB significant differences were observed between the two
(p<0.05).
Activation and maturation markers of NK cells
include CD69, an early activation marker on T lymphocytes and NK
cells (22–24), and CD11c (25), a member of β2 integrin family. CD45R
(B220) modulates many processes such as differentiation, division
and cellular development and is essential in the development of B
lymphocytes but is also present on T lymphocytes and NK cells.
Activation of NK cells leads to an increased expression of B220 on
the cell membrane. The main change observed in PB was the
significant decrease of CD11c in melanoma (p<0.05) while the
levels of expression for CD69 and B220 were similar for the two
experimental groups. In spleen cells suspension, an increased
expression of these activation markers was observed. Significant
differences (p<0.05) were obtained between the experimental
groups (MbM and control group) for all three activation/maturation
markers analyzed.
B220+CD11c+NK1.1+
cells show functional features similar to the human
CD56bright cell subset which is present in lymphoid
organs and produces IFN-γ (26). In
humans, the NK cell population is divided in two subsets:
CD56dim and CD56bright. The
CD56dim population predominates in the blood and on
inflammatory sites, displays high cytotoxic potential, expresses
MHC-I specific inhibitory receptors and is considered to consist of
generally terminally differentiated cells. CD56bright
cells are predominant in lymph nodes, secrete cytokines, have a low
cytotoxic potential, and are considered precursors of
CD56dim cells. The CD56bright subset in
humans represents >10% of NK cells in PB, while
CD56dim cells are predominant in spleen (over 85%).
Previous studies on breast, head and neck cancer showed a decrease
in the percentage of CD56bright in PB and an increase in
secondary lymphoid organs (27). Our
data obtained for tumor-bearing mice showed a decrease in the
percentage of B220+CD11c+NK1.1+
cells in PB, as well as a significant increase in the spleen
(p<0.01).
The development of NK cells occurs mainly in the
bone marrow. The NK cell lineage commitment occurs with the
increase of β-IL-2/IL-15R (CD122) chain expression followed by the
acquisition of the NK1.1 marker in B6 mice. NK maturation can be
discussed in terms of phenotype and functional capacity. During
this step-wise maturation, integrin expression CD49b (DX5) defines
the early stages. After CD49b acquisition, bone marrow NK cells
upregulate the expression of CD11b and CD43. This is strongly
correlated with the capacity of NK cells to produce large amounts
of IFN-γ (28). After maturation, NK
cells migrate to different lymphoid and non-lymphoid organs, where
most NK cells express high levels of CD49b, CD11b and CD43. After
migration, the NK cells continue to adapt to the environment,
downregulate CD27 expression and upregulate KLRG1 expression
(29). KLRG1 is another marker
associated with the maturation of NK cells (30). KLRG1 expression on mature NK cells
allows identification of their terminal differentiation status
associated with a reduction in proliferation and effector functions
(24). In this study, analysis of
CD49b, CD11b, CD43 and KLRG1 expression in PB revealed similar
levels for MbM and control mice. The expression of CD27 is
downregulated in MbM, and the differences between the experimental
groups are significant (p=0.001). Significantly lower levels of NK
cells expressing CD49b, CD11b, CD43 and KLRG1 were observed in
spleen cell suspension in MbM (p<0.05).
In mice, depending on the expression of CD11b, two
types of NK cell populations were identified: CD11b− and
CD11b+. CD11b− NK cell population is the
major population in new-born mice; in adults it is mainly found in
bone marrow, lymph nodes, liver and has a high proliferation rate.
The CD11b+ population is mainly present in PB, spleen
and lung. Compared to CD11b− NK cells, it has a more
powerful effector function.
CD27, a member of the TNFR superfamily, is an
important marker that can be used to define NK cell subsets. The
distribution of CD27 and CD11b markers can be used to divide murine
NK cells into 4 subsets defining different maturation stages
(31). The
CD27−CD11b− phenotype defines immature NK
cells, CD27+CD11b− suggest an early
maturation stage, while CD27+CD11b+ and
CD27−CD11b+ are specific to mature NK cells.
These phenotypic changes also reflect variations in the functional
activity of NK cells-double positive mature cells exhibit the
highest responsiveness, while
CD27−CD11b+KLRG1+ represents the
terminal stage. Our results suggested that the distribution of
mature NK cell subsets in tumor-bearing mice are different from
that of healthy mice. In PB we observed significant differences
between MbM and the control mice for immature
CD27−CD11b− NK cells (p<0.05), early
mature CD27+CD11b− (p<0.01) and mature
CD27+CD11b+ (p<0.01) NK cells. The
CD27−CD11b+ NK subset was increased in MbM,
but without statistical significance.
CD27−CD11b−,
CD27+CD11b− and
CD27+CD11b+ NK subsets in spleen cell
suspension revealed increased values for immature (p<0.05) and
early mature (p<0.01) NK cells, while the percentage of mature
NK cells is lower in MbM than in the control group (p<0.01).
CD27−CD11b+ NK subset was decreased in MbM,
but without statistical significance. Comparing the distribution of
NK cell subsets between healthy and MbM revealed a significant
decrease in mature NK cell subsets suggesting that the tumor
influences NK cell regulation of maturation and homeostasis. An
increase in the number of immature NK cells suggests that the tumor
can render NK cells less tumoricidal and thus contribute to cancer
progression.
The NK cell lineage commitment, proliferation,
activation, and functional capacity of NK cells are controlled by
various cytokines, among which the most important are IL-2, IL-7,
IL-15 and IL-21. All these cytokines share the γ-receptor (CD132)
chain, and IL-2 and IL-15 share the β receptor subunit (CD122). We
observed a decreased expression in the common γ receptor subunit,
both in blood and spleen cell suspension (p<0.001) in MbM, and a
decrease in receptor subunit β in spleen (p<0.01). Also, we
noted a decrease in IL-2α receptor subunit (CD25) expression in PB
(p<0.05) in MbM, and an increase in spleen cell suspension
(p<0.001).
Functional characterization of NK cells from cell
suspensions was performed by evaluating the degranulation of NK
cells (effector cells) after exposure to target tumor cells (YAC-1
cells) and the release of LDH to evaluate the cytotoxic effect that
NK cells exhibit on tumor cells. CD107a expression change is
reflecting NK cell degranulation and it was measured by flow
cytometry, assessing thus NK cytotoxicity. NK cell degranulation
capacity (expressed by the release of cytolytic granules containing
granzymes and perforins in the presence of tumor target cells) is a
demonstration of the NK effector function (i.e., the ability to
kill tumor cells). The surface of the cytolytic granules is covered
by CD107a or LAMP-1, a highly glycosylated protein that makes up
~50% of the lysosomal membrane proteins. After degranulation,
CD107a is exposed on the surface of cytotoxic cells.
Externalization of CD107a has been shown to be a marker of NK cell,
T-CD8+ and T-CD4+ cell degranulation
(32). In this study a significant
decrease in NK cells degranulation capacity (the effector function)
was detected in MbM and the differences between the experimental
groups were statistically significant (p<0.001). Regarding
cytotoxicity inflicted upon YAC-1 cells the actual differences
between studied groups can be seen only in the 10:1 (effector
cell:target cell) ratio, with a decrease in melanoma- bearing mice
compared to controls. NK cytotoxicity to tumor target cells was
assessed by measuring the release of LDH from dead cells in a
culture medium. MbM NK cells showed a lower cytotoxicity against
target cells (YAC-1) compared to the control group. Even though the
decrease in cytotoxicity is not statistically significant, the data
suggest a decrease of the tumoricidal efficiency of NK cells.
In patients diagnosed with melanoma evaluating the
NK subpopulation percentage should be combined with NK activity
evaluation (degranulation potential and cytolytic activity) to
additionally monitor therapy efficacy.
In conclusion, lymphocyte populations are
differently distributed in PB and in secondary lymphoid organs,
like the spleen in melanoma-bearing animals. Tumors induce the
decrease in the percentage of B lymphocytes and NK cells in the
spleen. Experimental data show a statistical significant reduction
of the percentage of NK cells in MbM compared to control animals.
Analysis of NK cell subsets, defined by the differential expression
of CD27 and CD11b demonstrated a significant difference in the
distribution of NK cell subsets, with the mature subset being
dominant in the healthy mice. Also, we found a decrease of both
CD43 and KLRG1 (markers commonly used for terminally differentiated
NK cells), and a downregulation of activating receptor NKp46 in
MbM.
The evaluation of the distribution of NK cells
revealed a significant decrease in mature NK cell subsets in MbM,
suggesting that the tumor alters NK cell maturation and
homeostasis. An increase of the number of immature NK cells
suggests that the tumor can cause NK cells to become less
tumoricidal and thus contributes to cancer progression.
These results are confirmed by a decrease in NK cell
functions in MbM compared to the control group. Cytokine secretion
is reduced for γ chains of IL-2, IL-7, IL-15 and IL-21 (33–38).
Degranulation is also impaired in MbM, with effect on the cytotoxic
function.
Overall, our findings highlight a complex functional
disorder of NK cells during tumor development, both in the terms of
composition of active cytotoxic NK cell subsets, and of tumoricidal
activity efficiency. We showed that NK cells from the spleen in a
MbM model are reduced as percentage and have different phenotypic
characteristics than NK cells from a healthy mouse. In case of MbM
CD11b−CD27− NK cells, which have been defined
as immature NK cells, they are increased both in PB and the
spleen.
It is clear that the decrease of NK activity is a
tumor development-associated phenomenon, which can be caused by
both tumoral factors and by dysfunctions in the development and
maturation of NK cells (39). This
could suggest that the tumor may render NK cells less cytotoxic and
thereby it contributes to cancer progression and moreover NK
populations can be used as tumor development cell biomarkers.
Hence, this study provided new insights into NK cell phenotypic
changes, proving that the analysis of NK cell phenotype and
cytokine secretion markers in PB may be useful in assessing the
impairment degree of the antitumor immune response. Re-adjusting
this response may become a target for new approaches to cancer
immunotherapy (40).
Acknowledgements
Not applicable.
Funding
This study was supported by Core Program,
implemented with the support NASR, projects nos. 18.21.02.01 and
18.21.02.02; grant PN-III-P1-1.2-PCCDI-2017-0341/2018 and
7PFE/16.10.2018.
Availability of data and materials
The data sets used and/or analysed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
GI, MS, RIH, CC, MN and CU contributed to the
conception and design of the study, the acquisition, analysis and
interpretation of the data, drafting the manuscript and revising it
critically for important intellectual content. GI, MS, ANM, IRP,
DC, OB and CC were responsible for the acquisition, analysis and
interpretation of the data, drafting the manuscript and revising it
critically for important intellectual content. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of ‘Victor Babeș’ Institute and by the National Sanitary
Veterinary and Food Safety Authority through project authorization
no. 388/22.03.2018 (Bucharest, Romania).
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
APC
|
allophycocyanin
|
|
BD
|
Becton-Dickinson
|
|
CD
|
cluster of differentiation
|
|
E:T
|
effector:target
|
|
FACS
|
fluorescence-activated cell
sorting
|
|
FBS
|
fetal bovine serum
|
|
FITC
|
fluorescein isothiocyanate
|
|
IFN
|
interferon
|
|
Ig
|
immunoglobulin
|
|
IL
|
interleukin
|
|
K2-EDTA
|
Kalium 2
ethylenediaminetetraacetate
|
|
KLRG1
|
killer cell lectin-like receptor
G1
|
|
LDH
|
lactate dehydrogenase
|
|
LAMP-1
|
lysosome-associated membrane
protein-1
|
|
MbM
|
melanoma-bearing mice
|
|
MHC
|
major histocompatibility complex
|
|
NK
|
natural killer
|
|
PB
|
peripheral blood
|
|
PBMCs
|
peripheral blood mononuclear
cells
|
|
PE
|
phycoerythrin
|
|
PE/Cy
|
phycoerythrin complex with
cyanine
|
|
PerCP/Cy
|
peridinin chlorophyll protein complex
with cyanine
|
|
R
|
receptor
|
|
RPMI
|
Roswell Park Memorial Institute
|
|
SD
|
standard deviation
|
|
TNFR
|
tumor necrosis factor receptor
|
References
|
1
|
Chin L, Garraway LA and Fisher DE:
Malignant melanoma: Genetics and therapeutics in the genomic era.
Genes Dev. 20:2149–2182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lakshmikanth T and Johansson MH: Current
perspectives on immunomodulation of NK cells in melanoma. Melanoma
in the Clinic - Diagnosis. Management and Complications of
Malignancy; Mandi M: InTech, Rijeka: pp. 133–162. 2011
|
|
3
|
Serban ED, Farnetani F, Pellacani G and
Constantin MM: Role of in vivo reflectance confocal microscopy in
the analysis of melanocytic lesions. Acta Dermatovenerol Croat.
26:64–67. 2018.PubMed/NCBI
|
|
4
|
Rangwala S and Tsai KY: Roles of the
immune system in skin cancer. Br J Dermatol. 165:953–965. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lupu M, Caruntu A, Caruntu C, Papagheorghe
LML, Ilie MA, Voiculescu V, Boda D, Constantin C, Tanase C, Sifaki
M, et al: Neuroendocrine factors: The missing link in non-melanoma
skin cancer (Review). Oncol Rep. 38:1327–1340. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caruntu C, Boda D, Constantin C, Caruntu A
and Neagu M: Catecholamines increase in vitro proliferation of
murine B16F10 melanoma cells. Acta Endocrinol (Copenh). 10:545–558.
2014.
|
|
7
|
Neagu M: The immune system - a hidden
treasure for biomarker discovery in cutaneous melanoma. Adv Clin
Chem. 58:89–140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holtan SG, Creedon DJ, Thompson MA, Nevala
WK and Markovic SN: Expansion of CD16-negative natural killer cells
in the peripheral blood of patients with metastatic melanoma. Clin
Dev Immunol. 2011:3163142011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Imai K, Matsuyama S, Miyake S, Suga K and
Nakachi K: Natural cytotoxic activity of peripheral-blood
lymphocytes and cancer incidence: An 11-year follow-up study of a
general population. Lancet. 356:1795–1799. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dons'koi BV, Chernyshov VP and Osypchuk
DV: Measurement of NK activity in whole blood by the CD69
upregulation after co-incubation with K562, comparison with NK
cytotoxicity assays and CD107a degranulation assay. J Immunol
Methods. 372:187–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fehniger TA, Cooper MA, Nuovo GJ, Cella M,
Facchetti F, Colonna M and Caligiuri MA: CD56bright
natural killer cells are present in human lymph nodes and are
activated by T cell-derived IL-2: A potential new link between
adaptive and innate immunity. Blood. 101:3052–3057. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Colucci F, Caligiuri MA and Di Santo JP:
What does it take to make a natural killer? Nat Rev Immunol.
3:413–425. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ursaciuc C, Surcel M, Huică R, Ciotaru D,
Dobre M, Pîrvu IR, Cirimbei C, Mischianu D, Bratu O, Isvoranu G, et
al: CD56dim/CD56bright NK cell subpopulations
and CD16/CD57 expression correlated with tumor development stages.
S East Eur J Immunol. 2016:1–5. 2016. View Article : Google Scholar
|
|
14
|
Hayakawa Y and Smyth MJ: CD27 dissects
mature NK cells into two subsets with distinct responsiveness and
migratory capacity. J Immunol. 176:1517–1524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Inngjerdingen M, Kveberg L, Naper C and
Vaage JT: Natural killer cell subsets in man and rodents. Tissue
Antigens. 78:81–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun JC and Lanier LL: NK cell development,
homeostasis and function: Parallels with CD8+ T cells.
Nat Rev Immunol. 11:645–657. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chiossone L, Chaix J, Fuseri N, Roth C,
Vivier E and Walzer T: Maturation of mouse NK cells is a 4-stage
developmental program. Blood. 113:5488–5496. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
National Research Council: Animal care and
use program. In: Guide for the Care and Use of Laboratory Animals.
8th. National Academy Press; Washington, DC: pp. 11–35. 2011
|
|
19
|
Faustino-Rocha A, Oliveira PA,
Pinho-Oliveira J, Teixeira-Guedes C, Soares-Maia R, da Costa RG,
Colaço B, Pires MJ, Colaço J, Ferreira R and Ginja M: Estimation of
rat mammary tumor volume using caliper and ultrasonography
measurements. Lab Animal. 42:217–224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Freud AG, Zhao S, Wei S, Gitana GM,
Molina-Kirsch HF, Atwater SK and Natkunam Y: Expression of the
activating receptor, NKp46 (CD335), in human natural killer and
T-cell neoplasia. Am J Clin Pathol. 140:853–866. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hadad U, Thauland TJ, Martinez OM, Butte
MJ, Porgador A and Krams SM: NKp46 clusters at the immune synapse
and regulates NK cell polarization. Front Immunol. 6:4952015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
North J, Bakhsh I, Marden C, Pittman H,
Addison E, Navarrete C, Anderson R and Lowdell MW: Tumor-primed
human natural killer cells lyse NK-resistant tumor targets:
Evidence of a two-stage process in resting NK cell activation. J
Immunol. 178:85–94. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iizuka K, Chaplin DD, Wang Y, Wu Q, Pegg
LE, Yokoyama WM and Fu YX: Requirement for membrane lymphotoxin in
natural killer cell development. Proc Natl Acad Sci USA.
96:6336–6340. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fogel LA, Sun MM, Geurs TL,
Carayannopoulos LN and French AR: Markers of nonselective and
specific NK cell activation. J Immunol. 190:6269–6276. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Burt BM, Plitas G, Stableford JA, Nguyen
HM, Bamboat ZM, Pillarisetty VG and DeMatteo RP: CD11c identifies a
subset of murine liver natural killer cells that responds to
adenoviral hepatitis. J Leukoc Biol. 84:1039–1046. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blasius AL, Barchet W, Cella M and Colonna
M: Development and function of murine
B220+CD11c+NK1.1+ cells identify
them as a subset of NK cells. J Exp Med. 204:2561–2568. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Michel T, Poli A, Cuapio A, Briquemont B,
Iserentant G, Ollert M and Zimmer J: Human CD56bright NK
cells: An update. J Immunol. 196:2923–2931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Richards JO, Chang X, Blaser BW, Caligiuri
MA, Zheng P and Liu Y: Tumor growth impedes natural-killer-cell
maturation in the bone marrow. Blood. 108:246–252. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Clinthorne JF, Beli E, Duriancik DM and
Gardner EM: NK cell maturation and function in C57BL/6 mice are
altered by caloric restriction. J Immunol. 190:712–722. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huntington ND, Tabarias H, Fairfax K,
Brady J, Hayakawa Y, Degli-Esposti MA, Smyth MJ, Tarlinton DM and
Nutt SL: NK cell maturation and peripheral homeostasis is
associated with KLRG1 upregulation. J Immunol. 178:4764–4770. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ballas ZK, Buchta CM, Rosean TR, Heusel JW
and Shey MR: Role of NK cell subsets in organ-specific murine
melanoma metastasis. PLoS One. 8:e655992013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Alter G, Malenfant JM and Altfeld M:
CD107a as a functional marker for the identification of natural
killer cell activity. J Immunol Methods. 294:15–22. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Neagu M, Constantin C and Longo C:
Chemokines in the melanoma metastasis biomarkers portrait. J
Immunoassay Immunochem. 36:559–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Neagu M, Constantin C and Tanase C:
Immune-related biomarkers for diagnosis/prognosis and therapy
monitoring of cutaneous melanoma. Expert Rev Mol Diagn. 10:897–919.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neagu M, Caruntu C, Constantin C, Boda D,
Zurac S, Spandidos DA and Tsatsakis AM: Chemically induced skin
carcinogenesis: Updates in experimental models (Review). Oncol Rep.
35:2516–2528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zurac S, Neagu M, Constantin C, Cioplea M,
Nedelcu R, Bastian A, Popp C, Nichita L, Andrei R, Tebeica T, et
al: Variations in the expression of TIMP1, TIMP2 and TIMP3 in
cutaneous melanoma with regression and their possible function as
prognostic predictors. Oncol Lett. 11:3354–3360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Isvoranu G, Marinescu B, Surcel M,
Ursaciuc C and Manda G: Immunotherapy in cancer - in vivo study of
the anti-tumor activity of the IL-15/IL-15R alfa combination in an
experimental model of melanoma. Farmacia. 63:631–636. 2015.
|
|
38
|
Isvoranu G: The memory activation of NK
cells: New methods in cancer immunotherapy. Immunotherapy: Myths,
Reality, Ideas, Future. Metodiev K: IntechOpen, London: pp.
201–219. 2017
|
|
39
|
Zaharescu I, Moldovan AD and Tanase C:
Natural killer (NK) cells and their involvement in different types
of cancer. Current status of clinical research. J Mind Med Sci.
4:31–37. 2017. View Article : Google Scholar
|
|
40
|
Tanase CP, Albulescu R and Neagu M:
Application of 3D hydrogel microarrays in molecular diagnostics:
Advantages and limitations. Expert Rev Mol Diagn. 11:461–464. 2011.
View Article : Google Scholar : PubMed/NCBI
|