Introduction
Various factors such as genetic variations and
changes in mRNA expression patterns are known to influence drug
efficacy, and such factors have been studied extensively. However,
drug responses are affected by changes in the conformation,
localization, and expression of numerous proteins, which are
regulated by mutations and mRNA expression levels. In 2014, Zhang
et al reported that only 32% of the genes showed
statistically significant positive mRNA-protein correlation in 86
CRC samples (1). Therefore,
proteomics analysis-the direct evaluation of the expression levels
and modifications of proteins-has been focused on recently as a
powerful exploration method for identifying predictive biomarkers
for the efficacy of chemotherapeutic drugs.
The expression levels of some serum proteins have
been reported to be useful indicators of sensitivity to
chemotherapy for cancer, and Li et al reported that
variation in serum LDH level was useful as a predictive biomarker
of efficacy of bevacizumab in non-small cell lung cancer (NSCLC)
patients (2). Furthermore, proteomic
studies analyzing several serum proteins using matrix-assisted
laser desorption/ionization mass spectrometry (VeriStrat; Biodesix,
Boulder, CO), classified NSCLC patients treated with erlotinib, an
epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor,
into two groups with good or poor prognosis (3–10).
However, serum proteins derived from tumor tissue and circulating
in blood stably represent only a small fraction of total protein
derived from a tumor. Thus, to find more suitable predictive
biomarkers for sensitivity to anticancer drugs, the analysis should
include not only proteins released into blood but all proteins
derived from a tumor.
In 2015, Sun et al (11) reported that a high level of L-lactate
dehydrogenase B (LDHB) expression in tumor tissue was associated
with poor overall survival in oral cancer patients treated with
paclitaxel, and Ferrer et al (12) also reported that 17 kDa
membrane-associated protein expression in tumor tissue could
predict sensitivity to platinum-based therapy, EGFR inhibitors, and
the proteasome inhibitor bortezomib in lung adenocarcinoma in 2018.
In a meta-analysis of clinical studies, Li et al (13) showed that aldehyde dehydrogenase 1
could be a predictor of response to neoadjuvant chemotherapy in
breast cancer. Furthermore, in recent years, proteins in tumor
cells have been comprehensively analyzed. Yu et al (14) identified some predictive marker
proteins for sensitivity to platinum-containing drugs in patients
with ovarian cancer. Moreover, Chauvin et al (15) detected predictive marker proteins for
the efficacy of 5-fluorouracil (5-FU) in patients with locally
advanced rectal cancer. As mentioned above, the usefulness of
proteome analysis of tumor tissues to develop predictive markers
for the efficacy of certain small molecule drugs has been
demonstrated. On the other hand, although L-lactate dehydrogenase A
(LDHA) expression levels in tumors have been reported to correlate
with cetuximab sensitivity in patients with Ewing's sarcoma
(16) and gankyrin has been reported
to contribute to resistance to chemotherapy containing bevacizumab
in CRC (17), comprehensive proteome
analysis to identify predictive biomarkers for the efficacy of
antibody drugs has not been conducted.
Anti-EGFR monoclonal antibodies (anti-EGFR mAbs),
including cetuximab and panitumumab, are key drugs in the treatment
of colorectal cancer (CRC) and are highly effective for some CRC
patients. On the other hand, anti-EGFR mAbs are very expensive, and
are also known to cause serious adverse effects such as an infusion
reaction or skin rash. Therefore, these drugs should be used only
for patients in which an effective response is expected. Many
clinical trials have concluded that variations in the KRAS
gene are a crucial factor affecting the clinical efficacy of
anti-EGFR mAbs. Anti-EGFR mAbs have been recommended to be used for
wild-type KRAS CRC patients, approximately 60% of all CRC
patients. However, more than 50% of patients with wild-type
KRAS tumors do not receive a therapeutic benefit from
anti-EGFR mAbs (18–22). Even when variations of NRAS,
BRAF, and PIK3CA mutations or EGFR overexpression are
taken into account, the primary cause of resistance to anti-EGFR
mAbs in more than half of wild-type KRAS CRC patients with
poor drug responses remains unknown (20,23–28). As
mentioned above, although genome research is widely performed and
has many advantages such as the ease of the procedure, it has
become clear that fluctuation in protein expression, which directly
affects drug efficacy, cannot be explained with genome information
alone. Thus, factors affecting drug efficacy other than genetic
factors need to be studied through a comprehensive analysis, such
as a proteomic approach.
In this study, we focused on time-of-flight (TOF)
mass spectrometry (MS) which can be used for non-target protein
analysis and comprehensive protein analysis, and performed
comprehensive analysis of proteins derived from CRC cell lines
without genetic mutations affecting sensitivity to anti-EGFR mAbs,
such as KRAS, NRAS, BRAF, and PIK3CA mutations, and
PTEN overexpression. Finally, we explored some tumor-specific
proteins that were correlated with sensitivity to anti-EGFR
mAbs.
Materials and methods
Materials
CACO2, C10, HT55, and C99 cell lines were purchased
from the European Collection of Cell Cultures (Salisbury, UK). The
COLO320DM cell line was purchased from the Japanese Collection of
Research Bioresources Cell Bank (Osaka, Japan). The SW48 cell line
was obtained from the American Type Culture Collection (Manassas,
VA, USA). These cell lines were classified into three groups
according to cetuximab sensitivity: C99 and SW48 in the
cetuximab-sensitive group (Cmab-S), C10 and HT55 in the
cetuximab-partial resistance group (Cmab-PR), and CACO2 and
COLO320DM in the cetuximab-resistance group (Cmab-R). Reagents for
culture [culture medium, fetal bovine serum (FBS), and supplements]
and sample preparation (dithiothreitol, iodoacetamide, and trypsin)
were purchased from Wako Pure Chemical Industries (Osaka, Japan).
All other reagents were obtained from commercial sources, and those
used to analyze peptides were graded for high-performance liquid
chromatography, liquid chromatography-tandem mass spectrometry
(LC-MS/MS), or analytical use. Standard peptides used for MS
analysis were synthesized by Eurofins Genomics Japan (Tokyo,
Japan).
Cell culture and sample
preparation
Six CRC cell lines with different sensitivities to
cetuximab were used in this study (Table
I) (29). CACO2, COLO320DM, C10,
HT55, and C99 cells were cultured in a humidified incubator at 37°C
in the presence of 5% CO2, while SW48 cells were
cultured in a 37°C incubator with no supplemental CO2.
Cetuximab-acquired resistance cell lines (SW48R and C99R) were
generated upon continuous exposure of SW48 and C99 cell lines to
cetuximab according to the method of Troiani et al (30). Briefly, for a period of 8 months,
SW48 and C99 cells were continuously exposed to cetuximab to
increase the inhibition of 50% of cancer cell growth
(IC50), and the final concentration was 12.8 µg/ml.
Cytoplasmic and membrane proteins were extracted from 80% confluent
cell lines using a Minute Plasma Membrane Protein Isolation kit
(Invent Biotechnologies, Inc., Plymouth, MN, USA), and the
concentrations of the extracted proteins were measured using a DC™
Protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cytoplasmic and membrane protein extracts were diluted to 2.0 and
1.0 mg/ml, respectively, and assayed immediately or stored at −80°C
until assay.
 | Table I.Cetuximab sensitivities and culture
of six CRC cell lines. |
Table I.
Cetuximab sensitivities and culture
of six CRC cell lines.
| Cell line | Cetuximab
sensitivity | Growth inhibition
relative to control (%) | Culture medium |
|---|
| CACO2 | Resistant | 0 | EMEM + 1% NEAA +
10% FBS |
| COLO320DM | Resistant | 0 | RPMI1640 + 10%
FBS |
| C10 | Partially
resistant | 5.9 | IMDM + 10% FBS |
| HT55 | Partially
resistant | 21.3 | EMEM + 1% NEAA +
20% FBS |
| SW48 | Sensitive | 69.1 | L15 + 10% FBS |
| C99 | Sensitive | 96.8 | IMDM + 10% FBS |
Mass spectrometry analysis
As a pretreatment for cytoplasmic and membrane
protein samples, protein samples (45 µl) were mixed with 1 mg/ml
infliximab (5 µl) and an internal standard (ISTD), and incubated at
37°C for 30 min with urea (41 mg) and 40 mg/ml dithiothreitol (7.7
µl) in 8 mol/l urea/0.5 mol/l Tris HCl (pH 8.5) to reduce disulfide
bonds. Reduced samples were alkylated by reaction with 40 mg/ml
iodoacetamide in 8 mol/l urea/0.5 mol/l Tris-HCl (19.2 µl; pH 8.5)
for 30 min at 37°C. Subsequently, to digest the proteins,
cytoplasmic and membrane protein samples (300 µl each, diluted
4-fold with Milli-Q water) were trypsinized by adding trypsin
solution (42 and 22 µl, respectively; 100 µg/ml in 20 mmol/l acetic
acid) and incubating at 37°C for 12 h. Surfactants in the membrane
protein samples were removed with Detergent OUT™ (Takara Bio, Inc.,
Shiga, Japan), and trypsinized samples were desalted using a
MonoSpin C18 column (GL Sciences, Inc., Tokyo, Japan).
Peptide fragments analysis was performed using
liquid chromatography-time-of-flight mass spectrometry (LC-TOF MS)
consisting of an ACQUITY UPLC (Waters, Milford, MA, USA) for LC and
LCT Premier XE (Waters) for TOF MS. The LC conditions were as
follows: Column, ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7
µm) (Waters); column temperature, 40°C; mobile phase, 0.1% formic
acid in Milli-Q water (A) and 0.1% formic acid in acetonitrile (B);
flow rate, 0.3 ml/min; and gradient program, 5 to 95% B in 55 min
and 95 to 5% B in 3 min. MS analysis was performed using an
electrospray ionization (ESI) source in positive ionization mode (W
mode). Survey scans were acquired in the range 100 to 2000 m/z.
Instrument settings were as follows: Capillary voltage, 3000 V;
sample cone voltage, 50 V; desolvation temperature, 350°C; source
temperature, 120°C; cone gas flow, 60 l/h; desolvation gas flow,
700 l/h; and aperture 1 voltage, 15 V.
Tandem quadrupole MS was used to analyze specific
peptide fragments. LC was performed with an ACQUITY UPLC system
(Waters). An ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 µm)
(Waters) was used as the LC column. The LC conditions were as
follows: Column temperature, 40°C; mobile phase, 0.1% formic acid
in Milli-Q water (A) and 0.1% formic acid in acetonitrile (B); flow
rate, 0.5 ml/min; and gradient program, 5 to 35% B in 6 min, 35 to
95% B in 1 min, and 95 to 5% B in 1 min. XevoTQ (Waters) with ESI
turbo spray in positive ionization mode was used, and the
ionization parameters were as follows: Capillary voltage, 1000 V;
desolvation temperature, 500°C; source temperature, 150°C;
desolvation gas flow, 1000 l/h; and cone gas flow, 70 l/h. The
transitions m/z 1176.53>130.85 for LDHB and m/z 835.44>175.20
for ISTD were monitored. Sample cone voltage and collision energy
were 74 and 80 V for LDHB and 35 and 55 V for ISTD,
respectively.
Proteomics analysis
Principal component analysis (PCA) and orthogonal
projections for latent structure-discriminant analysis (OPLS-DA)
using MarkerLynx XS software (Waters) were performed on all peaks
obtained from Cmab-R and Cmab-S cell lines by LC-TOF MS analysis
with the following parameters: Initial retention time, 0.1 min;
final retention time, 55.0 min; peak width at 5% height, 3.00 s;
peak-to-peak baseline noise, 50.00; mass tolerance, 0.05 Da;
intensity threshold, 10 counts; mass window, 0.05 Da; and retention
time window, 0.10 min. In OPLS-DA, the peaks were grouped into
Cmab-R and Cmab-S, and cut-off values for Pearson's correlation
coefficients ± 0.8 were used to define the peaks associated with
sensitivity to cetuximab. Among the selected peaks, the peaks
without isotopic peaks were excluded, and the peaks with
intensities that were significantly correlated with cetuximab
sensitivity in the six CRC cell lines, including Cmab-PR, were
finally extracted. Thereafter, the proteins composing the extracted
peaks were determined using MS fit analysis (ProteinProspector;
http://prospector.ucsf.edu/prospector/mshome.htm).
Statistical analysis
Correlations between the peak intensities and growth
inhibitory concentration of cetuximab, an index of cetuximab
sensitivity, were evaluated based on Pearson's correlation
coefficient. Differences in LDHB expression levels between groups
were evaluated by One-way analysis of variance (ANOVA) and
Mann-Whitney U test. The Tukey-Kramer test was used for post hoc
analysis. Statistical analysis was performed using SPSS software
version 24.0 (SPSS, Inc., Chicago, IL, USA). A P-value less than
0.05 was considered statistically significant.
Results
Exploration of predictive marker
proteins
PCA was performed on data of the MS peaks obtained
from Cmab-R and Cmab-S using LC-TOF MS. A total of 1,599 and 1,012
peaks that were specific to cytoplasmic proteins of Cmab-R and
Cmab-S, respectively, and 2,370 and 3,478 peaks that were specific
to membrane proteins of Cmab-R and Cmab-S, respectively, were
detected. Then, we extracted 271 and 197 peaks as monoisotopic ion
peaks that were specific to cytoplasmic proteins of Cmab-R and
Cmab-S, respectively, and excluded other peaks as isotopic peaks or
noise peaks. Similarly, 533 and 422 peaks were extracted as
monoisotopic peaks that were specific to membrane proteins of
Cmab-R and Cmab-S, respectively. Finally, 148 and 146 peaks from
cytoplasmic proteins and 363 and 267 peaks from membrane proteins
were extracted as candidate specific peaks to Cmab-R and Cmab-S,
respectively, the intensities of which were significantly
correlated with cetuximab sensitivity in the six CRC cell lines.
Candidate proteins composed of those peaks were identified by MS
fit analysis, and LDHB in the cytoplasmic fraction showed the
highest MOWSE score, an indicator of the probability of protein
identification (Table II).
 | Table II.Candidate marker proteins for
cetuximab sensitivity. |
Table II.
Candidate marker proteins for
cetuximab sensitivity.
| No. | MOWSE score | % Coverage | Protein name | Accession no. | Fraction | Marker |
|---|
| 1 | 7198 | 14.4 | L-lactate
dehydrogenase B chain | P07195 | Cytoplasm | Cmab-R |
| 2 | 911 | 10.4 | Uncharacterized
protein C18orf63 | Q68DL7 | Membrane | Cmab-S |
| 3 | 573 | 7.1 |
Serine/threonine-protein kinase TAO1 | Q7L7X3 | Membrane | Cmab-S |
| 4 | 303 | 4 | Protein phosphatase
Slingshot homolog 1 | Q8WYL5 | Membrane | Cmab-S |
| 5 | 273 | 4.9 | Coiled-coil
domain-containing protein 110 | Q8TBZ0 | Cytoplasm | Cmab-S |
| 6 | 242 | 5.4 | Pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX16 | O60231 | Membrane | Cmab-S |
| 7 | 240 | 4.6 | Exocyst complex
component 6B | Q9Y2D4 | Membrane | Cmab-S |
| 8 | 237 | 7.9 | Zinc finger protein
771 | Q7L3S4 | Membrane | Cmab-R |
| 9 | 224 | 13.5 | Protein LIAT1 | Q6ZQX7 | Membrane | Cmab-S |
| 10 | 223 | 8.7 | Kelch-like protein
41 | O60662 | Membrane | Cmab-S |
Evaluation of the correlation between
peak intensity of an LDHB-specific peptide and cetuximab
sensitivity
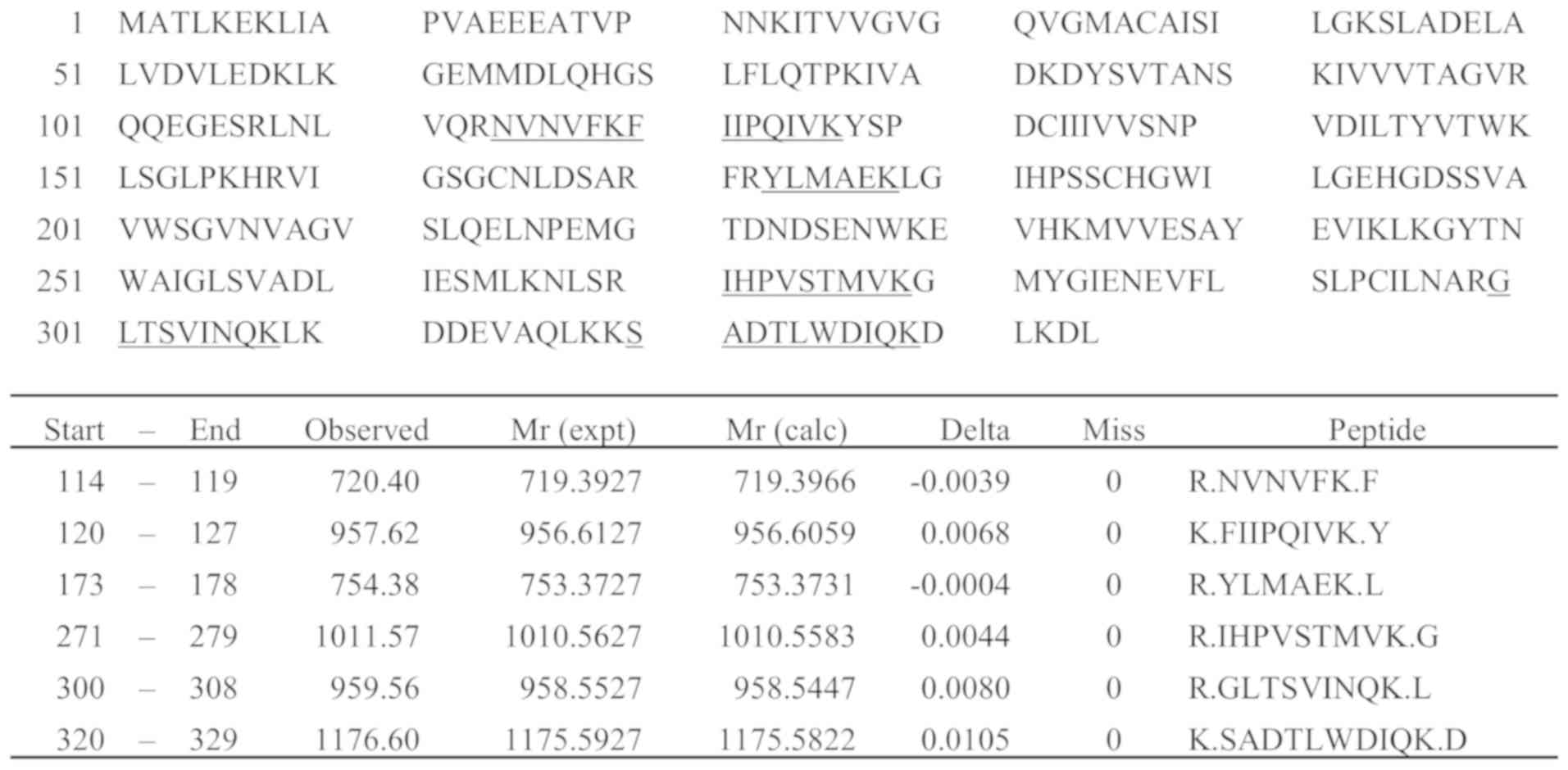
From the final candidate peaks, six (m/z 720.40,
957.62, 754.38, 1011.57, 959.56, and 1176.60) were extracted as
peptide peaks derived from LDHB by MS fit analysis, and an
LDHB-specific peptide, SADTLWDIQK m/z 1176.60, was analyzed by
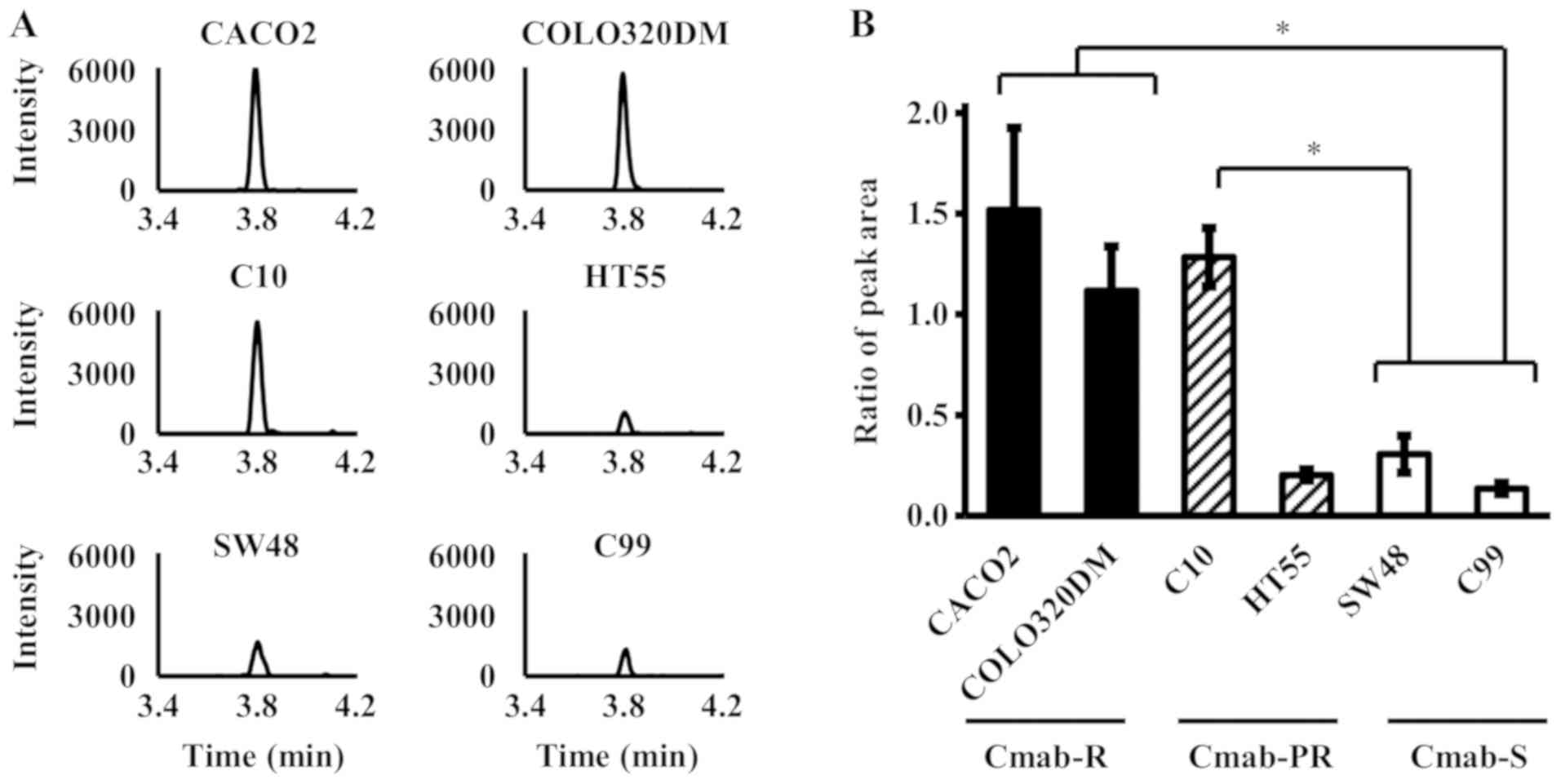
LC-MS/MS (Fig. 1). As a result, the
peak intensity of SADTLWDIQK was significantly higher in Cmab-R
than in Cmab-S (P<0.05, One-way ANOVA) (Fig. 2). Only C10, but not C99, was
significantly higher in Cmab-PR than in Cmab-S (P<0.05, One-way
ANOVA) (Fig. 2). In addition, the
data obtained by LC-MS/MS were consistent with data obtained by
analysis using enzyme-linked immunosorbent assay (ELISA) (data not
shown).
Relationship between acquired
cetuximab resistance and LDHB expression
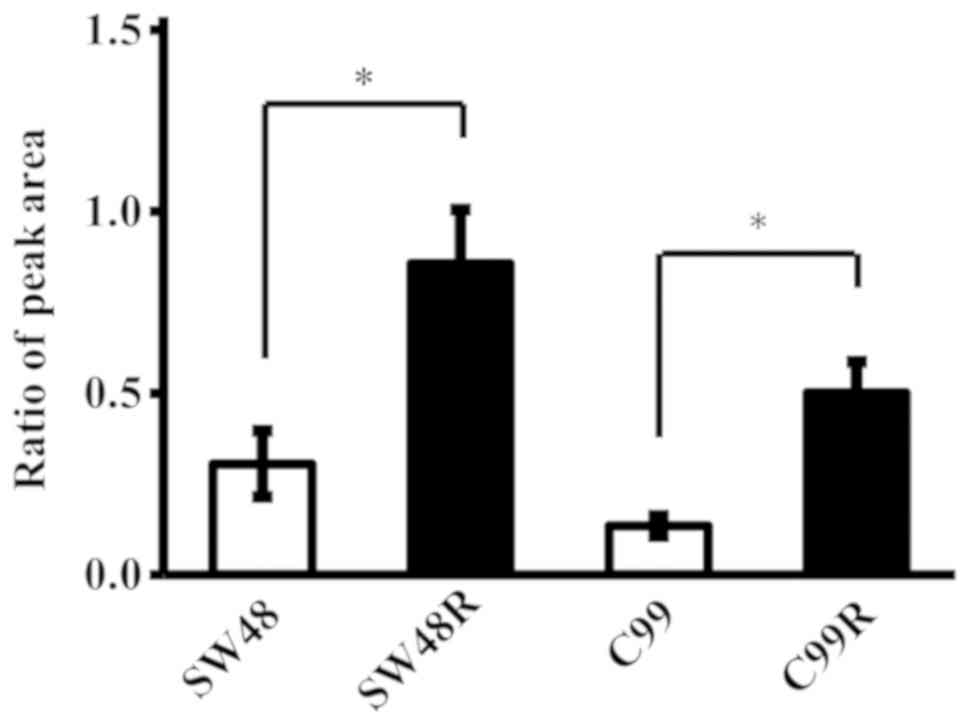
We compared the expression levels of LDHB in the
SW48R and C99R cell lines, which acquired resistance to cetuximab,
and in the original cell lines. LDHB expression levels were
significantly upregulated in the CRC cell lines that developed
cetuximab resistance when compared to the original cell lines
(P<0.05, Mann-Whitney U test) (Fig.
3). In addition, the data obtained using LC-MS/MS were
consistent with data obtained by analysis using ELISA (data not
shown).
Discussion
We showed that the expression level of LDHB may be a
predictive biomarker for sensitivity to anti-EGFR mAbs by proteome
analysis for the first time. Furthermore, our findings suggested
that LDHB may also be involved in acquired resistance to anti-EGFR
mAbs.
LDH is a key glycolytic enzyme catalyzing the
conversion of pyruvate acid to lactic acid, and is composed of two
major subunits, LDHA (muscle type, M subunit) and LDHB (heart type,
H subunit). LDHA is reported to be involved in cell proliferation
and metabolism in most cancers (31–34),
while LDHB expression varies greatly among cancer types (35–40). In
2012, LDHB expression was found to be correlated with cell growth
in lung cancer with KRAS mutations (41), and its usefulness as a prognostic
factor for triple-negative breast cancer, osteosarcoma and NSCLC
was also reported by Li et al (35), McCleland et al (38), and Koh et al (42). Moreover, the effect of LDHB on the
efficacy of some anticancer drugs has been investigated, and LDHB
expression levels are reported to be correlated with the
pathological complete response ratio, disease-free survival, and
poor overall survival in patients treated with small molecule
anticancer drugs, such as anthracycline and taxane (11,43).
The activation of LDH has been reported to enhance
the Warburg effect (11,44,45). The
Warburg effect causes the activation of intracellular stress
signaling pathways by increasing glucose consumption and inducing
hypoglycemia, and its activation is also known to lead to the
development of resistance against anticancer drugs, such as 5-FU,
carboplatin, and topoisomerase inhibitors (46–48).
Furthermore, enhanced heat shock protein 90 activation, a molecular
chaperone evoked during stress reactions, by the Warburg effect has
also been reported to be involved in the development of resistance
to anticancer drugs by activating client proteins, such as PI3K/Akt
(49–51).
Unlike many previous studies of the effects of LDHB
expression on prognosis or response to chemotherapy, which focused
on LDHB alone, we demonstrated that the level of LDHB expression
correlated with sensitivity to anti-EGFR mAbs by comprehensive
analysis of proteins derived from CRC cell lines. Furthermore, LDHB
expression levels were significantly upregulated with the
acquisition of resistance to cetuximab in Cmab-S CRC cell lines.
These results indicated that high LDHB expression was particularly
important for the acquisition of drug resistance among many key
factors involved in the development of drug resistance. In
contrast, the LDHB expression level was low in the partially
resistant HT55 cell line, and these data indicated that cetuximab
sensitivity could not be evaluated with LDHB expression level
alone. Although the cause of partial resistance to cetuximab in
HT55 has not been clarified, various factors have been reported to
be associated with resistance to cancer chemotherapy. Since primary
resistance to drugs is known to be caused by so many factors,
primary resistance to cetuximab in HT55 could be caused by many
factors in addition to the increase of LDHB. Further detailed
investigations are needed to examine drug resistance in this cell
line.
One limitation of our study is that we could not
clarify whether the drug resistance caused the increase in LDHB
levels or whether increased levels of LDHB led to drug resistance.
As mentioned above, LDHB is known to be an important factor related
to the Warburg effect, and LDHB expression in tumor cells is
reported to be increased to facilitate survival in hypoxic and
low-energy conditions associated with insufficient angiogenesis.
Since our results are based on an in vitro study, the
survival environment of cancer cells is considered to be unchanged
at least with respect to oxygen concentration and nutritional
condition. In addition, LDHB protein level was low in the partially
resistant HT55 cell line. According to these data, resistance to
cetuximab was considered to be developed following changes
associated with increased LDHB expression levels but LDHB protein
level did not increase with the acquisition of resistance to
cetuximab. The report by Sun et al (11) showed that LDHB deletion sensitized
oral squamous cell carcinoma cell lines to taxane, whereas the
introduction of LDHB decreased sensitivity to taxane. Furthermore,
Lu et al (52) also reported
that cetuximab showed antitumor efficacy due to downregulation of
the α subunit of HIF-1 (HIF-1α). This in turn regulated the
expression of LDHA and inhibition of glycolysis in
cetuximab-sensitive head and neck squamous cell carcinoma cells in
a HIF-1α downregulation-dependent manner; these reports support our
hypothesis. However, we could not prove our hypothesis with our
data, and so a more detailed study of the role of LDHB in
resistance to cetuximab is needed. In addition, because the
mRNA-protein correlation with pyruvate metabolism in the Kyoto
Encyclopedia of Genes and Genomes pathway, including LDHB, was
reported to be low (1), we consider
that development of an easy method to detect fluctuation in LDHB
protein levels, other than analysis of mRNA, is needed for clinical
application.
In conclusion, we found that LDHB may be an
important factor affecting cetuximab sensitivity using
comprehensive proteome analysis for the first time. We believe that
these data could contribute to the improvement of chemotherapy for
CRC patients and promote the development of a method to overcome
resistance to anti-EGFR mAbs.
Acknowledgements
Not applicable.
Funding
This work was supported by JSPS KAKENHI (grant nos.
JP16H00504 and JP15H00498).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
KY, TA and DN conceived and designed the
experiments. AN and MM performed the experiments. AN and TA
analyzed the data and wrote the paper. KY, TA and DN revised the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi
Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al:
Proteogenomic characterization of human colon and rectal cancer.
Nature. 513:382–387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li B, Li C, Guo M, Shang S, Li X, Xie P,
Sun X, Yu J and Wang L: Predictive value of LDH kinetics in
bevacizumab treatment and survival of patients with advanced NSCLC.
Onco Targets Ther. 11:6287–6294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gregorc V, Novello S, Lazzari C, Barni S,
Aieta M, Mencoboni M, Grossi F, De Pas T, de Marinis F, Bearz A, et
al: Predictive value of a proteomic signature in patients with
non-small-cell lung cancer treated with second-line erlotinib or
chemotherapy (PROSE): A biomarker-stratified, randomised phase 3
trial. Lancet Oncol. 15:713–721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lazzari C, Spreafico A, Bachi A, Roder H,
Floriani I, Garavaglia D, Cattaneo A, Grigorieva J, Viganò MG,
Sorlini C, et al: Changes in plasma mass-spectral profile in course
of treatment of non-small cell lung cancer patients with epidermal
growth factor receptor tyrosine kinase inhibitors. J Thorac Oncol.
7:40–48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garrisi VM, Bongarzone I, Mangia A,
Cremona M, De Bortoli M, Vaghi E, Galetta D, Pastorino U, Quaranta
M, Abbate I and Paradiso A: Characterization of a serum protein
pattern from NSCLC patients treated with Gefitinib. Clin Biochem.
44:936–940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pitteri SJ, Amon LM, Busald Buson T, Zhang
Y, Johnson MM, Chin A, Kennedy J, Wong CH, Zhang Q, Wang H, et al:
Detection of elevated plasma levels of epidermal growth factor
receptor before breast cancer diagnosis among hormone therapy
users. Cancer Res. 70:8598–8606. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chung CH, Seeley EH, Roder H, Grigorieva
J, Tsypin M, Roder J, Burtness BA, Argiris A, Forastiere AA,
Gilbert J, et al: Detection of tumor epidermal growth factor
receptor pathway dependence by serum mass spectrometry in cancer
patients. Cancer Epidemiol Biomarkers Prev. 19:358–365. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taguchi F, Solomon B, Gregorc V, Roder H,
Gray R, Kasahara K, Nishio M, Brahmer J, Spreafico A, Ludovini V,
et al: Mass spectrometry to classify non-small-cell lung cancer
patients for clinical outcome after treatment with epidermal growth
factor receptor tyrosine kinase inhibitors: A multicohort
cross-institutional study. J Natl Cancer Inst. 99:838–846. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grossi F, Genova C, Rijavec E, Barletta G,
Biello F, Dal Bello MG, Meyer K, Roder J, Roder H and Grigorieva J:
Prognostic role of the VeriStrat test in first line patients with
non-small cell lung cancer treated with platinum-based
chemotherapy. Lung Cancer. 117:64–69. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fidler MJ, Fhied CL, Roder J, Basu S,
Sayidine S, Fughhi I, Pool M, Batus M, Bonomi P and Borgia JA: The
serum-based VeriStrat® test is associated with
proinflammatory reactants and clinical outcome in non-small cell
lung cancer patients. BMC Cancer. 18:3102018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun W, Zhang X, Ding X, Li H, Geng M, Xie
Z, Wu H and Huang M: Lactate dehydrogenase B is associated with the
response to neoadjuvant chemotherapy in oral squamous cell
carcinoma. PLoS One. 10:e01259762015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferrer I, Quintanal-Villalonga Á,
Molina-Pinelo S, Garcia-Heredia JM, Perez M, Suárez R, Ponce-Aix S,
Paz-Ares L and Carnero A: MAP17 predicts sensitivity to
platinum-based therapy, EGFR inhibitors and the proteasome
inhibitor bortezomib in lung adenocarcinoma. J Exp Clin Cancer Res.
37:1952018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li J, Zhang B, Yang YF, Jin J and Liu YH:
Aldehyde dehydrogenase 1 as a predictor of the neoadjuvant
chemotherapy response in breast cancer: A meta-analysis. Medicine
(Baltimore). 97:e120562018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu KH, Levine DA, Zhang H, Chan DW, Zhang
Z and Snyder M: Predicting ovarian cancer patients' clinical
response to platinum-based chemotherapy by their tumor proteomic
signatures. J Proteome Res. 15:2455–2465. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chauvin A, Wang CS, Geha S, Garde-Granger
P, Mathieu AA, Lacasse V and Boisvert FM: The response to
neoadjuvant chemoradiotherapy with 5-fluorouracil in locally
advanced rectal cancer patients: A predictive proteomic signature.
Clin Proteomics. 15:162018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu J, Jiang H, Wu C, Jiang Y, Xiao L and
Tian Y: Overcoming cetuximab resistance in Ewing's sarcoma by
inhibiting lactate dehydrogenase-A. Mol Med Rep. 14:995–1001. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sakurai T, Komeda Y, Nagai T, Kamata K,
Minaga K, Yamao K, Takenaka M, Hagiwara S, Watanabe T, Nishida N,
et al: Gankyrin contributes to tumorigenesis and chemoresistance in
sporadic colorectal cancer. Digestion. 4:1–9. 2018. View Article : Google Scholar
|
|
18
|
Kawazoe A, Shitara K, Fukuoka S, Kuboki Y,
Bando H, Okamoto W, Kojima T, Fuse N, Yamanaka T, Doi T, et al: A
retrospective observational study of clinicopathological features
of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with
metastatic colorectal cancer. BMC Cancer. 15:2582015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leto SM and Trusolino L: Primary and
acquired resistance to EGFR-targeted therapies in colorectal
cancer: Impact on future treatment strategies. J Mol Med (Berl).
92:709–722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong R and Cunningham D: Using predictive
biomarkers to select patients with advanced colorectal cancer for
treatment with epidermal growth factor receptor antibodies. J Clin
Oncol. 26:5668–5670. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khambata-Ford S, Garrett CR, Meropol NJ,
Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin
AK, et al: Expression of epiregulin and amphiregulin and K-ras
mutation status predict disease control in metastatic colorectal
cancer patients treated with cetuximab. J Clin Oncol. 25:3230–3237.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Price T, Kim TW, Li J, Cascinu S, Ruff P,
Suresh AS, Thomas A, Tjulandin S, Guan X and Peeters M: Final
results and outcomes by prior bevacizumab exposure, skin toxicity,
and hypomagnesaemia from ASPECCT: Randomized phase 3
non-inferiority study of panitumumab versus cetuximab in
chemorefractory wild-type KRAS exon 2 metastatic colorectal cancer.
Eur J Cancer. 68:51–59. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Molinari F, Felicioni L, Buscarino M, De
Dosso S, Buttitta F, Malatesta S, Movilia A, Luoni M, Boldorini R,
Alabiso O, et al: Increased detection sensitivity for KRAS
mutations enhances the prediction of anti-EGFR monoclonal antibody
resistance in metastatic colorectal cancer. Clin Cancer Res.
17:4901–4914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sartore-Bianchi A, Moroni M, Veronese S,
Carnaghi C, Bajetta E, Luppi G, Sobrero A, Barone C, Cascinu S,
Colucci G, et al: Epidermal growth factor receptor gene copy number
and clinical outcome of metastatic colorectal cancer treated with
panitumumab. J Clin Oncol. 25:3238–3245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cunningham D, Humblet Y, Siena S, Khayat
D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype
C, et al: Cetuximab monotherapy and cetuximab plus irinotecan in
irinotecan-refractory metastatic colorectal cancer. N Engl J Med.
351:337–345. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rankin A, Klempner SJ, Erlich R, Sun JX,
Grothey A, Fakih M, George TJ Jr, Lee J, Ross JS, Stephens PJ, et
al: Broad detection of alterations predicted to confer lack of
benefit from EGFR antibodies or sensitivity to targeted therapy in
advanced colorectal cancer. Oncologist. 21:1306–1314. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pietrantonio F, Petrelli F, Coinu A, Di
Bartolomeo M, Borgonovo K, Maggi C, Cabiddu M, Iacovelli R, Bossi
I, Lonati V, et al: Predictive role of BRAF mutations in patients
with advanced colorectal cancer receiving cetuximab and
panitumumab: A meta-analysis. Eur J Cancer. 51:587–594. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rowland A, Dias MM, Wiese MD, Kichenadasse
G, McKinnon RA, Karapetis CS and Sorich MJ: Meta-analysis of BRAF
mutation as a predictive biomarker of benefit from anti-EGFR
monoclonal antibody therapy for RAS wild-type metastatic colorectal
cancer. Br J Cancer. 112:1888–1894. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashraf SQ, Nicholls AM, Wilding JL,
Ntouroupi TG, Mortensen NJ and Bodmer WF: Direct and immune
mediated antibody targeting of ERBB receptors in a colorectal
cancer cell-line panel. Proc Natl Acad Sci USA. 109:21046–21051.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Troiani T, Martinelli E, Napolitano S,
Vitagliano D, Ciuffreda LP, Costantino S, Morgillo F, Capasso A,
Sforza V, Nappi A, et al: Increased TGF-α as a mechanism of
acquired resistance to the anti-EGFR inhibitor cetuximab through
EGFR-MET interaction and activation of MET signaling in colon
cancer cells. Clin Cancer Res. 19:6751–6765. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou X, Chen R, Xie W, Ni Y, Liu J and
Huang G: Relationship between 18F-FDG accumulation and lactate
dehydrogenase A expression in lung adenocarcinomas. J Nucl Med.
55:1766–1771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP
and Huang G: Knockdown of lactate dehydrogenase A suppresses tumor
growth and metastasis of human hepatocellular carcinoma. FEBS J.
279:3898–3910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Le A, Cooper CR, Gouw AM, Dinavahi R,
Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL and Dang
CV: Inhibition of lactate dehydrogenase A induces oxidative stress
and inhibits tumor progression. Proc Natl Acad Sci USA.
107:2037–2042. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fantin VR, St-Pierre J and Leder P:
Attenuation of LDH-A expression uncovers a link between glycolysis,
mitochondrial physiology, and tumor maintenance. Cancer Cell.
9:425–434. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li C, Chen Y, Bai P, Wang J, Liu Z, Wang T
and Cai Q: LDHB may be a significant predictor of poor prognosis in
osteosarcoma. Am J Transl Res. 8:4831–4843. 2016.PubMed/NCBI
|
|
36
|
Chen R, Zhou X, Yu Z, Liu J and Huang G:
Low expression of LDHB correlates with unfavorable survival in
hepatocellular carcinoma: Strobe-compliant article. Medicine
(Baltimore). 94:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cui J, Quan M, Jiang W, Hu H, Jiao F, Li
N, Jin Z and Wang L, Wang Y and Wang L: Suppressed expression of
LDHB promotes pancreatic cancer progression via inducing glycolytic
phenotype. Med Oncol. 32:1432015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McCleland ML, Adler AS, Shang Y, Hunsaker
T, Truong T, Peterson D, Torres E, Li L, Haley B, Stephan JP, et
al: An integrated genomic screen identifies LDHB as an essential
gene for triple-negative breast cancer. Cancer Res. 72:5812–5823.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Leiblich A, Cross SS, Catto JW, Phillips
JT, Leung HY, Hamdy FC and Rehman I: Lactate dehydrogenase-B is
silenced by promoter hypermethylation in human prostate cancer.
Oncogene. 25:2953–2960. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Maekawa M, Taniguchi T, Ishikawa J,
Sugimura H, Sugano K and Kanno T: Promoter hypermethylation in
cancer silences LDHB, eliminating lactate dehydrogenase isoenzymes
1–4. Clin Chem. 49:1518–1520. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McCleland ML, Adler AS, Deming L, Cosino
E, Lee L, Blackwood EM, Solon M, Tao J, Li L, Shames D, et al:
Lactate dehydrogenase B is required for the growth of
KRAS-dependent lung adenocarcinomas. Clin Cancer Res. 19:773–784.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koh YW, Lee SJ and Park SY: Prognostic
significance of lactate dehydrogenase B according to histologic
type of non-small-cell lung cancer and its association with serum
lactate dehydrogenase. Pathol Res Pract. 213:1134–1138. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dennison JB, Molina JR, Mitra S,
González-Angulo AM, Balko JM, Kuba MG, Sanders ME, Pinto JA, Gómez
HL, Arteaga CL, et al: Lactate dehydrogenase B: A metabolic marker
of response to neoadjuvant chemotherapy in breast cancer. Clin
Cancer Res. 19:3703–3713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hirschhaeuser F, Sattler UG and
Mueller-Klieser W: Lactate: A metabolic key player in cancer.
Cancer Res. 71:6921–6925. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou M, Zhao Y, Ding Y, Liu H, Liu Z,
Fodstad O, Riker AI, Kamarajugadda S, Lu J, Owen LB, et al: Warburg
effect in chemosensitivity: Targeting lactate dehydrogenase-A
re-sensitizes taxol-resistant cancer cells to taxol. Mol Cancer.
9:332010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bhattacharya B, Mohd Omar MF and Soong R:
The Warburg effect and drug resistance. Br J Pharmacol.
173:970–979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bhattacharya B, Low SH, Soh C, Kamal
Mustapa N, Beloueche-Babari M, Koh KX, Loh J and Soong R: Increased
drug resistance is associated with reduced glucose levels and an
enhanced glycolysis phenotype. Br J Pharmacol. 171:3255–3267. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hwang JH, Kim JY, Cha MR, Ryoo IJ, Choo
SJ, Cho SM, Tsukumo Y, Tomida A, Shin-Ya K, Hwang YI, et al:
Etoposide-resistant HT-29 human colon carcinoma cells during
glucose deprivation are sensitive to piericidin A, a GRP78
down-regulator. J Cell Physiol. 215:243–250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jacobson C, Kopp N, Layer JV, Redd RA,
Tschuri S, Haebe S, van Bodegom D, Bird L, Christie AL,
Christodoulou A, et al: HSP90 inhibition overcomes ibrutinib
resistance in mantle cell lymphoma. Blood. 128:2517–2526. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Flandrin P, Guyotat D, Duval A, Cornillon
J, Tavernier E, Nadal N and Campos L: Significance of heat-shock
protein (HSP) 90 expression in acute myeloid leukemia cells. Cell
Stress Chaperones. 13:357–364. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sain N, Krishnan B, Ormerod MG, De Rienzo
A, Liu WM, Kaye SB, Workman P and Jackman AL: Potentiation of
paclitaxel activity by the HSP90 inhibitor
17-allylamino-17-demethoxygeldanamycin in human ovarian carcinoma
cell lines with high levels of activated AKT. Mol Cancer Ther.
5:1197–1208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lu H, Li X, Luo Z, Liu J and Fan Z:
Cetuximab reverses the Warburg effect by inhibiting HIF-1-regulated
LDH-A. Mol Cancer Ther. 12:2187–2199. 2013. View Article : Google Scholar : PubMed/NCBI
|