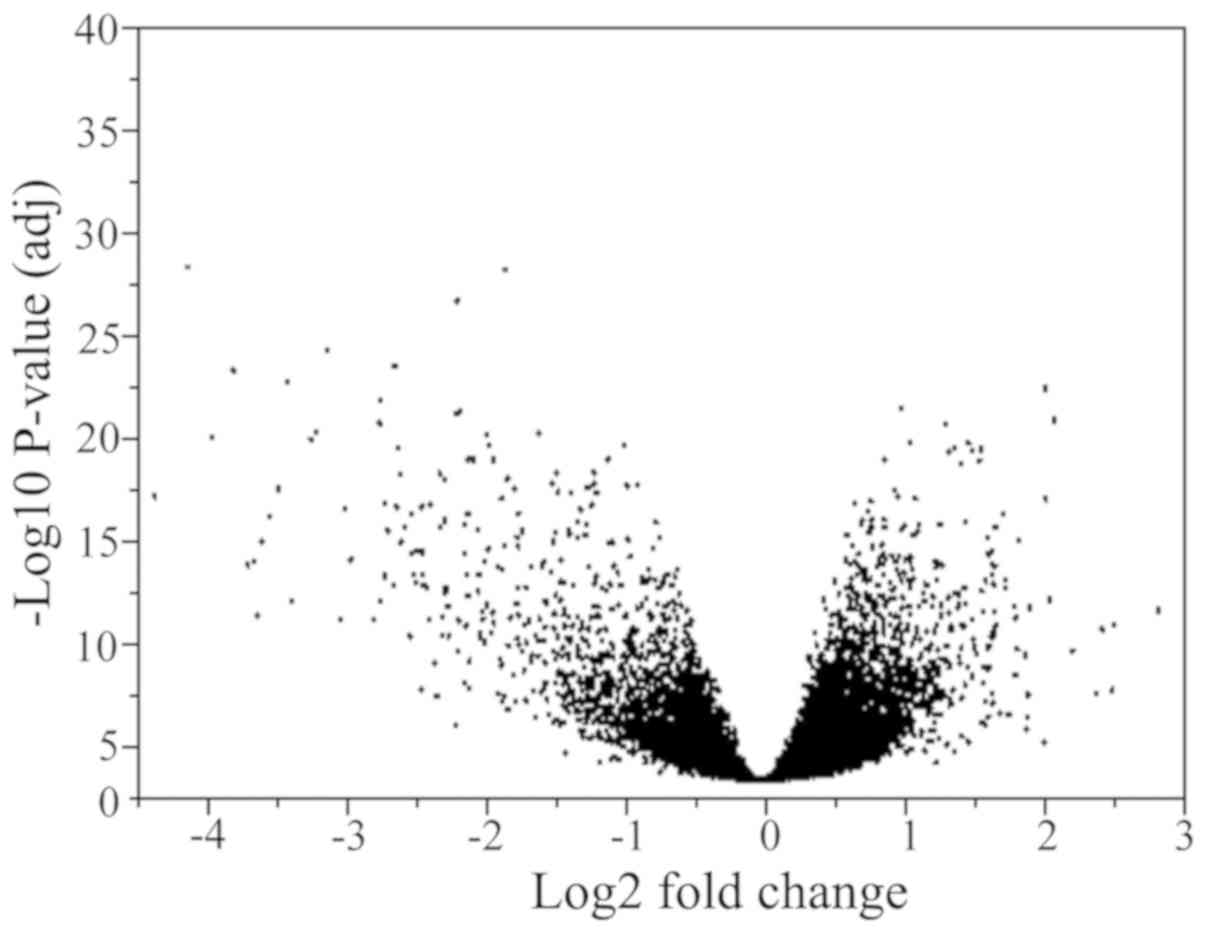
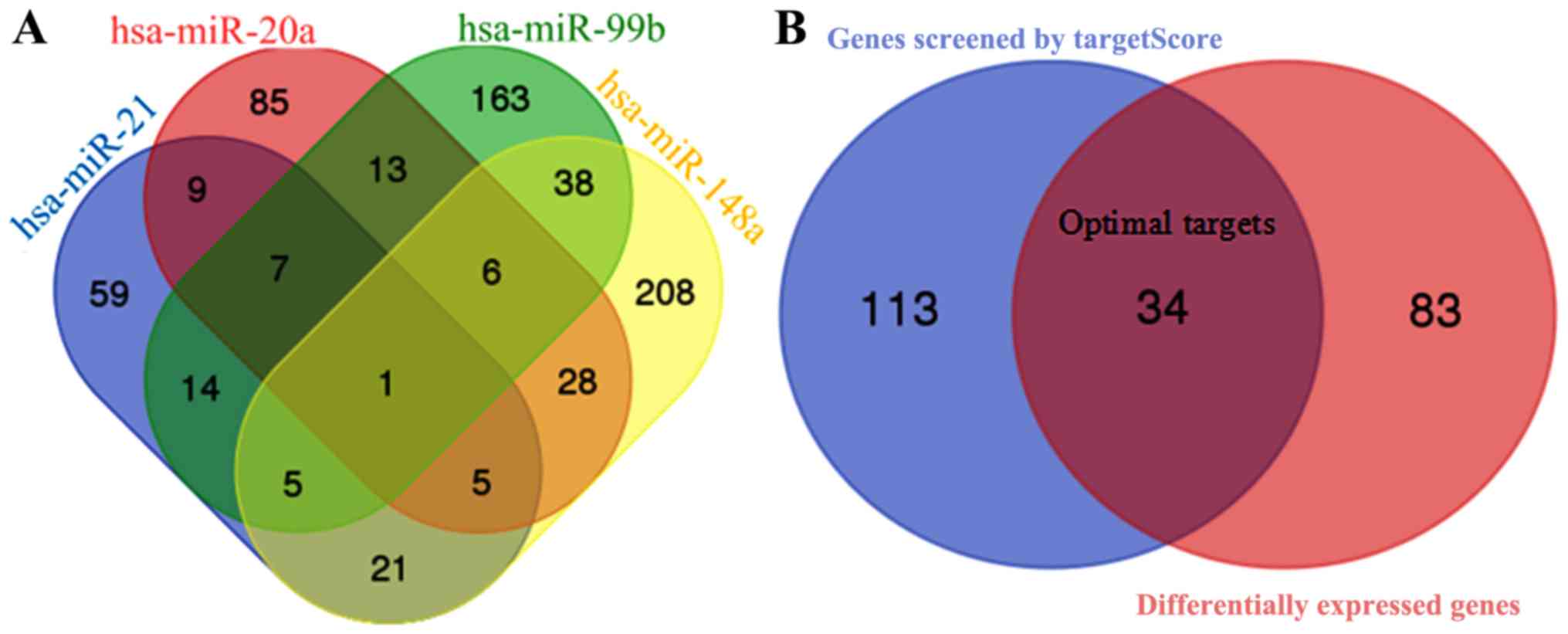
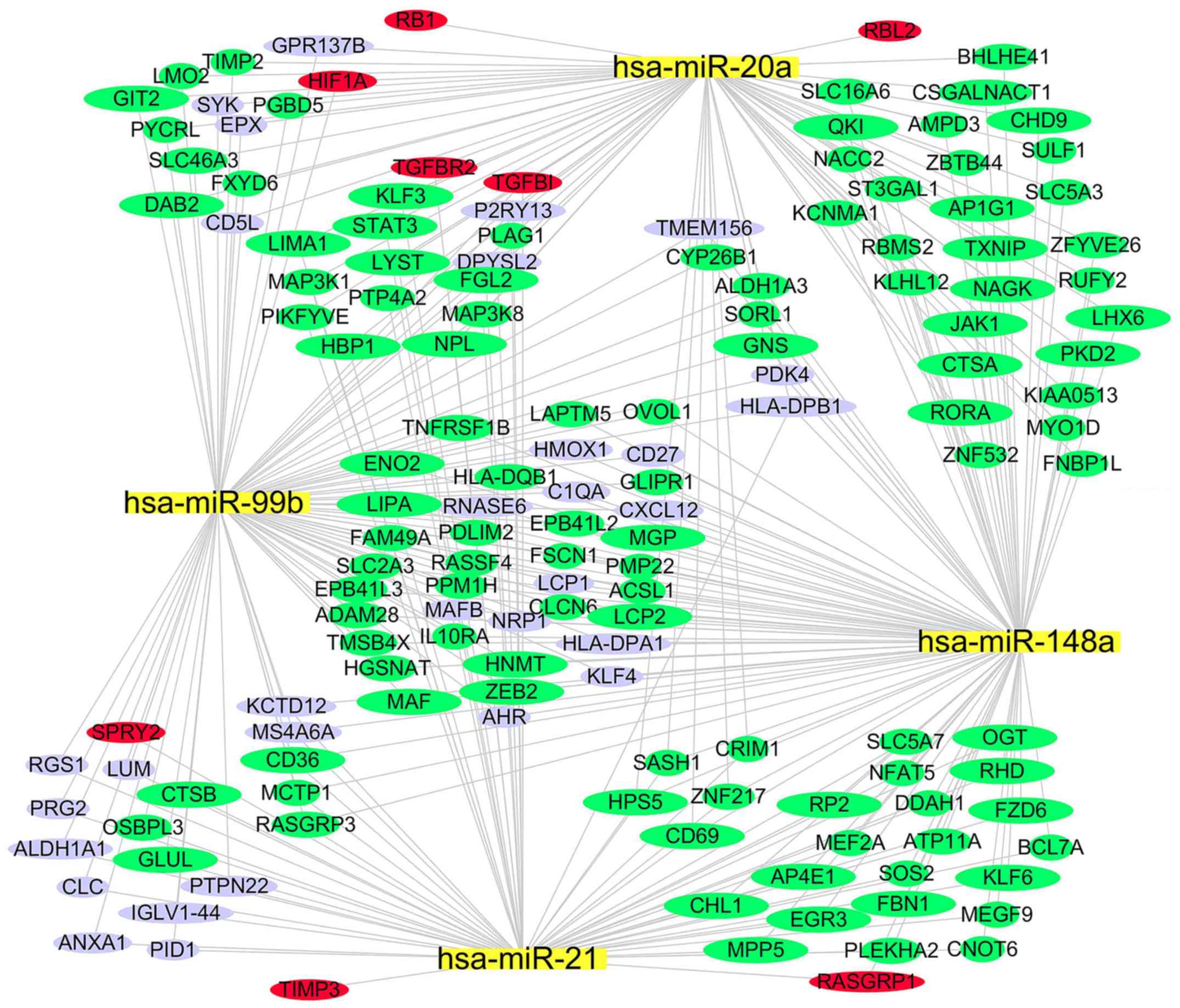
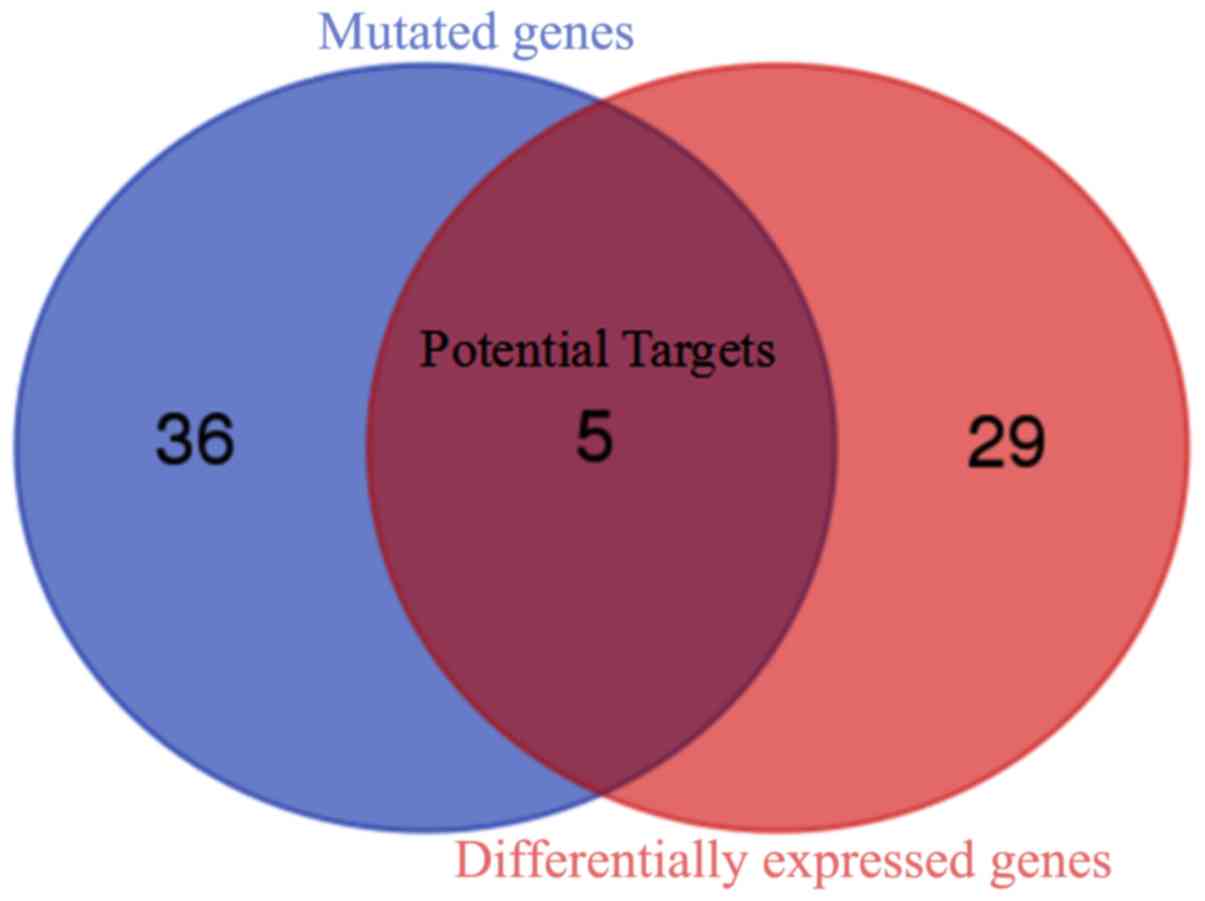
|
1
|
Eslick R and Talaulikar D: Multiple
myeloma: From diagnosis to treatment. Aust Fam Physician.
42:684–688. 2013.PubMed/NCBI
|
|
2
|
Falank C, Fairfield H and Reagan MR:
Signaling interplay between bone marrow adipose tissue and multiple
myeloma cells. Front Endocrinol (Lausanne). 7:672016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Mel S, Lim SH, Tung ML and Chng WJ:
Implications of heterogeneity in multiple myeloma. BioMed Res Int.
2014:2325462014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xie S, Zhang Y, Qu L and Xu H: A Helm
model for microRNA regulation in cell fate decision and conversion.
Sci China Life Sci. 56:897–906. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng L, Li Y, Zhang L and Yu W: Moving RNA
moves RNA forward. Sci China Life Sci. 56:914–920. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang J, Xiao X and Liu J: The role of
circulating miRNAs in multiple myeloma. Sci China Life Sci.
58:1262–1269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J, Deng S, Zhang S, Chen Z, Wu S, Cai
X, Yang X, Guo B and Peng Q: The role of miRNAs in the
differentiation of adipose-derived stem cells. Curr Stem Cell Res
Ther. 9:268–279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137 and miR-197 induce apoptosis and suppress
tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer
Res. 21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alexiou P, Maragkakis M, Papadopoulos GL,
Reczko M and Hatzigeorgiou AG: Lost in translation: An assessment
and perspective for computational microRNA target identification.
Bioinformatics. 25:3049–3055. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chng WJ, Kumar S, Vanwier S, Ahmann G,
Price-Troska T, Henderson K, Chung TH, Kim S, Mulligan G, Bryant B,
et al: Molecular dissection of hyperdiploid multiple myeloma by
gene expression profiling. Cancer Res. 67:2982–2989. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tiedemann RE, Zhu YX, Schmidt J, Yin H,
Shi CX, Que Q, Basu G, Azorsa D, Perkins LM, Braggio E, et al:
Kinome-wide RNAi studies in human multiple myeloma identify
vulnerable kinase targets, including a lymphoid-restricted kinase,
GRK6. Blood. 115:1594–1604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khan AA, Betel D, Miller ML, Sander C,
Leslie CS and Marks DS: Transfection of small RNAs globally
perturbs gene regulation by endogenous microRNAs. Nat Biotechnol.
27:549–555. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Goldenberg A, Wong KC and Zhang Z: A
probabilistic approach to explore human miRNA targetome by
integrating miRNA-overexpression data and sequence information.
Bioinformatics. 30:621–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Palumbo A and Anderson K: Multiple
myeloma. N Engl J Med. 364:1046–1060. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mahindra A, Laubach J, Raje N, Munshi N,
Richardson PG and Anderson K: Latest advances and current
challenges in the treatment of multiple myeloma. Nat Rev Clin
Oncol. 9:135–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lohr JG, Stojanov P, Carter SL,
Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B,
Gould J, Saksena G, et al Multiple Myeloma Research Consortium, :
Widespread genetic heterogeneity in multiple myeloma: Implications
for targeted therapy. Cancer Cell. 25:91–101. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Augustson BM, Begum G, Dunn JA, Barth NJ,
Davies F, Morgan G, Behrens J, Smith A, Child JA and Drayson MT:
Early mortality after diagnosis of multiple myeloma: Analysis of
patients entered onto the United Kingdom Medical Research Council
trials between 1980 and 2002 - Medical Research Council Adult
Leukaemia Working Party. J Clin Oncol. 23:9219–9226. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lawrie CH, Soneji S, Marafioti T, Cooper
CD, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood
J, et al: MicroRNA expression distinguishes between germinal center
B cell-like and activated B cell-like subtypes of diffuse large B
cell lymphoma. Int J Cancer. 121:1156–1161. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Munker R, Liu CG, Taccioli C, Alder H and
Heerema N: MicroRNA profiles of drug-resistant myeloma cell lines.
Acta Haematol. 123:201–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma J, Liu S and Wang Y: MicroRNA-21 and
multiple myeloma: Small molecule and big function. Med Oncol.
31:942014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang JJ, Yu J, Li JY, Liu YT and Zhong
RQ: Circulating microRNA expression is associated with genetic
subtype and survival of multiple myeloma. Med Oncol. 29:2402–2408.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z
and Yang B: A single anti-microRNA antisense
oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers
an improved approach for microRNA interference. Nucleic Acids Res.
37:e242009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mulligan G, Lichter DI, Di Bacco A,
Blakemore SJ, Berger A, Koenig E, Bernard H, Trepicchio W, Li B,
Neuwirth R, et al: Mutation of NRAS but not KRAS significantly
reduces myeloma sensitivity to single-agent bortezomib therapy.
Blood. 123:632–639. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van Andel H, Kocemba KA, de Haan-Kramer A,
Mellink CH, Piwowar M, Broijl A, van Duin M, Sonneveld P, Maurice
MM, Kersten MJ, et al: Loss of CYLD expression unleashes Wnt
signaling in multiple myeloma and is associated with aggressive
disease. Oncogene. 36:2105–2115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Trompeter HI, Abbad H, Iwaniuk KM, Hafner
M, Renwick N, Tuschl T, Schira J, Müller HW and Wernet P: MicroRNAs
MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F
activity on cell cycle arrest during neuronal lineage
differentiation of USSC. PLoS One. 6:e161382011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim YJ, Hwang SJ, Bae YC and Jung JS:
MiR-21 regulates adipogenic differentiation through the modulation
of TGF-β signaling in mesenchymal stem cells derived from human
adipose tissue. Stem cells. 27:3093–3102. 2009.PubMed/NCBI
|
|
29
|
He M, Wang QY, Yin QQ, Tang J, Lu Y, Zhou
CX, Duan CW, Hong DL, Tanaka T, Chen GQ, et al: HIF-1α
downregulates miR-17/20a directly targeting p21 and STAT3: a role
in myeloid leukemic cell differentiation. Cell Death Differ.
20:408–418. 2013. View Article : Google Scholar : PubMed/NCBI
|