Introduction
Since the 1990s, the incidence of prostate cancer
(PCa) has increased considerably due to population growth,
increased age expectancy and prevalent prostate-specific antigen
(PSA) screening (1). Active
surveillance may effectively manage patients with low-risk PCa
(2,3)
while those suffering from aggressive PCa require efficient
intervention to reduce the development of advanced disease and
PCa-associated mortality (4,5). Therefore, distinguishing low-risk
disease from more aggressive forms of PCa, and subsequently
reducing the rate of overtreatment, is of great clinical relevance.
At present, various combinations of PSA, Gleason score, tumor,
node, metastasis stage, and surgical margin are used as prognosis
predictors (6,7). The use of PSA screening not only
reduces the development of advanced and metastatic PCa and
PCa-associated mortality, but also assists in the detection of
indolent PCa and consequential overtreatment (5). Therefore, determining the molecular
mechanisms of aggressive and indolent PCa and investigating novel
prognostic biomarkers to predict clinical outcome is crucial.
The polycomb group (PcG) regulates transcription
through nucleosome modification and chromatin remodeling, and
interacts with other transcription factors (8). Malignant brain tumor domain containing
protein 1 (MBTD1), encoded by the MBTD1 gene on chromosome
17q21.33 is one such PcG protein, consisting of 628 amino acids and
four malignant brain tumor (MBT) repeats (9). The MBT repeat is a structural motif of
100 amino acids conserved from Caenorhabditis elegans, and
originally discovered in Drosophila as L(3)MBT. L(3)MBT
mutations cause neoplastic translations of optic neuroblasts
(10). The PcG proteins have been
recognized for their role in tumorigenesis (8). MBTD1 has been reported to be a subunit
of the histone acetyltransferase TIP60/NuA4
acetyltransferase complex, which promotes the repair of DNA
double-strand breaks through homologous recombination (11). Mutations in MBTD1 have been
identified in endometrial stromal sarcoma and leukemia (12–14).
Other MBT repeat proteins have been reported to control the
transcriptional state of chromatin regions (15,16).
However, the role of MBTD1 in PCa is yet to be ascertained.
Therefore, the present study investigated the expression of MBTD1
in PCa to determine the association between expression levels of
MBTD1, clinicopathological characteristics and PCa prognosis.
Materials and methods
Patients and tissue samples
A total of 71 PCa tissues were selected, along with
three adjacent and four normal prostate tissues, from a tissue
microarray (TMA) (Xi'an Alenabio Co., Ltd. Xi'an, China; no.
PR803c), and the clinical characteristics were assessed using IHC
analysis. Patients who had undergone radiotherapy or chemotherapy
prior to surgery were excluded. The clinical parameters, from The
Cancer Genome Atlas (TCGA) database, of 499 PCa tissues and 52
normal prostate tissues were collected to study the expression
level of MBTD1, and to perform survival analysis. The clinical
characteristics of all patients are summarized in Table I.
 | Table I.Correlation of MBTD1 expression with
clinicopathologic characteristics in patients with prostate
cancer. |
Table I.
Correlation of MBTD1 expression with
clinicopathologic characteristics in patients with prostate
cancer.
|
| TMA | TCGA |
|---|
|
|
|
|
|---|
| Clinical
features | Case | Low, n (%) | High, n (%) | P-value | Case | x±s | P-value |
|---|
| Tissue |
|
|
|
|
|
|
|
|
Cancer | 71 | 32 (45.1) | 39 (54.9) | 0.006b | 499 | 520.20±8.31 |
<0.001b |
|
Benign | 7 | 7 (100.0) | 0 (0.0) |
| 52 | 448.3±15.21 |
|
| Age, years |
|
|
|
|
|
|
|
|
<60 | 12 | 7 (58.3) | 5 (41.7) | 0.536 | 201 (<60) | 533.67±209.88 | 0.189 |
|
≥60 | 66 | 32 (48.5) | 34 (51.5) |
| 296 (≥60) | 511.38±166.72 |
|
| PSA level |
|
|
|
|
|
|
|
| ≤4 | – | – | – | – | 413 | 520.59±179.19 | 0.023a |
|
>4 | – | – | – |
| 27 | 605.62±289.09 |
|
| Gleason score |
|
|
|
|
|
|
|
| ≤7 | – | – | – | – | 291 | 502.56±178.27 | 0.011a |
|
>7 | – | – | – |
| 206 | 545.60±192.93 |
|
| Pathological
grade |
|
|
|
|
|
|
|
| ≤2 | 23 | 9 (39.1) | 14 (60.9) | 0.397 | – | – | – |
|
>2 | 44 | 22 (50.0) | 22 (50.0) |
| – | – |
|
| Clinical stage |
|
|
|
|
|
|
|
|
I–II | 44 | 26 (59.1) | 21 (40.9) | 0.003b | – | – | – |
|
III–IV | 26 | 6 (23.1) | 20 (76.9) |
| – | – |
|
| Tumor invasion |
|
|
|
|
|
|
|
|
T1-T2 | 46 | 26 (56.5) | 20 (43.5) | 0.012a | 351 | 497.24±171.27 | 0.110 |
|
T3-T4 | 24 | 6 (25.0) | 18 (75.0) |
| 55 | 556.07±260.38 |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
| N0 | 58 | 30 (51.7) | 28 (48.3) | 0.026a | 344 | 520.81±181.97 | 0.307 |
| N1 | 12 | 2 (16.7) | 10 (83.3) |
| 80 | 543.69±172.95 |
|
| Distant
metastasis |
|
|
|
|
|
|
|
| M0 | 56 | 29 (51.8) | 27 (48.2) | 0.039a | 455 | 517.20±178.44 |
<0.001b |
| M1 | 14 | 3 (21.4) | 11 (78.6) |
| 3 | 1041.11±691.26 |
|
IHC analysis
The tissue microarray (Xi'an Alenabio Co., Ltd.) was
used in IHC analysis. Specimens were fixed in 10% neutral-buffered
formalin solution overnight at room temperature and embedded in
paraffin. The paraffin-embedded tissues were then cut into 4
µm-thick slices. The tissue slices were subsequently deparaffinized
by immersing in xylene and rehydrated using a graded ethanol series
at room temperature. The tissues were washed in PBS and distilled
water for further peroxidase (3′-diaminobenzidine) IHC staining
using the DAKO EnVision System (Dako; Agilent Technologies, Inc.,
Santa Clara, CA, USA). Following proteolytic digestion with
peroxidase with 0.01 M citrate (catalog no. AR0024; Boster
Biological Technology, Pleasanton, CA, USA) in a microwave for ~6
min, blocking was performed with goat serum (catalog no. KIT-0305;
MX Biotechnologies, Fuzhou, China) at room temperature for 30 min.
IHC staining was conducted using the UltraSensitive™ SP
(Mouse/Rabbit) IHC kit (catalog no. KIT-0305; MX Biotechnologies),
which contained endogenous peroxidase blocking solution, serum,
secondary antibody, streptavidin-perosidase and
substrate-chromogen. The tissues sections were incubated overnight
at 4°C with a rabbit anti-human polyclonal antibody against MBTD1
(cat. no. ab170848; Abcam, Cambridge, UK), at a dilution of 1:80.
Following washing, the sections were incubated with an
avidin-conjugated secondary antibody (catalog no. KIT-0305; MX
Biotechnologies, Fuzhou) for 30 min at room temperature.
Streptavidin-peroxidase (50 µl for 15 min at room temperature) and
substrate-chromogen (100 µl for 2 min at room temperature) were
used to observe the staining of the target protein. For negative
controls, the primary antibody was omitted in each IHC run
(17).
Evaluation of immunostaining
results
The intensity of immunostaining was scored by two
experienced pathologists with no prior knowledge of the clinical
and pathological details of the patients. The scores were compared,
and any conflict was reevaluated by regrading of the immunostaining
by the same pathologists. A total of five representative fields
were microscopically observed (magnification, ×400) and the number
of positively-stained cells was enumerated. Cytoplasmic and nuclear
staining was regarded as positive according to the antibody
specification sheet (18). The
expression level of MBTD1 in each sample was semi quantitatively
scored depending on the staining intensity and percentage of
stained cells. The staining intensity was classified based on the
following criteria: None (0), mild (1), moderate (2) and strong (3). The percentage of positive cells was
stratified as follows: <5% (0), 6–25% (1), 26–50% (2), 51–75% (3) and >75% (4). The final immunoreactivity scores (IRS)
were calculated by adding the immunostaining percentages and
immunostaining intensity scores. Using the median score as the
cutoff point, an IRS ≥4 and <4 was defined as high and low
expression, respectively (19,20).
Assays of protein expression of MBTD1
in cell lines
PCa cells 22RV1, LNCap, DU-145, PC3 and the benign
prostatic hyperplasia epithelial cell line (BPH1) were obtained
from the Center of Experiment Animal of Sun Yat-sen University
(Guangzhou, China). The cells were cultured in a humidified
CO2 incubator at 37°C with RPMI 1640 medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum and 1% penicillin-streptomycin (both from
Invitrogen; Thermo Fisher Scientific, Inc.), The cell lines were
used to assess the protein expression level of MBTD1. A total of
1×107 cells/sample were washed twice using PBS, prior to
lysis using radioimmunoprecipitation buffer (Beyotime Institute of
Biotechnology, Haimen, China) for 30 min, and subsequently
centrifuged at 17,225.6 × g for 30 min at 4°C. A bicinchoninic acid
assay kit was employed to measure the total protein concentration
of each extract. The proteins were transferred to a PVDF membrane
(EMD Millipore, Billerica, MA, USA) following 10% SDS-PAGE
separation with 20 µg protein per lane. The membranes were blocked
using 5% nonfat dry milk for 1 h in TBS with Tween-20 (TBST) at
room temperature and subsequently probed with rabbit anti-human
polyclonal antibody against MBTD1 (cat. no. ab170848; 1:500; Abcam)
and β-actin (cat. no. BM0627; 1:4,000; Boster Biological
Technology) overnight at 4°C. After washing three times with TBST,
the membrane was incubated with horseradish peroxidase-conjugated
secondary antibody (cat. no. BA1054; 1:5,000; Boster Biological
Technology) for a further 2 h. Finally, the protein bands were
detected using a Chemiluminescence imaging analysis system (catalog
no. 5200; Tanon Science and Technology Co., Ltd., Tokyo, Japan).
The relative density of protein expression was quantified using
Image J 1.8.0 software (National Institutes of Health, Bethesda,
MD, USA) (21).
Statistical analysis
SPSS 22.0 software (IBM Corp., Armonk, NY, USA) was
used for statistical analysis. Patient characteristics are
presented as counts and percentages for categorical data, and mean
± standard deviation for continuous data. Fisher's exact test and
Pearson's χ2 tests were employed to evaluate the
correlation between MBTD1 expression levels and clinicopathological
parameters. For independent samples, Student's t-tests for normally
distributed data or the Mann-Whitney U test for non-normally
distributed data were employed to evaluate continuous data.
Analysis of variance was employed in the quantification analysis of
western blots (Fig. 1H). The overall
survival was analyzed using the Kaplan-Meier method and the
differences were evaluated using the log-rank test (22). The Cox proportional hazards
regression model was employed for univariate analysis comparisons
and multivariate survival comparisons. The corresponding 95%
confidence interval and the adjusted hazard ratios (HRs) were used
to represent the relative risk of mortality. P<0.05 was
considered to indicate a statistically significant difference.
Results
Overexpression of MBTD1 protein in
human PCa tissues and cell lines and its association with
clinicopathological features
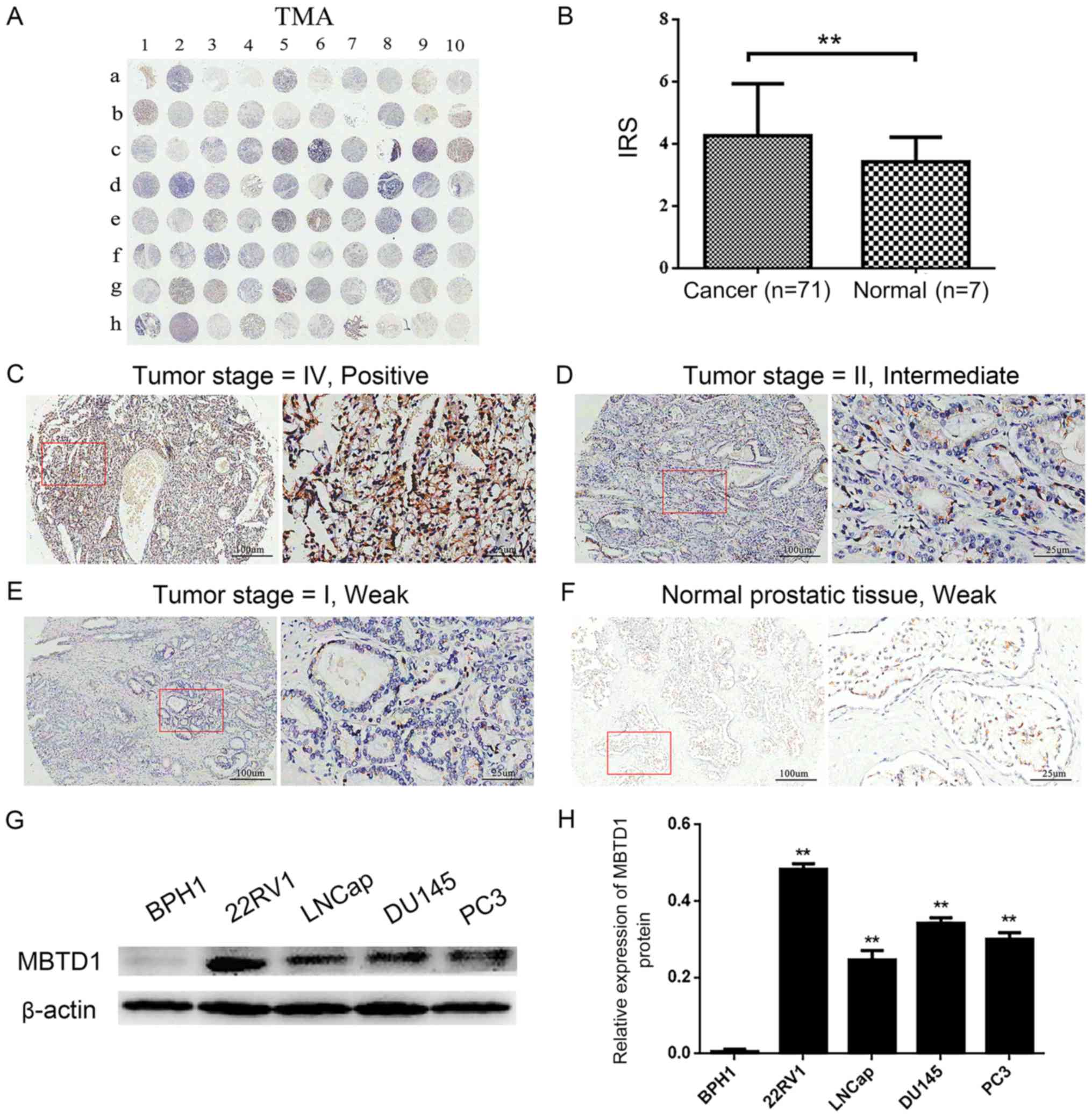
First, the IHC analysis was preformed to evaluate
the expression level of MBTD1 in 71 PCa tissues and three adjacent
normal and four normal prostate tissues (Table I). As presented in Fig. 1A-F, a strong MBTD1 immunostaining
signal was present in the cytoplasm of PCa cells, while normal
tissues stained weakly. The expression of MBTD1 was significantly
higher in PCa tissues compared with normal tissues (Fig. 1B). The expression of MBTD1 protein
was high in 39 (54.9%) patients and low in 32 (45.1%) patients out
of the 71 patients with PCa. Analysis of the association between
MBTD1 levels and clinicopathological characteristics of PCa
demonstrated that the expression level of MBTD1 was significantly
correlated with aggressive clinical stage, tumor invasion, lymph
node metastasis, and advanced distant metastasis, while the
expression level of MBTD1 was not associated with age and
pathological grade (Table I).
Furthermore, western blot analysis indicated that the expression
level of MBTD1 in PCa cells, 22RV1, LNCap, DU-145 and PC3, was
significantly higher compared with that in benign prostatic
hyperplasia BPH1 cells (Fig. 1G and
H).
Correlation between overexpression of
MBTD1, enhanced progression and poor prognosis of PCa in human
patients, as documented in the TCGA database
To further confirm the results of the TMA, the MBTD1
mRNA expression levels of 499 PCa tissues and 52 normal prostate
tissues deposited in the TCGA dataset were investigated. In PCa
tissues, the expression levels of MBTD1 mRNA were upregulated and
positively correlated with PSA levels, Gleason score, and distant
metastasis, but not with age, tumor invasion, and lymph node
metastasis (Table I).
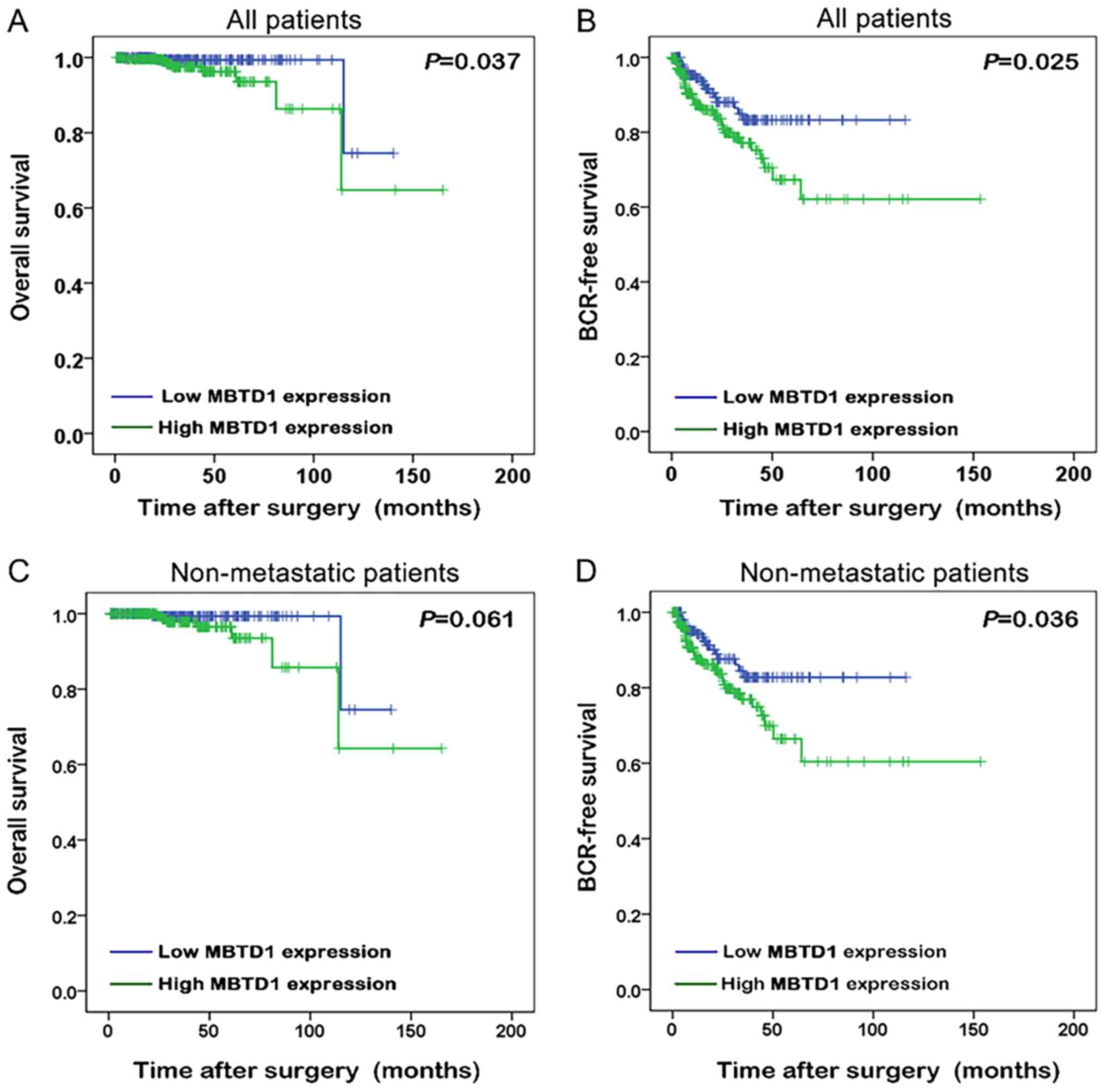
The Kaplan-Meier method was used to analyze the
association between the expression of MBTD1 and the survival time
of patients with PCa. Improved biochemical recurrence (BCR)-free
survival and overall survival were presented in patients with low
expression level of MBTD1 (Fig. 2A and
B). Among non-metastatic patients, low expression of MBTD1 was
associated with longer BCR-free survival (Fig. 2C and D).
MBTD1 is an independent prognostic
factor for the survival of patients with PCa
The Cox proportional hazards model was applied to
assess whether MBTD1 acted as an independent prognostic factor for
PCa survival in the TCGA dataset. The expression of MBTD1
correlated with BCR-free survival in patients with PCa with an HR
of 1.884. The PSA level, Gleason score, tumor invasion and lymph
node stage exhibited the same trends, as demonstrated by univariate
analysis (Table II). Multivariate
analysis with the Cox proportional hazards model was employed to
confirm that the MBTD1 level acted as a significant factor for PCa
(Table II).
 | Table II.Prognostic value of MBTD1 expression
for BCR-free survival revealed by Cox proportional hazards
model. |
Table II.
Prognostic value of MBTD1 expression
for BCR-free survival revealed by Cox proportional hazards
model.
| A, Univariate
analysis |
|---|
|
|
|
|---|
|
| BCR-free
survival |
|---|
|
|
|
|---|
| Variable | HR (95%CI) | P-value |
|---|
| Age (≥60 vs.
<60) | 1.319
(0.773–2.250) | 0.310 |
| PSA level (>4
vs. ≤4) | 10.426
(5.309–20.474) |
<0.001b |
| Gleason score
(<7 vs. 7 vs. >7) | 3.175
(1.881–5.360) |
<0.001b |
| Clinical stage (I
vs. II vs. III vs. IV) | 1.451
(0.822–2.559) | 0.199 |
| Tumor invasion
(T1-T2 vs. T3-T4) | 3.950
(2.226–7.008) |
<0.001b |
| Lymph node
metastasis (N0 vs. N1) | 1.879
(1.049–3.365) | 0.034a |
| Distant metastasis
(M0 vs. M1) | 3.536
(0.488–25.641) | 0.212 |
| MBTD1 expression
(low vs. high) | 1.884
(1.072–3.312) | 0.028a |
|
| B, Multivariate
analysis |
|
|
| BCR-free
survival |
|
|
|
|
Variable | HR
(95%CI) | P-value |
|
| Age (≥60 vs.
<60) | 1.193
(0.683–2.085) | 0.536 |
| Lymph node
metastasis (N0 vs. N1) | 1.716
(0.951–3.098) | 0.073 |
| MBTD1 expression
(low vs. high) | 1.840
(1.007–3.362) | 0.047a |
Discussion
Although the diagnostic and treatment methodologies
for PCa have advanced, and the survival rate has increased,
efficient risk attribution and prognosis prediction of PCa remains
a crucial issue. Discovering novel biomarkers to predict recurrence
and metastatic potential may aid patient management and decrease
the morbidity associated with PCa. In the present study, MBTD1 was
overexpressed in PCa tissues and was significantly associated with
the advanced clinicopathological characteristics of PCa.
Furthermore, MBTD1 was recognized as an independent prognostic
factor in PCa, and higher expression of MBTD1 predicted poorer
BCR-free survival.
The role of PcG as an oncogenic factor has been
confirmed (8), and is consistent
with the findings of the present study, that MBTD1 was
overexpressed in PCa tissues. Like other PcG proteins, MBTD1 may
regulate the transcription of developmentally-associated genes
through chromatin remodeling, nucleosome modification and
interaction with other transcription factors (8,9). It
interrupts signaling pathways that govern cellular behavior,
reduces the activity of tumor suppressors, activates
proto-oncogenes and ultimately influences tumorigenesis. The
present study demonstrated that the overexpression of MBTD1 was
associated with progressive disease and consequently, poor
prognosis and shorter patient survival time.
The mechanisms underlying the contribution of MBTD1
to the progression of PCa are elusive and complex. MBTD1 acts as a
subunit of the NuA4/TIP60 acetyltransferase complex (9,23) and
allows TIP60 to interact with specific promoters for the activation
of homologous recombination (via histone reader domain H4K20me1/2)
(11). In malignant tumor cells, the
DNA damage repair system is usually upregulated to provide the
optimal environment for cell proliferation and survival. Defects in
this system are associated with the sensitivity of chemotherapy
drugs in tumor cells and considered as targets for tumor therapy
(24,25). The overexpression of MBTD1 in PCa may
be associated with the upregulation of DNA damage repair and
consequently, with poor prognosis (26). Since PcG proteins have been
recognized as candidates for targeted therapy (27), MBTD1 may also serve as a candidate
for PCa therapy following further investigation. In addition to
regulating DNA damage repair, the MBTD1-containing TIP60 complex
regulates the transcription/protein expression of the Myc
proto-oncogene protein signaling pathway (11). Studies have suggested that Myc
suppresses tumor invasion and cell migration by inhibiting c-Jun
N-terminal kinase signaling (28).
The present study demonstrated that the overexpression of MBTD1
positively correlated with distant metastasis and tumor invasion;
whether it is associated with Myc pathway regulation remains to be
explored.
In conclusion, the present study revealed that MBTD1
was overexpressed in PCa tissues and significantly correlated with
advanced clinicopathological characteristics of PCa. Furthermore,
MBTD1 may act as a novel prognostic factor and diagnostic marker in
PCa.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (nos. 81472382 and 81672550); the
Guangdong Province Natural Science Foundation (no. 2014A030313079);
Guangdong Province Science and Technology for Social Development
Project (no. 2017A020215018); Guangzhou Science and Technology
Cooperation Program (Foreign research and development cooperation)
(no. 201807010087); Guangzhou City in 2015 Scientific Research
Project (no. 201510010298); International Science and Technology
Cooperation Project of Guangdong Province Science and Technology
Plan (no. 2016A050502020), Grant [2013]163 from the Key Laboratory
of Malignant Tumor Molecular Mechanism and Translational Medicine
of Guangzhou Bureau of Science and Information Technology and the
Chinese National Scholarship to HH. National Natural Science
Foundation of China (no. 81772733); Guangdong Province Science and
Technology for Social Development Project (nos. 2014A020212018 and
2016A020215011); Guangzhou Science and Technology Plan (no.
201707010371) to ZHG and the National Natural Science Foundation of
China for Young Scientists Grant (no. 81802527) to YML.
Availability of data and materials
All data generated during the presente study are
included in this published article.
Authors' contributions
WHW and SMB were responsible for the study concept
and design. DJZ and KWL acquired the tissue samples. WD and SMP
conducted the immunohistochemistry analysis. YML and WH were
responsible for the statistical analysis. QW, DJZ and KWL helped to
perform the analysis and interpretation of data. HH, ZHG and LYL
were involved in the design of the study, and the writing, review
and revision of the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Ethical approval for this investigation was attained
from the Research Ethics Committee, The Sun Yat-Sen Memorial
Hospital, Sun Yat-Sen University (reference no.
sysec-ky-ks-2018-011). Written informed consent was obtained from
all participants at the time of original tissue collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BCR
|
biochemical recurrence
|
|
IHC
|
immunohistochemistry
|
|
MBTD1
|
malignant brain tumor domain
containing protein 1
|
|
PCa
|
prostate cancer
|
|
PcG
|
polycomb group of genes
|
|
PSA
|
prostate-specific antigen
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TMA
|
tissue microarray
|
References
|
1
|
Dy GW, Gore JL, Forouzanfar MH, Naghavi M
and Fitzmaurice C: Global burden of urologic cancers, 1990–2013.
Eur Urol. 71:437–446. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fenner A: Prostate cancer:
AS-contemplation, not intervention. Nat Rev Oncol. 12:5952015.
|
|
3
|
Tosoian JJ, Mamawala M, Epstein JI, Landis
P, Wolf S, Trock BJ and Carter HB: Intermediate and longer-term
outcomes from a prospective active-surveillance program for
favorable-risk prostate cancer. J Clin Oncol. 33:3379–3385. 2016.
View Article : Google Scholar
|
|
4
|
Attard G, Parker C, Eeles RA, Schröder F,
Tomlins SA, Tannock I, Drake CG and de Bono JS: Prostate cancer.
Lancet. 387:70–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heidenreich A, Abrahamsson PA, Artibani W,
Catto J, Montorsi F, Van Poppel H, Wirth M and Mottet N; European
Association of Urology, : Early detection of prostate cancer:
European Association of Urology recommendation. Eur Urol.
64:347–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Center MM, Jemal A, Lortet-Tieulent J,
Ward E, Ferlay J, Brawley O and Bray F: International variation in
prostate cancer incidence and mortality rates. Eur Urol.
61:1079–1092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kattan MW, Vickers AJ, Yu C, Bianco FJ,
Cronin AM, Eastham JA, Klein EA, Reuther AM, Edson Pontes J and
Scardino PT: Preoperative and postoperative nomograms incorporating
surgeon experience for clinically localized prostate cancer.
Cancer. 115:1005–1010. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mills AA: Throwing the cancer switch:
Reciprocal roles of polycomb and trithorax proteins. Nat Rev
Cancer. 10:669–682. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eryilmaz J, Pan P, Amaya MF,
Allali-Hassani A, Dong A, Adams-Cioaba MA, Mackenzie F, Vedadi M
and Min J: Structural studies of a four-MBT repeat protein MBTD1.
PLoS One. 4:e72742009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wismar J, Loffler T, Habtemichael N, Vef
O, Geissen M, Zirwes R, Altmeyer W, Sass H and Gateff E: The
Drosophila melanogaster tumor suppressor gene lethal(3)malignant
brain tumor encodes a proline-rich protein with a novel zinc
finger. Mech Dev. 53:141–154. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jacquet K, Fradet-Turcotte A, Avvakumov N,
Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC,
Pandita TK, et al: The TIP60 complex regulates bivalent chromatin
recognition by 53BP1 through direct H4K20me binding and H2AK15
acetylation. Mol Cell. 62:409–421. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dewaele B, Przybyl J, Quattrone A, Finalet
Ferreiro J, Vanspauwen V, Geerdens E, Gianfelici V, Kalender Z,
Wozniak A, Moerman P, et al: Identification of a novel, recurrent
MBTD1-CXorf67 fusion in low-grade endometrial stromal sarcoma. Int
J Cancer. 134:1112–1122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Rooij JD, van den Heuvel-Eibrink MM,
Kollen WJ, Sonneveld E, Kaspers GJ, Beverloo HB, Fornerod M,
Pieters R and Zwaan CM: Recurrent translocation t(10;17)(p15;q21)
in minimally differentiated acute myeloid leukemia results in
ZMYND11/MBTD1 fusion. Genes Chromosomes Cancer. 55:237–241. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
De Braekeleer E, Auffret R, Douet-Guilbert
N, Basinko A, Le Bris MJ, Morel F and De Braekeleer M: Recurrent
translocation (10;17)(p15;q21) in acute poorly differentiated
myeloid leukemia likely results in ZMYND11-MBTD1 fusion. Leuk
Lymphoma. 55:1189–1190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Klymenko T, Papp B, Fischle W, Köcher T,
Schelder M, Fritsch C, Wild B, Wilm M and Müller J: A Polycomb
group protein complex with sequence-specific DNA-binding and
selective methyl-lysine-binding activities. Genes Dev.
20:1110–1122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grimm C, de Ayala Alonso AG, Rybin V,
Steuerwald U, Ly-Hartig N, Fischle W, Müller J and Müller CW:
Structural and functional analyses of methyl-lysine binding by the
malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep.
8:1031–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Xie W, Gu P, Cai Q, Wang B, Xie Y,
Dong W, He W, Zhong G, Lin T and Huang J: Upregulated WDR5 promotes
proliferation, self-renewal and chemoresistance in bladder cancer
via mediating H3K4 trimethylation. Sci Rep. 5:82932015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma X, Du T, Zhu D, Chen X, Lai Y, Wu W,
Wang Q, Lin C, Li Z, Liu L and Huang H: High levels of glioma tumor
suppressor candidate region gene 1 predicts a poor prognosis for
prostate cancer. Oncol Lett. 16:6749–6755. 2018.PubMed/NCBI
|
|
19
|
Cimino-Mathews A, Subhawong AP, Illei PB,
Sharma R, Halushka MK, Vang R, Fetting JH, Park BH and Argani P:
GATA3 expression in breast carcinoma: utility in triple-negative,
sarcomatoid, and metastatic carcinomas. Hum Pathol. 44:1341–1349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bachmann IM, Halvorsen OJ, Collett K,
Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP and
Akslen LA: EZH2 expression is associated with high proliferation
rate and aggressive tumor subgroups in cutaneous melanoma and
cancers of the endometrium, prostate, and breast. J Clin Oncol.
24:268–273. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu P, Chen X, Xie R, Han J, Xie W, Wang B,
Dong W, Chen C, Yang M, Jiang J, et al: lncRNA HOXD-AS1 regulates
proliferation and chemo-resistance of castration-resistant prostate
cancer via recruiting WDR5. Mol Ther. 25:1959–1973. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Gu P, Xie R, Han J, Liu H, Wang B,
Xie W, Xie W, Zhong G, Chen C, et al: Heterogeneous nuclear
ribonucleoprotein K is associated with poor prognosis and regulates
proliferation and apoptosis in bladder cancer. J Cell Mol Med.
21:1266–1279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nady N, Krichevsky L, Zhong N, Duan S,
Tempel W, Amaya MF, Ravichandran M and Arrowsmith CH: Histone
recognition by human malignant brain tumor domains. J Mol Biol.
423:702–718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang YQ, Wang PY, Wang YT, Yang GF, Zhang
A and Miao ZH: An update on Poly(ADP-ribose)polymerase-1 (PARP-1)
inhibitors: Opportunities and challenges in cancer therapy. J Med
Chem. 59:9575–9598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kluzek K, Bialkowska A, Koczorowska A and
Zdzienicka MZ: Poly(ADP-ribose) polymerase (PARP) inhibitors in
BRCA1/2 cancer therapy. Postepy Hig Med Dosw (Online). 66:372–384.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao D, Herman JG and Guo M: The clinical
value of aberrant epigenetic changes of DNA damage repair genes in
human cancer. Oncotarget. 7:37331–37346. 2016.PubMed/NCBI
|
|
27
|
Kirmizis A, Bartley SM and Farnham PJ:
Identification of the polycomb group protein SU(Z)12 as a potential
molecular target for human cancer therapy. Mol Cancer Ther.
2:113–121. 2003.PubMed/NCBI
|
|
28
|
Ma X, Huang J, Tian Y, Chen Y, Yang Y,
Zhang X, Zhang F and Xue L: Myc suppresses tumor invasion and cell
migration by inhibiting JNK signaling. Oncogene. 36:3159–3167.
2017. View Article : Google Scholar : PubMed/NCBI
|