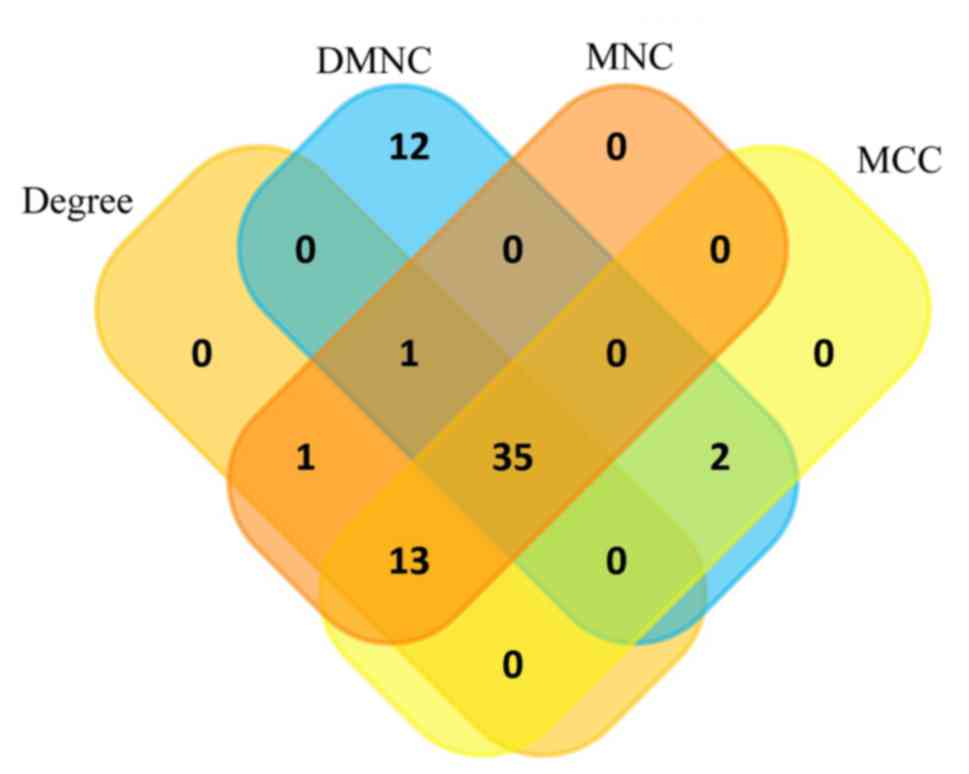
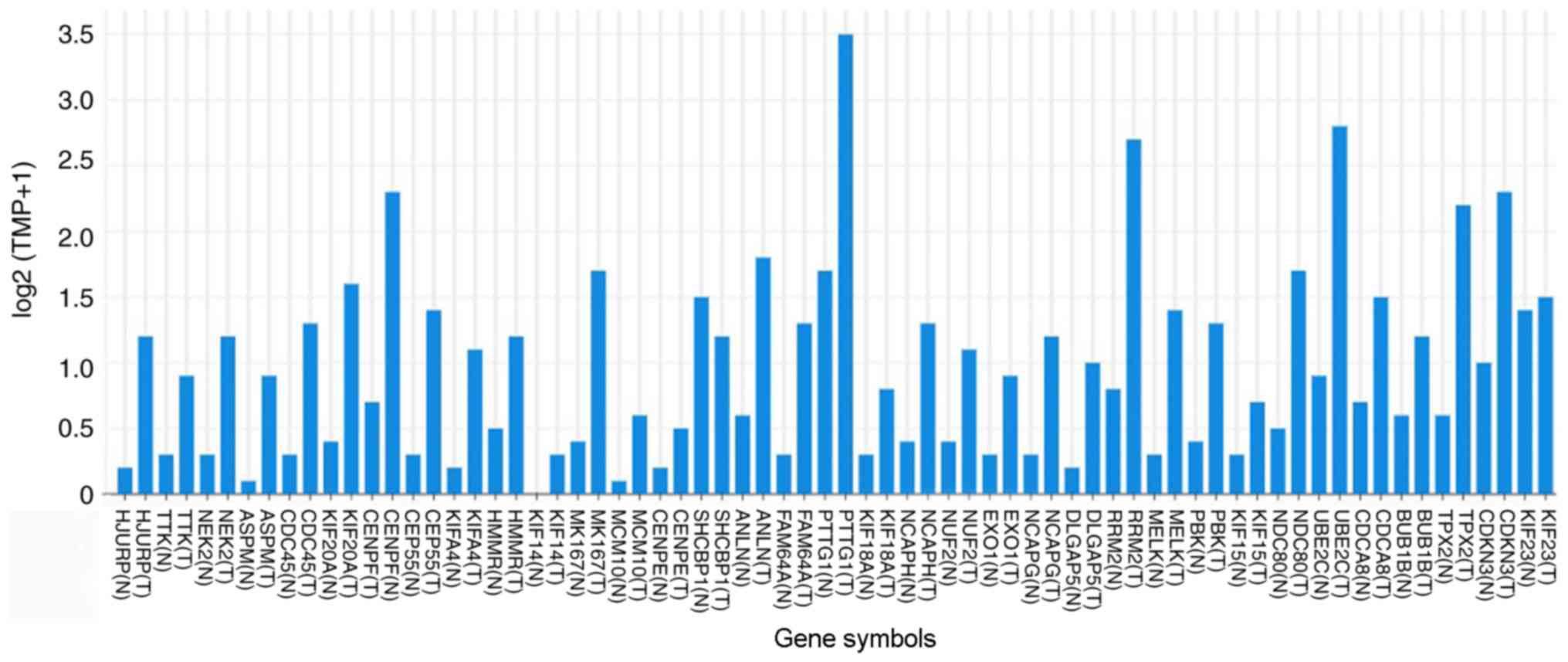
|
1
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Clark PE: The role of VHL in clear-cell
renal cell carcinoma and its relation to targeted therapy. Kidney
Int. 76:939–945. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wolff I, May M, Hoschke B, Zigeuner R,
Cindolo L, Hutterer G, Schips L, De Cobelli O, Rocco B, De Nunzio
C, et al: Do we need new high-risk criteria for surgically treated
renal cancer patients to improve the outcome of future clinical
trials in the adjuvant setting? Results of a comprehensive analysis
based on the multicenter CORONA database. Eur J Surg Oncol.
42:744–750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frank I, Blute ML, Cheville JC, Lohse CM,
Weaver AL and Zincke H: An outcome prediction model for patients
with clear cell renal cell carcinoma treated with radical
nephrectomy based on tumor stage, size, grade and necrosis: The
SSIGN score. J Urol. 168:2395–2400. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pal SK, Williams S, Josephson DY,
Carmichael C, Vogelzang NJ and Quinn DI: Novel therapies for
metastatic renal cell carcinoma: Efforts to expand beyond the
VEGF/mTOR signaling paradigm. Mol Cancer Ther. 11:526–537. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Courtney KD and Choueiri TK: Updates on
novel therapies for metastatic renal cell carcinoma. Ther Adv Med
Oncol. 2:209–219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lv Y, Wei W, Huang Z, Chen Z, Fang Y, Pan
L, Han X and Xu Z: Long non-coding RNA expression profile can
predict early recurrence in hepatocellular carcinoma after curative
resection. Hepatol Res. 48:1140–1148. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin Y, Lv Y, Liang R, Yuan C and Zhang J,
He D, Zheng X and Zhang J: Four-miRNA signature as a prognostic
tool for lung adenocarcinoma. Onco Targets Ther. 11:29–36. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu L, Xiang L, Feng J, Li B, Zhou Z, Li J,
Lin Y, Lv Y, Zou D, Lei Z and Zhang J: miRNA-21 and miRNA-223
expression signature as a predictor for lymph node metastasis,
distant metastasis and survival in kidney renal clear cell
carcinoma. J Cancer. 9:3651–3659. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Idris SF, Ahmad SS, Scott MA, Vassiliou GS
and Hadfield J: The role of high-throughput technologies in
clinical cancer genomics. Expert Rev Mol Diagn. 13:167–181. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Le Gallo M, Lozy F and Bell DW:
Next-generation sequencing. Adv Exp Med Biol. 943:119–148. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldman M, Craft B, Kamath A, Brooks AN,
Zhu J and Haussler D: The UCSC Xena platform for cancer genomics
data visualization and interpretation. BioRxiv. May 18–2018.(Epub
ahead of print). doi: https://doi.org/10.1101/326470.
|
|
15
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Torto-Alalibo T, Purwantini E, Lomax J,
Setubal JC, Mukhopadhyay B and Tyler BM: Genetic resources for
advanced biofuel production described with the Gene Ontology. Front
Microbiol. 5:5282014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–101. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: CytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 4:S112014. View Article : Google Scholar
|
|
22
|
Jeong H, Mason SP, Barabasi AL and Oltvai
ZN: Lethality and centrality in protein networks. Nature.
411:41–42. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin CY, Chin CH, Wu HH, Chen SH, Ho CW and
Ko MT: Hubba: Hub objects analyzer-A framework of interactome hubs
identification for network biology. Nucleic Acids Res.
36:W438–W443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gerlinger M, Horswell S, Larkin J, Rowan
AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos
CR, et al: Genomic architecture and evolution of clear cell renal
cell carcinomas defined by multiregion sequencing. Nat Genet.
46:225–233. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Champeris Tsaniras S, Kanellakis N,
Symeonidou IE, Nikolopoulou P, Lygerou Z and Taraviras S: Licensing
of DNA replication, cancer, pluripotency and differentiation: An
interlinked world? Semin Cell Dev Biol. 30:174–180. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Askeland EJ, Chehval VA, Askeland RW,
Fosso PG, Sangale Z, Xu N, Rajamani S, Stone S and Brown JA: Cell
cycle progression score predicts metastatic progression of clear
cell renal cell carcinoma after resection. Cancer Biomark.
15:861–867. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ho TH, Serie DJ, Parasramka M, Cheville
JC, Bot BM, Tan W, Wang L, Joseph RW, Hilton T, Leibovich BC, et
al: Differential gene expression profiling of matched primary renal
cell carcinoma and metastases reveals upregulation of extracellular
matrix genes. Ann Oncol. 28:604–610. 2017.PubMed/NCBI
|
|
30
|
Ghatalia P, Yang ES, Lasseigne BN, Ramaker
RC, Cooper SJ, Chen D, Sudarshan S, Wei S, Guru AS, Zhao A, et al:
Kinase gene expression profiling of metastatic clear cell renal
cell carcinoma tissue identifies potential new therapeutic targets.
PLoS One. 11:e01609242016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Makhov P, Joshi S, Ghatalia P, Kutikov A,
Uzzo RG and Kolenko VM: Resistance to systemic therapies in clear
cell renal cell carcinoma: Mechanisms and management strategies.
Mol Cancer Ther. 17:1355–1364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Massari F, Di Nunno V, Ciccarese C, Graham
J, Porta C, Comito F, Cubelli M, Iacovelli R and Heng DYC: Adjuvant
therapy in renal cell carcinoma. Cancer Treat Rev. 60:152–157.
2017. View Article : Google Scholar : PubMed/NCBI
|