Introduction
Clear cell renal cell carcinoma (ccRCC) is the third
most common adult genitourinary cancer, accounting for ~3% of all
malignancies, with ~209,000 newly diagnosed cases and 14,970 cases
of ccRCC-associated mortality worldwide in 2017 (1–3). ccRCC
exhibits radiotherapy and chemotherapy resistance, but surgical
tumor resection is an effective treatment strategy for ccRCC at
present (4–6). However, there are certain limitations
to the surgical resection treatments (7,8).
Following surgery, 25–30% of patients with ccRCC exhibit distant
metastasis occurrence and 40% of patients with ccRCC exhibit local
recurrence (2,9). The prognosis of ccRCC is unfavorable
and hard to predict due to the complicated biological behavior and
unclear molecular mechanism of ccRCC (10). In summary, a novel method for early
detection and advanced treatment strategies for ccRCC are urgently
required.
Tumor suppressor candidate 3 (TUSC3) is considered
to be a promising anti-oncogene (11). TUSC3 expression has been demonstrated
to be downregulated in various malignant tumors with a poor
prognosis, including prostate cancer, colorectal cancer and
pancreatic carcinoma (12–15). TUSC3 is a homolog of the yeast Ost3p
subunit of the oligosaccharyltransferase (OST) complex, which
promotes N-linked glycation of proteins in the endoplasmic
reticulum (16). Peng et al
(17) reported that the autophagy of
human non-small cell lung cancer cells may be induced by TUSC3 via
the activation of the Wnt/β-catenin signaling pathway. An
experiment conducted by Liu et al (18) suggested that the proliferation and
migration of breast cancer cells were inhibited by the upregulation
of TUSC3 expression. Although TUSC3 has been identified to be
crucial for tumorigenesis, TUSC3 expression in ccRCC and its
clinical significance remain unclear. The purpose of the present
study was to compare the expression levels of TUSC3 in ccRCC and
normal renal tissues, and to assess the prognostic value of TUSC3
expression in patients with ccRCC.
Materials and methods
Clinical specimens
Surgical specimens were obtained from 54 patients
with ccRCC during a nephrectomy at Jingzhou Central Hospital
(Jingzhou, China) between September 2014 and January 2017. The mean
age of the 54 patients was 61.5±6.2 years (range, 48–76 years). A
total of 20 patients were female and 34 were male. The present
study was approved by the Institutional Ethics Committee of
Jingzhou Central Hospital. All patients provided relevant clinical
information and written informed consent. Half of each specimen was
preserved by fixation using 4% paraformaldehyde at room temperature
for 1–2 weeks, followed by routine paraffin embedding. The
remaining half of each sample was preserved in liquid nitrogen at
−196°C. All specimens were identified to be ccRCC tissues or normal
tissues by histological identification, and tumor stage and grade
were evaluated according to the American Joint Committee on Cancer
guidelines (19).
Cell lines and cell culture
The human renal proximal tubular epithelial HKC8
cell line, human ccRCC A498, 786-O and OS-RC-2 cell lines, and the
papillary renal cell carcinoma ACHN cell line (Type Culture
Collection of the Chinese Academy of Sciences, Shanghai, China;
cat. nos. GNHu12, TCHu158, TCHu186, TCHu40 and TCHu199) were
cultured at 37°C with 5% CO2 in Dulbecco's modified
Eagle's medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA),
supplemented with 10% fetal bovine serum (ScienCell Research
Laboratories, Inc., San Diego, CA, USA), 0.1 mg/ml streptomycin and
100 U/ml penicillin. The medium was replaced every 24 h.
Immunohistochemistry
Immunohistochemical staining was used to evaluate
the expression levels of TUSC3. The tissues were cut into 4-µm
thick sections. Endogenous peroxidase activity was inhibited with
3% hydrogen peroxide at 37°C for 10 min. Subsequently, the sections
were incubated in normal horse serum (1:50) in Tris-buffered saline
(TBS) for 30 min at 37°C. Next, rabbit polyclonal anti-TUSC3
antibody (1:1,000; cat. no. ab77600; Abcam, Cambridge, MA, USA) was
applied, followed by overnight incubation at 4°C. The sections were
washed with PBS three times. Subsequently, the sections were
incubated with horseradish peroxidase-conjugated secondary antibody
(1:2,000; cat. no. SA00001-9; ProteinTech Group, Inc., Chicago, IL,
USA) for 30 min at 20°C. The sections were incubated with the color
reagent 3,3′-diaminobenzidine for 2 min at 20°C. Tissues were
observed under a light microscope (magnification, ×400) and images
were captured. The integrated optical density (IOD) was calculated
from five random fields of view per slide using Image-Pro Plus
software version 5.0 (Media Cybernetics, Inc., Rockville, MD, USA),
and the IOD was presented as the mean value of three detections for
each sample.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Extraction of total RNA from clinical specimens and
cancer cells was performed using the TRIzol® RNA Reagent
kit (Takara Bio, Inc., Otsu, Japan). RT of RNA into cDNA was
conducted using the Applied Biosystems SYBR Green mix kit (cat. no.
163795-75-3; Shanghai Aladdin Biochemical Company, Shanghai,
China), according to the manufacturer's protocol. The primer
sequences for TUSC3 and GAPDH are shown in Table I. A total of 5 µl DNA Marker was
used, and 1.5% agarose gel electrophoresis was performed using 5 µl
RT-PCR product. GAPDH was used as an endogenous reference gene to
analyze the relative gene expression levels. The thermocycling
conditions were as follows: One cycle of 95°C for 3 min, followed
by 35 cycles of 95°C for 5 sec, 58°C for 30 sec and 72°C for 30
sec. The expression levels were analyzed according to the
2−ΔΔCq method (20). All
experiments were performed in triplicate. The appearance of a
single peak in the melting curve implicated the specificity of the
PCR products.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction analysis of
mRNA levels. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction analysis of
mRNA levels.
| Gene | Primer sequence
(5′-3′) |
|---|
| TUSC3 | F:
GGCTCAGTTTGTGGCAGAATC |
|
| R:
CATCGCCTTTCGAAGTTGCT |
| GAPDH | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
Western blotting
HKC8, A498, 786-O, OS-RC-2 and ACHN cells were lysed
using a Total Protein Extraction kit (Wuhan Goodbio Technology Co.,
Ltd., Wuhan, China), according to the manufacturer's protocol.
Protein concentrations were assessed using a Bicinchoninic Acid
assay prior to loading the samples. Briefly, 40 µg/lane total
protein from each sample was separated by 10% SDS-PAGE and
transferred to a nitrocellulose membrane. The membranes were
blocked for 2 h at room temperature with 5% milk dissolved in TBS
containing 0.05% Tween-20 (TBST)), and incubated with the primary
polyclonal antibodies anti-TUSC3 (1:500; cat. no. ab77600; Abcam)
and anti-β-actin (1:500; cat. no. SA00001-9; ProteinTech Group
Inc.), at 4°C overnight. Subsequently, the membranes were washed
three times with TBST, and incubated with horseradish
peroxidase-conjugated secondary antibodies (1:1,000; cat. no.
SA00001-9; ProteinTech Group, Inc.) in 5% non-fat milk at room
temperature for 1 h. Following three washes with TBST, the
membranes were developed with enhanced chemiluminescence western
blotting detection kit (EMD Millipore, Billerica, MA, USA),
Expression of TUSC3 protein in each group was semi-quantified using
ImageJ software (National Institutes of Health, Bethesda, MD) and
Image Pro Plus v6.0 software (Media Cybernetics, Inc.).
Statistical analysis
Experiments were repeated at least three times. All
data are presented as the mean ± standard deviation and were
analyzed using SPSS v11.0 (SPSS, Inc., Chicago, IL, USA).
Differences in values and percentages among groups were compared
using a paired t-test, χ2 test, Fisher's exact test or
one-way analysis of variance followed by Dunnett's multiple
comparison test, respectively. Survival length was calculated from
the date of surgery to the date of mortality or last follow-up.
Survival curves and univariate analysis were estimated using the
Kaplan-Meier method and the log-rank test. Parameters that
demonstrated a statistically significant effect on overall survival
in the univariate analysis were included in a Cox multivariate
proportional hazards regression model. P<0.05 was considered to
indicate a statistically significant difference.
Results
TUSC3 expression in clinical
specimens
To clarify whether TUSC3 expression is associated
with ccRCC progression, carcinoma tissues and adjacent
non-neoplastic parenchyma were analyzed using RT-qPCR and
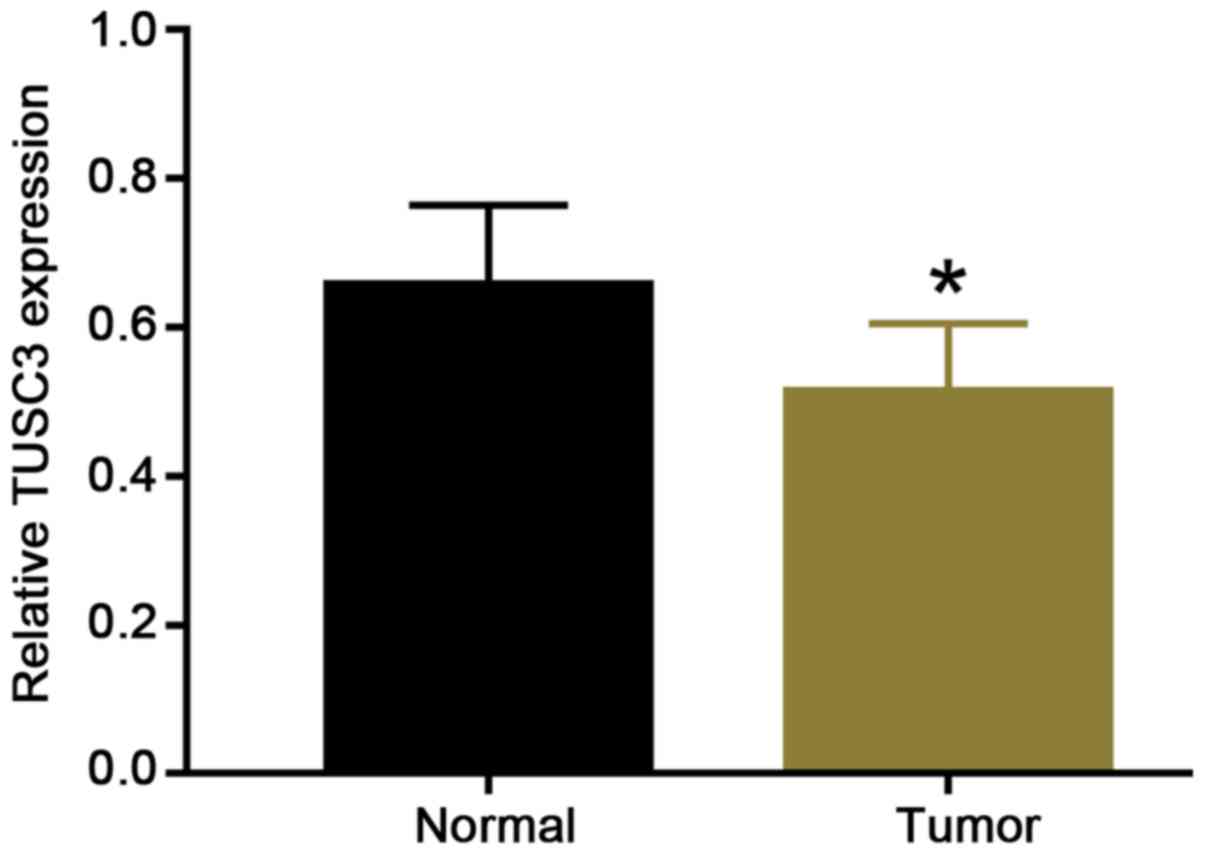
immunohistochemical staining. The RT-qPCR data revealed that the
relative expression of TUSC3 in adjacent normal tissues was
0.657±0.101 and the relative expression of TUSC3 in ccRCC tissues
was 0.512±0.087. Therefore, compared with that in adjacent normal
tissues, TUSC3 expression was significantly reduced in ccRCC
tissues (Fig. 1). The mean (0.5845)
of the relative expression of TUSC3 in tissues of all paired
samples was used as a cut-off to determine high and low expression
groups. In 54 paired specimens from patients with ccRCC, 35.2% of
tissues exhibited high expression of TUSC3, and 64.8% of tissues
exhibited low expression of TUSC3 (Table II). TUSC3 expression in adjacent
normal tissues was distinctly higher than that in ccRCC tissues
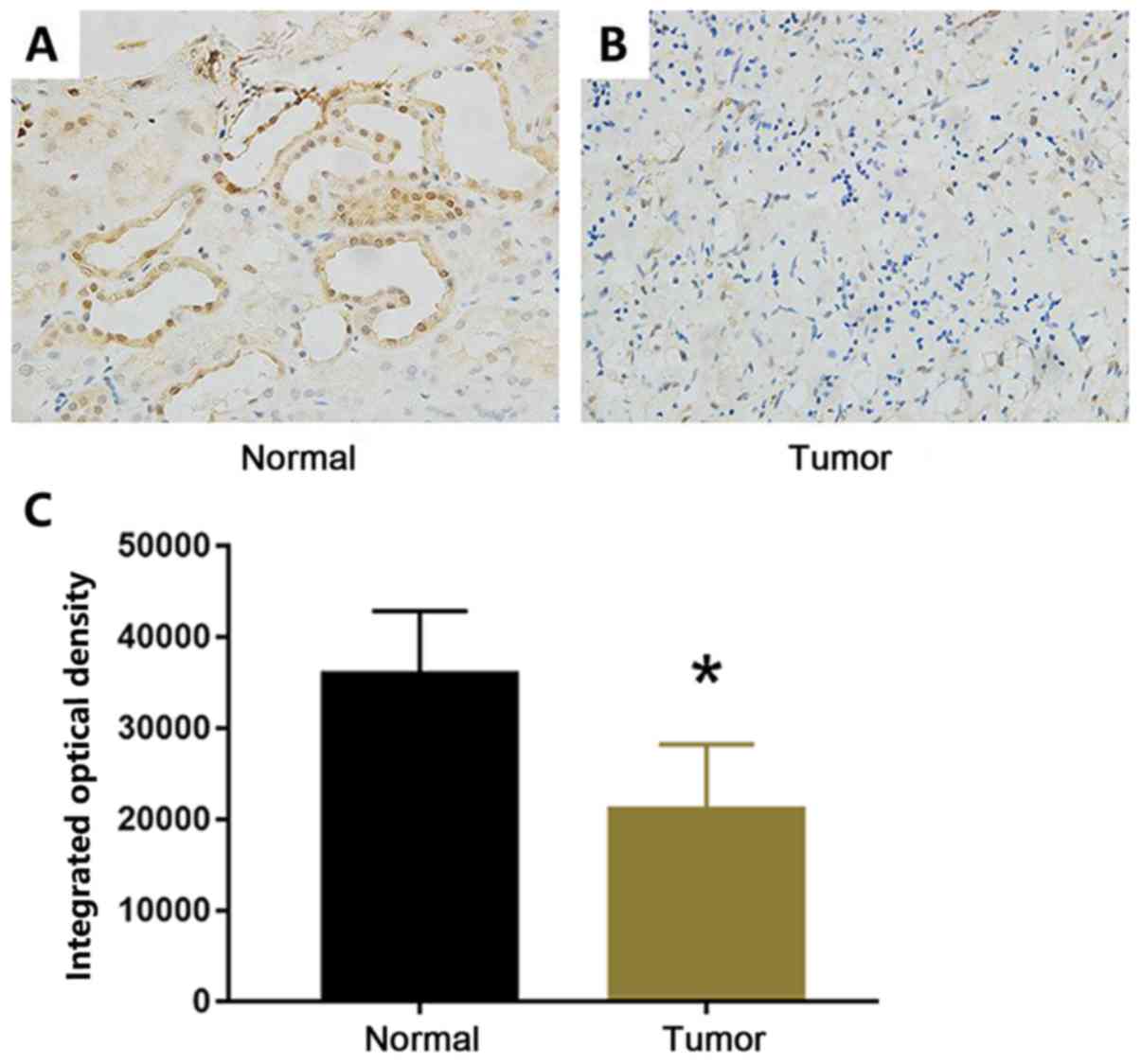
(P=0.007; Table II). As shown in
Fig. 2, TUSC3 expression levels were
decreased in tumor tissues compared with those in adjacent normal
tissues. In tumor tissues, TUSC3 was expressed in the cytoplasm and
nucleus of renal epithelial cells of proximal and distal tubules
(Fig. 2). Furthermore, tumor stage
and grade were classified according to the Tumor-Node-Metastasis
(TNM) staging system of the American Joint Committee on Cancer
guidelines (19) (Table III). TUSC3 levels were
significantly associated with primary tumor size (P=0.030), tumor
thrombus (P=0.044), clinical tumor stage (P=0.039), regional lymph
node involvement (P=0.040), distant metastasis (P=0.044), TNM stage
(P=0.039) and nuclear grade (P=0.021). TUSC3 expression in tumor
tissues was significantly decreased in higher TNM stages. These
results indicated that TUSC3 expression was closely associated with
ccRCC progression.
 | Table II.Expression levels of TUSC3 in ccRCC
and adjacent tissues. |
Table II.
Expression levels of TUSC3 in ccRCC
and adjacent tissues.
|
|
| TUSC3
expression |
|---|
|
|
|
|
|---|
| Tissues | n | Low, n (%) | High, n (%) |
|---|
| ccRCC | 54 | 35 (64.8) | 19 (35.2) |
| Adjacent normal
tissues | 54 | 21 (38.9) | 33 (61.1) |
| χ2 |
|
| 7.269 |
| P-value |
|
|
0.007a |
 | Table III.Clinicopathological characteristics
of 54 patients with clear cell renal cell carcinoma and their
association with TUSC3 expression. |
Table III.
Clinicopathological characteristics
of 54 patients with clear cell renal cell carcinoma and their
association with TUSC3 expression.
|
|
| TUSC3
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | n | Low, n (%) | High, n (%) | P-value |
|---|
| Age at surgery,
years |
|
|
| 0.999 |
|
<65 | 27 | 18 (66.6) | 9 (33.3) |
|
|
≥65 | 27 | 17 (63.0) | 10 (37.0) |
|
| Sex |
|
|
| 0.572 |
|
Male | 34 | 21 (61.8) | 13 (38.2) |
|
|
Female | 20 | 14 (70.0) | 6 (30.0) |
|
| Primary tumor size,
cm |
|
|
| 0.030a |
|
<7 | 38 | 21 (60.5) | 17 (39.5) |
|
| ≥7 | 16 | 14 (87.5) | 2 (12.5) |
|
| Tumor thrombus |
|
|
| 0.044a |
|
None | 47 | 28 (59.6) | 19 (40.4) |
|
| Level
0-IV | 7 | 7 (100.0) | 0 (0.0) |
|
| Primary tumor
classification |
|
|
| 0.039a |
|
T1+T2 | 42 | 24 (57.1) | 18 (42.9) |
|
|
T3+T4 | 12 | 11 (91.7) | 1 (8.3) |
|
| Regional lymph node
involvement |
|
|
| 0.040a |
| N0 | 43 | 26 (60.5) | 17 (39.5) |
|
|
N1+N2 | 11 | 11 (100.0) | 0 (0.0) |
|
| Distant
metastasis |
|
|
| 0.044a |
| M0 | 47 | 28 (59.6) | 19 (40.4) |
|
| M1 | 7 | 7 (100.0) | 0 (0.0) |
|
| TNM stage |
|
|
| 0.039a |
|
I+II | 42 | 24 (57.1) | 18 (42.9) |
|
|
III+IV | 12 | 11 (91.7) | 1 (8.3) |
|
| Nuclear grade |
|
|
| 0.021a |
|
1+2 | 41 | 23 (56.1) | 18 (43.9) |
|
|
3+4 | 13 | 12 (92.3) | 1 (7.7) |
|
Association between TUSC3 expression
and survival rate in patients
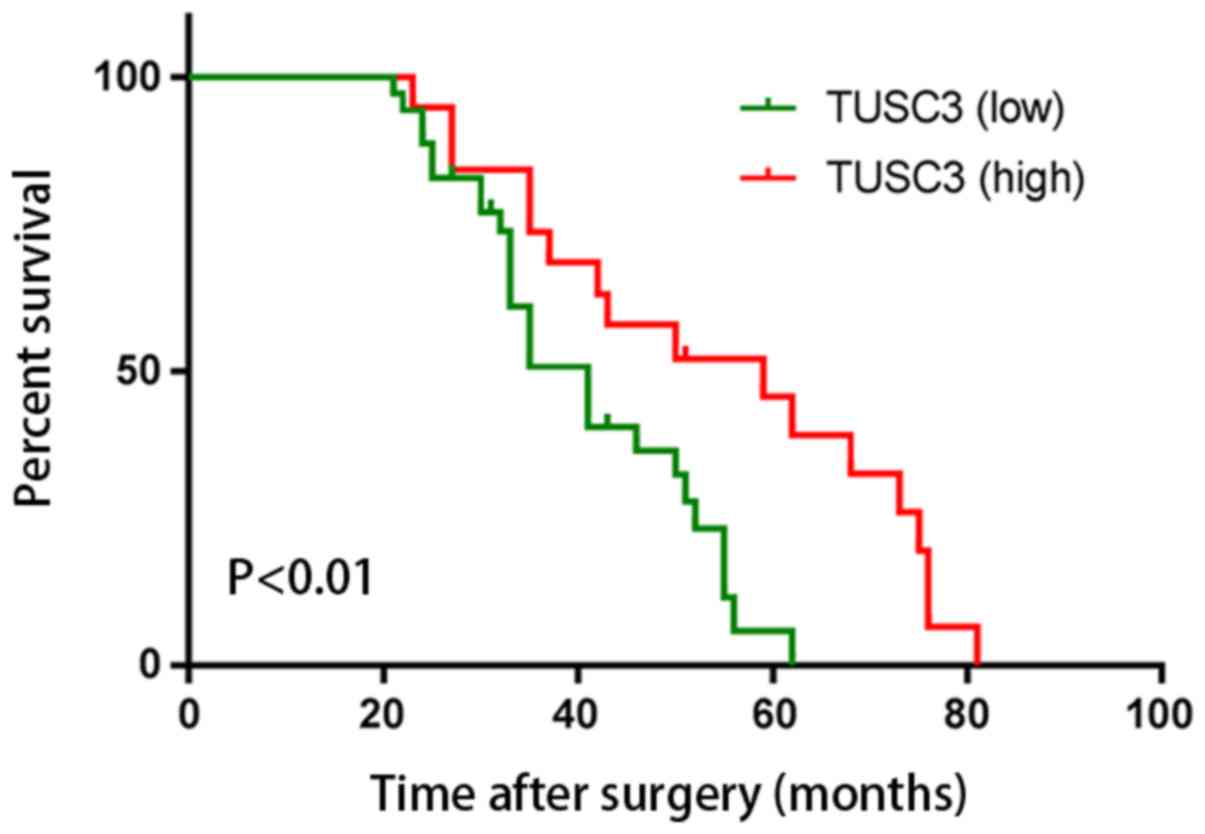
To the best of our knowledge, no study has
demonstrated that the level of TUSC3 expression is a good predictor
of survival in patients with ccRCC. In the present study, to
determine the prognostic value of TUSC3, overall survival rates of
54 patients with ccRCC were explored using Kaplan-Meier survival
curves. Postoperatively, patients with a high expression level of
TUSC3 exhibited a higher overall survival rate compared with that
of patients with a high expression level of TUSC3 (P<0.01;
Fig. 3). This indicated that TUSC3
expression was a prognostic factor in patients with ccRCC. Cox
univariate analysis revealed that primary tumor size (P=0.016),
tumor thrombus (P=0.020), primary tumor classification (P=0.022),
regional lymph node involvement (P=0.019), distant metastasis
(P=0.026), TNM stage group (P=0.020), nuclear grade (P=0.015) and
TUSC3 expression (P<0.001) were all significantly associated
with overall survival (Table IV).
Furthermore, Cox multivariate regression analysis suggested that
primary tumor size (P=0.032), tumor thrombus (P=0.018), primary
tumor classification (P=0.036), regional lymph node involvement
(P=0.039), distant metastasis (P=0.044), TNM stage group (P=0.037),
nuclear grade (P=0.012) and TUSC3 expression (P=0.015) were all
independent prognostic factors for patients with ccRCC. These data
indicated that low intratumoral TUSC3 expression may be used as a
novel marker for ccRCC progression and a poor prognosis.
 | Table IV.Univariate and multivariate analyses
of overall survival of patients with clear cell renal cell
carcinoma. |
Table IV.
Univariate and multivariate analyses
of overall survival of patients with clear cell renal cell
carcinoma.
|
| Overall
survival |
|---|
|
|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
Characteristics | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age at surgery
(<65 years vs. ≥65 years) | 1.784
(1.077–3.531) | 0.062 | 1.580
(1.012–3.230) | 0.079 |
| Sex (male vs.
female) | 0.728
(0.358–1.369) | 0.236 | 0.625
(0.310–1.158) | 0.291 |
| Primary tumor size
(<7 cm vs. ≥7 cm) | 4.632
(2.653–6.758) | 0.016a | 5.220
(2.793–9.365) | 0.032a |
| Tumor thrombus
(None vs. level 0-IV) | 9.458
(4.635–21.325) | 0.020a | 8.457
(3.656–17.632) | 0.018a |
| Primary tumor
classification (T1+T2 vs. T3+T4) | 4.320
(2.542–9.326) | 0.022a | 4.550
(2.860–11.736) | 0.036a |
| Regional lymph node
involvement (NX+N0 vs. N1+N2) | 7.236
(4.335–26.324) | 0.019a | 6.358
(3.886–23.656) | 0.039a |
| Distant metastasis
(M0 vs. M1) | 8.873
(2.365–24.325) | 0.026a | 5.637
(1.875–17.639) | 0.044a |
| TNM stage (I+II vs.
III+IV) | 3.369
(2.859–9.635) | 0.020a | 3.963
(3.582–12.362) | 0.037a |
| Nuclear grade (1+2
vs. 3+4) | 5.238
(3.596–11.387) | 0.015a | 4.698
(3.157–9.692) | 0.012a |
| TUSC3 expression
(low vs. high) | 0.185
(0.102–3.112) | <0.001 | 0.326
(1.786–5.447) | 0.015a |
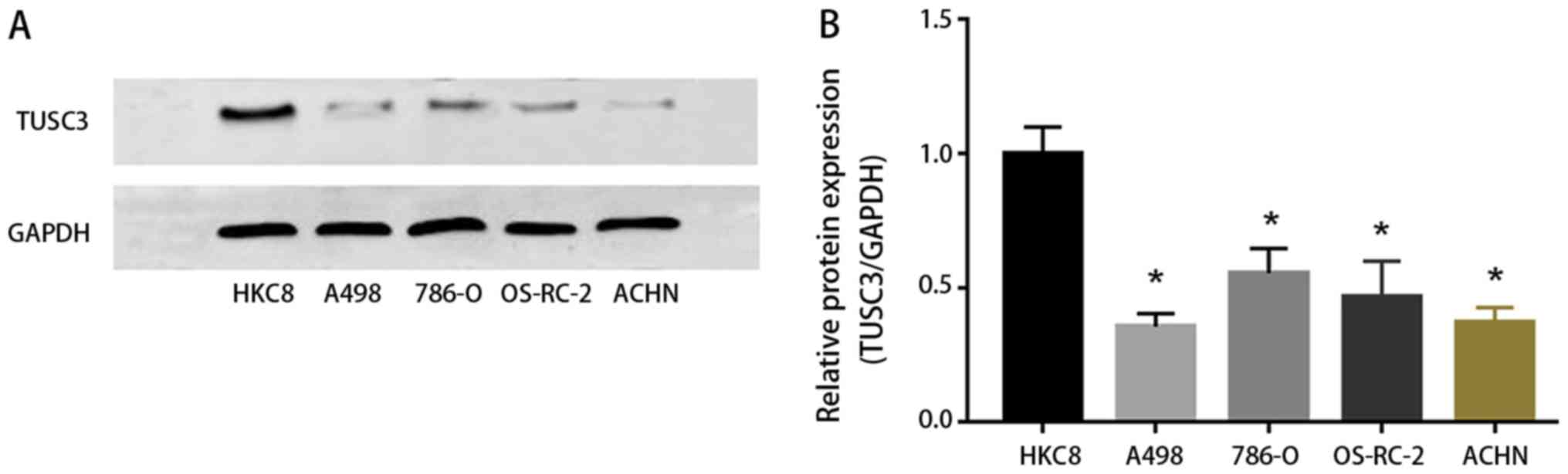
TUSC3 expression in ccRCC cells
To explore the influence of TUSC3 on RCC cell lines
(A498, 786-O, OS-RC-2 and ACHN), expression levels of TUSC3 were
detected by western blotting. Compared with the control group (HKC8
cells), A498, 786-O, OS-RC-2 and ACHN cells exhibited lower protein
expression levels of TUSC3 (P<0.05; Fig. 4). This supported our in vivo
data demonstrating that TUSC3 expression inhibits ccRCC
tumorigenesis.
Discussion
ccRCC accounts for 2–3% of adult tumors,
representing 90% of renal malignancies (21,22).
Despite the development of diagnostic techniques in recent years,
~30% of patients with ccRCC are diagnosed with metastases at the
first diagnosis, and 30–40% of patients exhibit localized ccRCC
recurrence and metastasis following surgical resection (23,24).
Surgery is the most common primary therapeutic method for ccRCC;
however, ccRCC cannot be treated completely by radical surgery.
Recent studies have focused on the possibility of combining
modalities for improving the therapeutic value of existing standard
therapies, including chemotherapy and radiotherapy (25,26);
however, ccRCC is not sensitive to radiotherapy and chemotherapy.
Patients with ccRCC continue to have extremely poor outcomes
(27,28). The genesis and progression of ccRCC
involve various factors, including carcinogenic substances and
environmental factors (29,30). The mortality rate of patients with
metastatic ccRCC is high, although novel targeted therapies have
been developed. Therefore, determining prognostic markers to more
accurately select patients with ccRCC with poor survival is
becoming increasingly important.
TUSC3, also known as N33, is a gene segment with a
length of ~349,435 bp that is composed of 11 exons (31). The OST complex, which is a component
of the endoplasmic reticulum, promotes N-linked glycation of
proteins during the protein folding process (32,33). The
human OST complex is composed of seven elements, including OST
complex subunit 4, dolichyl-diphosphooligosaccharide protein
glycosyltransferase non-catalytic subunit, defender against cell
death 1, ribophorin I, ribophorin II, STT3
oligosaccharyltransferase complex catalytic subunit A or STT3
oligosaccharyltransferase complex catalytic subunit B, TUSC3/N33
and magnesium transporter 1 (16,34).
TUSC3 has been demonstrated to exert various biological functions
in human learning and memory processes, and its mechanism is
associated with the alteration of the magnesium ion transport
system (35). Numerous studies have
demonstrated that TUSC3 is mainly expressed in the epithelium of
the liver, lung, placenta, prostate, ovary, colon, testis and
adipose tissues (13,18,36–40).
TUSC3 was identified as a potential anti-oncogene when it was first
identified in the 1990s, and deletion of TUSC3 expression is
associated with the malignant transformation of cells (37). A large number of studies reported
that the carcinogenesis of pancreatic, gastric, ovarian and
prostate cancer may be associated with the mutation or deletion of
TUSC3 (11,41,42).
Furthermore, the loss of TUSC3 expression may result in an increase
in cancer cell growth, migration and invasion (43,44).
In the present study, TUSC3 protein was identified
to be downregulated in human ccRCC tissues, and its expression was
significantly associated with clinical stage and lymph node
metastasis. Patients with ccRCC with low expression of TUSC3
exhibited a higher TNM stage, and TUSC3 was a prognostic factor for
the overall postoperative survival of patients. Numerous studies
have demonstrated the anticancer properties of TUSC3 (11,13,42,44);
however, the precise molecular mechanism of the function of TUSC3
in the development of tumor remains poorly understood. The sequence
of the chromosomal band 8p22, where TUSC3 is located, has been
revealed to be lost in human prostate cancer (45). Subsequent studies indicated that the
proliferation, migration and invasion of prostate and ovarian
cancer cells can be increased due to decreased TUSC3 expression
(40,46). A previous study demonstrated that
TUSC3 expression is downregulated in higher grades of ovarian
cancer (46). Subsequently, a study
explored the molecular mechanism of TUSC3 in ovarian cancer, which
revealed that the hypermethylation of the TUSC3 promoter could lead
to low expression of TUSC3, and the methylation of promoter is a
prognostic indicator of patients with cancer (37). The present study explored TUSC3
expression in ccRCC specimens and its association with TNM staging
and overall survival of patients with ccRCC, and demonstrated that
immunohistochemical staining for TUSC3 served an important role in
the prediction of ccRCC progression. Furthermore, the data
suggested that low expression of TUSC3 in ccRCC cells was
associated with the poor prognosis of patients, revealing that
TUSC3 expression may be associated with the progression of ccRCC.
In addition, the functions and biological mechanisms of TUSC3 are
currently being elucidated (33).
N-linked glycosylation of proteins serves an important role in
protein synthesis, suggesting that TUSC3 may block tumor
progression by transforming the glycosylation reaction in various
types of carcinoma (33).
Dysfunction of TUSC3 may result in improper protein glycosylation,
potentially leading to disorders of cellular biological function
(42). The present study revealed
that TUSC3 expression may inhibit tumor progression in patients
with ccRCC, and TUSC3 may serve as a biomarker to identify tumor
progression and the prognosis of patients with ccRCC.
There were several limitations to the present study.
One was the relatively small sample size. Second, the biomarker
role of TUSC3 was only tested in ccRCC cell lines and a papillary
renal cell carcinoma cell line, thus further verification in other
types of RCC cell lines is required. Third, the present study did
not investigate the antitumor mechanism of TUSC3 in ccRCC.
In conclusion, the present study demonstrated that
TUSC3 is associated with the progression of ccRCC and the prognosis
of patients with ccRCC. However, the antitumor mechanism of TUSC3
in ccRCC requires further investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JJZ conceived, designed and supervised the study.
YJY drafted the manuscript. YJY and ZJC conducted the experiment.
YXL contributed to statistical analysis. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee of Jingzhou Central Hospital, Jingzhou, China. All
patients enrolled in the present study signed informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barata PC and Rini BI: Treatment of renal
cell carcinoma: Current status and future directions. CA Cancer J
Clin. 67:507–524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cavaliere C, D'Aniello C, Pepa CD,
Pisconti S, Berretta M and Facchini G: Current and emerging
treatments for metastatic renal cell carcinoma. Curr Cancer Drug
Targets. 18:468–479. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miao D, Margolis CA, Gao W, Voss MH, Li W,
Martini DJ, Norton C, Bossé D, Wankowicz SM, Cullen D, et al:
Genomic correlates of response to immune checkpoint therapies in
clear cell renal cell carcinoma. Science. 359:801–806. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhai W, Ma J, Zhu R, Xu C, Zhang J, Chen
Y, Chen Z, Gong D, Zheng J, Chen C, et al: MiR-532-5p suppresses
renal cancer cell proliferation by disrupting the ETS1-mediated
positive feedback loop with the KRAS-NAP1L1/P-ERK axis. Br J
Cancer. 119:591–604. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu KG, Gupta S and Goel S: Immunotherapy:
Incorporation in the evolving paradigm of renal cancer management
and future prospects. Oncotarget. 8:17313–17327. 2017.PubMed/NCBI
|
|
8
|
Du Y, Pahernik S, Hadaschik B, Teber D,
Duensing S, Jäger D, Hohenfellner M and Grüllich C: Survival and
prognostic factors of patients with renal cell cancer with bone
metastasis in the era of targeted therapy: A single-institution
analysis. Urol Oncol. 34:433.e1–e8. 2016. View Article : Google Scholar
|
|
9
|
Escudier B, Eisen T, Stadler WM, Szczylik
C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA,
et al: Sorafenib in advanced clear-cell renal-cell carcinoma. N
Engl J Med. 356:125–134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vašíčková K, Horak P and Vaňhara P: TUSC3:
Functional duality of a cancer gene. Cell Mol Life Sci. 75:849–857.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan X, Zhang X, Shen J, Zhao H, Yu X, Chen
Y, Zhuang Z, Deng X, Feng H, Wang Y and Peng L: Decreased TUSC3
promotes pancreatic cancer proliferation, invasion and metastasis.
PLoS One. 11:e01490282016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu YF and Dong M: Expression of TUSC3 and
its prognostic significance in colorectal cancer. Pathol Res Pract.
214:1497–1503. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Birnbaum DJ, Adélaïde J, Mamessier E,
Finetti P, Lagarde A, Monges G, Viret F, Gonçalvès A, Turrini O,
Delpero JR, et al: Genome profiling of pancreatic adenocarcinoma.
Genes Chromosomes Cancer. 50:456–465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Arbieva ZH, Banerjee K, Kim SY, Edassery
SL, Maniatis VS, Horrigan SK and Westbrook CA: High-resolution
physical map and transcript identification of a prostate cancer
deletion interval on 8p22. Genome Res. 10:244–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mohorko E, Glockshuber R and Aebi M:
Oligosaccharyltrans-ferase: The central enzyme of N-linked protein
glycosylation. J Inherit Metab Dis. 34:869–878. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peng Y, Cao J, Yao XY, Wang JX, Zhong MZ,
Gan PP and Li JH: TUSC3 induces autophagy in human non-small cell
lung cancer cells through Wnt/beta-catenin signaling. Oncotarget.
8:52960–52974. 2017.PubMed/NCBI
|
|
18
|
Liu K, Xie F, Gao A, Zhang R, Zhang L,
Xiao Z, Hu Q, Huang W, Huang Q, Lin B, et al: SOX2 regulates
multiple malignant processes of breast cancer development through
the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol Cancer. 16:622017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amin MB and Edge SB: American Joint
Committee on Cancer. AJCC cancer staging manual. Eighth.
Switzerland: Springer; 2017, View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hirata H, Hinoda Y, Ueno K, Majid S, Saini
S and Dahiya R: Role of secreted frizzled-related protein 3 in
human renal cell carcinoma. Cancer Res. 70:1896–1905. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan X, Liu Y, Hou J and Cao G: Targeted
therapies for renal cell carcinoma in Chinese patients: Focus on
everolimus. Onco Targets Ther. 8:313–321. 2015.PubMed/NCBI
|
|
23
|
Pei X, Li M, Zhan J, Yu Y, Wei X, Guan L,
Aydin H, Elson P, Zhou M, He H and Zhang H: Enhanced IMP3
expression activates NF-κB pathway and promotes renal cell
carcinoma progression. PLoS One. 10:e01243382015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sanchez-Gastaldo A, Kempf E, González Del
Alba A and Duran I: Systemic treatment of renal cell cancer: A
comprehensive review. Cancer Treat Rev. 60:77–89. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miao J, Wang L, Zhu M, Xiao W, Wu H, Di M,
Huang Y, Huang S, Han F, Deng X, et al: Long-term survival and late
toxicities of elderly nasopharyngeal carcinoma (NPC) patients
treated by high-total- and fractionated-dose simultaneous modulated
accelerated radiotherapy with or without chemotherapy. Oral Oncol.
89:40–47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng R, Lian S, Huang X, Guan G, Li X,
Chi P and Xu B: The survival benefit of intensified full-dose XELOX
chemotherapy concomitant to radiotherapy and then resting-period
consolidation chemotherapy in locally advanced rectal cancer. J
Cancer. 10:730–736. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie J, Lin W, Huang L, Xu N, Xu A, Chen B,
Watanabe M, Liu C and Huang P: Bufalin suppresses the proliferation
and metastasis of renal cell carcinoma by inhibiting the
PI3K/Akt/mTOR signaling pathway. Oncol Lett. 16:3867–3873.
2018.PubMed/NCBI
|
|
28
|
Chandrasekar T, Klaassen Z, Goldberg H,
Kulkarni GS, Hamilton RJ and Fleshner NE: Metastatic renal cell
carcinoma: Patterns and predictors of metastases-A contemporary
population-based series. Urol Oncol. 35:661.e7–661.e14. 2017.
View Article : Google Scholar
|
|
29
|
Melkonian SC, Daniel CR, Ye Y, Tannir NM,
Karam JA, Matin SF, Wood CG and Wu X: Gene-environment interaction
of genome-wide association study-identified susceptibility loci and
meat-cooking mutagens in the etiology of renal cell carcinoma.
Cancer. 122:108–115. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deckers IA, van den Brandt PA, van
Engeland M, van Schooten FJ, Godschalk RW, Keszei AP, Hogervorst JG
and Schouten LJ: Potential role of gene-environment interactions in
ion transport mechanisms in the etiology of renal cell cancer. Sci
Rep. 6:342622016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang MJ, Xing LX, Cui M, Yang X, Shi JG,
Li J, Zhang KJ, Zheng ZJ, Zhang FC, Li JL and Gao XC: Association
of TUSC3 gene polymorphisms with non-syndromic mental retardation
based on nuclear families in the Qinba mountain area of China.
Genet Mol Res. 14:5022–5030. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Molinari F, Foulquier F, Tarpey PS,
Morelle W, Boissel S, Teague J, Edkins S, Futreal PA, Stratton MR,
Turner G, et al: Oligosaccharyltransferase-subunit mutations in
nonsyndromic mental retardation. Am J Hum Genet. 82:1150–1157.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mohorko E, Owen RL, Malojčić G, Brozzo MS,
Aebi M and Glockshuber R: Structural basis of substrate specificity
of human oligosaccharyl transferase subunit N33/Tusc3 and its role
in regulating protein N-glycosylation. Structure. 22:590–601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vandewynckel YP, Laukens D, Geerts A,
Bogaerts E, Paridaens A, Verhelst X, Janssens S, Heindryckx F and
Van Vlierberghe H: The paradox of the unfolded protein response in
cancer. Anticancer Res. 33:4683–4694. 2013.PubMed/NCBI
|
|
35
|
Zhou H and Clapham DE: Mammalian MagT1 and
TUSC3 are required for cellular magnesium uptake and vertebrate
embryonic development. Proc Natl Acad Sci USA. 106:15750–15755.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
PLOS ONE: Staff: Correction: Decreased
TUSC3 promotes pancreatic cancer proliferation, invasion and
metastasis. PLoS One. 11:e01517522016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pils D, Horak P, Vanhara P, Anees M, Petz
M, Alfanz A, Gugerell A, Wittinger M, Gleiss A, Auner V, et al:
Methylation status of TUSC3 is a prognostic factor in ovarian
cancer. Cancer. 119:946–954. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo J, Zhu H, Jiang H, Cui Y, Wang M, Ni X
and Ma C: The effects of aberrant expression of LncRNA
DGCR5/miR-873-5p/TUSC3 in lung cancer cell progression. Cancer Med.
2018. View Article : Google Scholar
|
|
39
|
Feng S, Zhai J, Lu D, Lin J, Dong X, Liu
X, Wu H, Roden AC, Brandi G, Tavolari S, et al: TUSC3 accelerates
cancer growth and induces epithelial-mesenchymal transition by
upregulating claudin-1 in non-small-cell lung cancer cells. Exp
Cell Res. 373:44–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Horak P, Tomasich E, Vaňhara P,
Kratochvílová K, Anees M, Marhold M, Lemberger CE, Gerschpacher M,
Horvat R, Sibilia M, et al: TUSC3 loss alters the ER stress
response and accelerates prostate cancer growth in vivo. Sci Rep.
4:37392014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gu Y, Pei X, Ren Y, Cai K, Guo K, Chen J,
Qin W, Lin M, Wang Q, Tang N, et al: Oncogenic function of TUSC3 in
non-small cell lung cancer is associated with Hedgehog signalling
pathway. Biochim Biophys Acta Mol Basis Dis. 1863:1749–1760. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kratochvílová K, Horak P, Ešner M, Souček
K, Pils D, Anees M, Tomasich E, Dráfi F, Jurtíková V, Hampl A, et
al: Tumor suppressor candidate 3 (TUSC3) prevents the
epithelial-to-mesenchymal transition and inhibits tumor growth by
modulating the endoplasmic reticulum stress response in ovarian
cancer cells. Int J Cancer. 137:1330–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Duppel U, Woenckhaus M, Schulz C, Merk J
and Dietmaier W: Quantitative detection of TUSC3 promoter
methylation-a potential biomarker for prognosis in lung cancer.
Oncol Lett. 12:3004–3012. 2017. View Article : Google Scholar
|
|
44
|
Gu Y, Wang Q, Guo K, Qin W, Liao W, Wang
S, Ding Y and Lin J: TUSC3 promotes colorectal cancer progression
and epithelial-mesenchymal transition (EMT) through WNT/β-catenin
and MAPK signalling. J Pathol. 239:60–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bova GS, Carter BS, Bussemakers MJ, Emi M,
Fujiwara Y, Kyprianou N, Jacobs SC, Robinson JC, Epstein JI, Walsh
PC, et al: Homozygous deletion and frequent allelic loss of
chromosome 8p22 loci in human prostate cancer. Cancer Res.
53:3869–3873. 1973.
|
|
46
|
Vaňhara P, Horak P, Pils D, Anees M, Petz
M, Gregor W, Zeillinger R and Krainer M: Loss of the oligosaccharyl
transferase subunit TUSC3 promotes proliferation and migration of
ovarian cancer cells. Int J Oncol. 42:1383–1389. 2013. View Article : Google Scholar : PubMed/NCBI
|