Introduction
Lung cancer is the most common malignancy and the
leading cause of mortality among cancer-associated diseases
worldwide (1). Non-small cell lung
cancer (NSCLC) accounts for >85% of lung cancer cases (2). The pathologic types of NSCLC include
large-cell carcinoma, squamous cell carcinoma and adenocarcinoma
(3). In spite of recent advances in
surgical resection, chemotherapy and radiotherapy for lung cancer,
the survival rate of patients remains unsatisfactory, with a 5-year
survival rate as low as 15%, as it is often diagnosed at an
advanced stage (4,5). Therefore, there is an urgent need to
identify effective molecular diagnostic markers for early detection
of lung cancer (6).
Circular RNAs (circRNAs) are a class of endogenous
non-coding RNAs generated from back-splicing and characterized by a
covalent closed loop without 3′ and 5′ ends (7). In the past two decades, circRNAs have
been hypothesized to be non-functional due to errors in splicing
(8). In 2011, Salmena et al
(9) presented the competing
endogenous RNA hypothesis, which suggests that mRNAs, transcribed
pseudogenes and long non-coding RNAs can suppress microRNA (miRNA)
expression and function by competitive binding to miRNA response
elements. Furthermore, it was demonstrated that non-coding RNAs are
involved in the initiation and progression of malignant tumors
(10). As another class of
non-coding RNAs, circRNAs are more stable than linear RNAs due to
their covalently closed structures, and the sponge effect of
circRNAs may be more efficient in comparison with that of linear
RNAs (11). Recently, a number of
circRNAs have been demonstrated to regulate the function of miRNAs,
serving as miRNA sponges, as well as be involved in
post-transcriptional regulation in cancer (12). For instance, Zhong et al
(13) reported that circTCF25 was
able to downregulate miR-103a-3p and miR-107, increase
cyclin-dependent kinase-6 expression, and promote the proliferation
and migration of bladder cancer cells. However, as an emerging type
of non-coding RNAs, the role of circRNAs in NSCLC has rarely been
reported (14).
Our previous study investigated circRNA expression
in NSCLC by ribosomal RNA depletion and RNA sequencing (15). Therefore, the present study was
designed to analyze the expression profile of circATXN7 in NSCLC
and examine its correlation with clinicopathological
characteristics. In addition, the effect of circATXN7 on NSCLC cell
proliferation and invasion was investigated.
Materials and methods
Patients and tissue samples
A total of 57 pairs of NSCLC tissues and adjacent
normal tissues (≥3 cm away from the tumor) were obtained from
patients who underwent surgery at the Department of Thoracic
Surgery, Peking University People's Hospital (Beijing, China)
between January 2012 and December 2013. These tissues were used for
validation of circRNAs by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). All surgical specimens were
snap-frozen and stored in liquid nitrogen immediately following
resection until the extraction of total RNA. The clinical and
pathological characteristics of each patient were collected. The
Tumor-Node-Metastasis (TNM) stage was determined according to The
8th Edition Lung Cancer Stage Classification (16). The present study was approved and
supervised by the Ethics Committee of Peking University People's
Hospital. Written informed consent was obtained from all the
patients for research purposes.
Cell culture and small interfering RNA
(siRNA) transfection
The NSCLC cell lines A549 and SPC-A1 were kindly
provided by Professor Xu (Jiangsu Key Laboratory of Molecular and
Translational Cancer Research, Nanjing Medical University
Affiliated Cancer Hospital, Nanjing, China). A549 cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), while SPC-A1 cells were grown in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin. All cells were cultured at 37°C in a humidified
atmosphere with 5% CO2. For siRNA transfection, NSCLC
cells were seeded in 6-well plates (6×104 cells/well)
and cultured at 37°C until the cell confluence reached 60–70%.
Subsequently, cells were transfected with specific siRNA (100 nM)
or control siRNA (100 nM) using Lipofectamine® RNAi MAX
according to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequent experiments were performed 24 h after
transfection. The following siRNA sequences were used: siRNA-1 for
circATXN7 sense, 5′-GGAAGGGAGCGGAAAGAAUGU-3′, and antisense,
5′-AUUCUUUCCGCUCCCUUCCCG-3′; siRNA-2 for circATXN7 sense,
5′-GGGAAGGGAGCGGAAAGAAUG-3′, and antisense,
5′-UUCUUUCCGCUCCCUUCCCGA-3′.
RNA extraction and RT-qPCR
analyses
Total RNA was extracted from the tumor tissues,
adjacent normal tissues and cultured cells using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol. Next, the RNA concentration was measured
by Uv-vis spectrophotometry (Evolution 60S; Thermo Fisher
Scientific, Inc.) and 1,000 ng total RNA was reverse transcribed in
a final volume of 20 µl using random primers under standard
conditions with the PrimeScript RT Reagent kit with gDNA Eraser
(cat. no. RR047A; Takara Biotechnology Co., Ltd., Dalian, China).
qPCR was then performed using SYBR Select Master Mix (cat. no.
4472908; Applied Biosystems; Thermo Fisher Scientific, Inc.) with
0.5 µl cDNA on a CFX96 Real-Time system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) according to the manufacturer's protocol.
β-actin was used as an internal control to determine the levels of
circATXN7 expression. The primer sequences used were as follows:
β-actin forward, 5′-GAAATCGTGCGTGACATTAA-3′, and reverse,
5′-AAGGAAGGCTGGAAGAGTG-3′; circATXN7 forward,
5′-CCTAGGGACAGAATTGGACGA-3′, and reverse, 5′-GCCCGCTCCGACATTCTT-3′.
The qPCR reaction included an initial denaturation step in a
96-well optical plate at 95°C for 10 min, followed by 40 cycles of
92°C for 15 sec and 60°C for 1 min. The relative levels of gene
expression were normalized to β-actin and calculated using the
2−ΔΔCq method to determine fold changes of circATXN7
expression (tumor vs. normal) (17).
The PCR products were subjected to Sanger sequencing by TSINGKE
Biological Technology Co., Ltd. (Beijing, China).
Nucleic acid electrophoresis
The cDNA and gDNA PCR products were investigated
using 4% agarose gel electrophoresis with TBE running buffer. The
primer sequences used were as follows: divergent forward,
5′-CCTAGGGACAGAATTGGACGA-3′, and reverse, 5′-GCCCGCTCCGACATTCTT-3′;
convergent forward, 5′-CTATCGTTTGCTGGGTTGCG-3′, and reverse,
5′-ACTTTCGTCCAATTCTGTCCCT-3′.
Cell proliferation assay
Cell proliferation was detected using an MTT assay
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China) according to the manufacturer's protocol. Briefly, the
transfected cells were seeded into 96-well plates (3,000
cells/well), and the proliferation rates were measured at 0, 24,
48, 72 and 96 h after transfection. A total of 20 µl MTT solution
was added to each well and incubated for 2 h at 37°C. Next, the
medium in each well was discarded, and 150 µl dimethyl sulfoxide
was added. Subsequent to shaking the plate for 15 min, the
absorbance was measured spectrophotometrically at 490 nm. The data
are representative of three individual experiments, where were
conducted in triplicate.
For the EdU assays, at 24 h after transfection,
6×104 cells/well were seeded in a 24-well plate. After a
further 24 h of incubation, EdU was added to a final concentration
of 50 µM per well. Following incubation with EdU for 2 h, the cells
were fixed for 20 min with 150 µl 4% formaldehyde solution in PBS.
EdU labelling with the kFluor488-azide was performed using a
kFluor488 Click-It EdU Cell Proliferation Assay kit (Nanjing KeyGen
Biotech Co., Ltd., Nanjing, China). For excitation of
kFluor488-azide, a 495-nm laser was used under a fluorescence
microscope (Olympus Corporation, Tokyo, Japan).
Clonogenic assay
A total of 600 cells/well were transfected in a
6-well plate with 2 ml medium containing 10% FBS. RPMI-1640 medium
and DMEM were used for A549 and SPC-A1 cells, respectively. The
medium was changed every 3 days. After 10 days, the cells were
immobilized using 4% paraformaldehyde and stained with 0.1% crystal
violet for 15 min at room temperature. The visible colonies of each
well were then manually counted (11).
Invasion assay
A Transwell assay was performed to investigate the
migration and invasion of human lung cancer SPC-A1 and A549 cells.
Briefly, the cells were transfected with 100 nM siRNA-circATXN7 or
siRNA-NC for 24 h, and then plated at a density of 1×105
cells/well in the upper Transwell chamber (pore size, 8 µm;
Corning, Inc., Corning, NY, USA), which contained 200 µl of the
corresponding serum-free medium. RPMI-1640 medium or DMEM
containing 10% FBS was added to the lower chamber for A549 and
SPC-A1 cells, respectively. After 48 h of incubation at 37°C with
5% CO2, the migrated cells on the filter surface were
fixed with methanol and stained with 0.01% crystal violet for 15
min at room temperature, while the cells remaining in the upper
membrane were discarded. The cell numbers were calculated in five
random fields under a microscope (CTR6000; Leica Microsystems GmbH,
Wetzlar, Germany). Each experiment was performed in triplicate.
Statistical analysis
All statistical analyses in the present study were
performed using the SPSS software package (version 20.0; IBM
Corporation, Armonk, NY, USA) and GraphPad Prism software (version
7.0; GraphPad software Inc., La Jolla, CA, USA). All quantitative
data are presented as the mean ± standard deviation from at least
three independent experiments. Two-tailed Student's t-test was used
for the analysis of continuous variables. Differences among the
three groups were analyzed by analysis of variance and Scheffe's
test. The prognostic value of circATXN7 expression was further
analyzed using the Kaplan-Meier method. P<0.05 was considered to
indicate a statistically significant difference.
Results
circRNAs in NSCLC
In our previous study, a number of circRNAs were
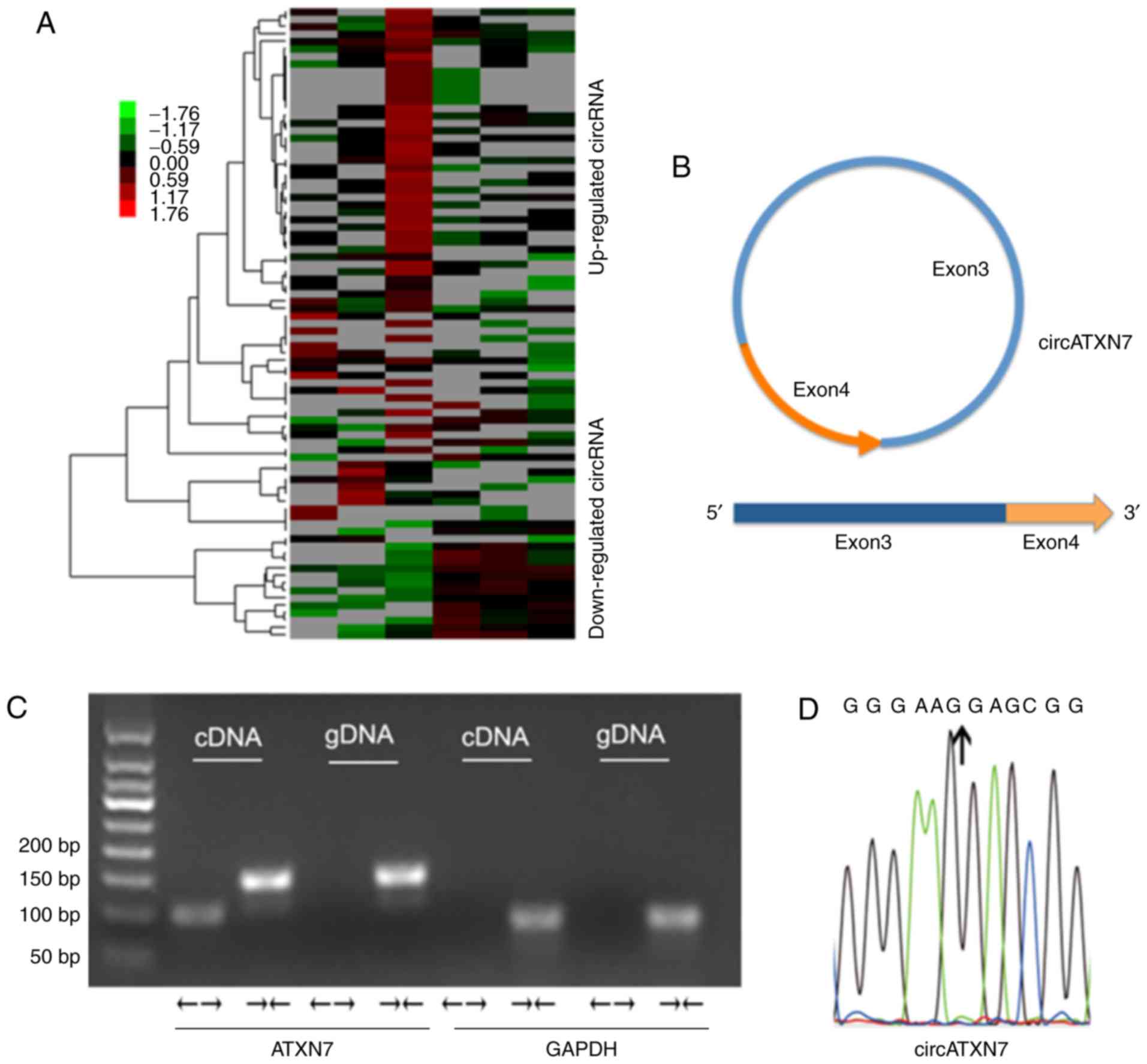
detected to be overexpressed in NSCLC (15) (Fig.
1A). In particular, one of the circRNAs was significantly
overexpressed, which was subsequently defined as circATXN7, since
it is derived from the ATXN7 gene. circATXN7 is back-spliced by
exons 3 and 4 of ATXN7 (Fig. 1B). To
characterize the circular form of circATXN7, convergent primers
that amplify the linear transcript of ATXN7 and divergent primers
that amplify circATXN7 were designed. The genomic DNA (gDNA) and
complementary DNA (cDNA) of A549 cells were amplified by divergent
and convergent primers, respectively. As indicated, the cDNA of the
PCR products was amplified by the divergent primers, while both
cDNA and gDNA of the PCR products were amplified by the convergent
primers. GAPDH served as a negative control (Fig. 1C). Sanger sequencing confirmed the
splicing junction site (Fig.
1D).
Aberrant expression of circATXN7 in
NSCLC
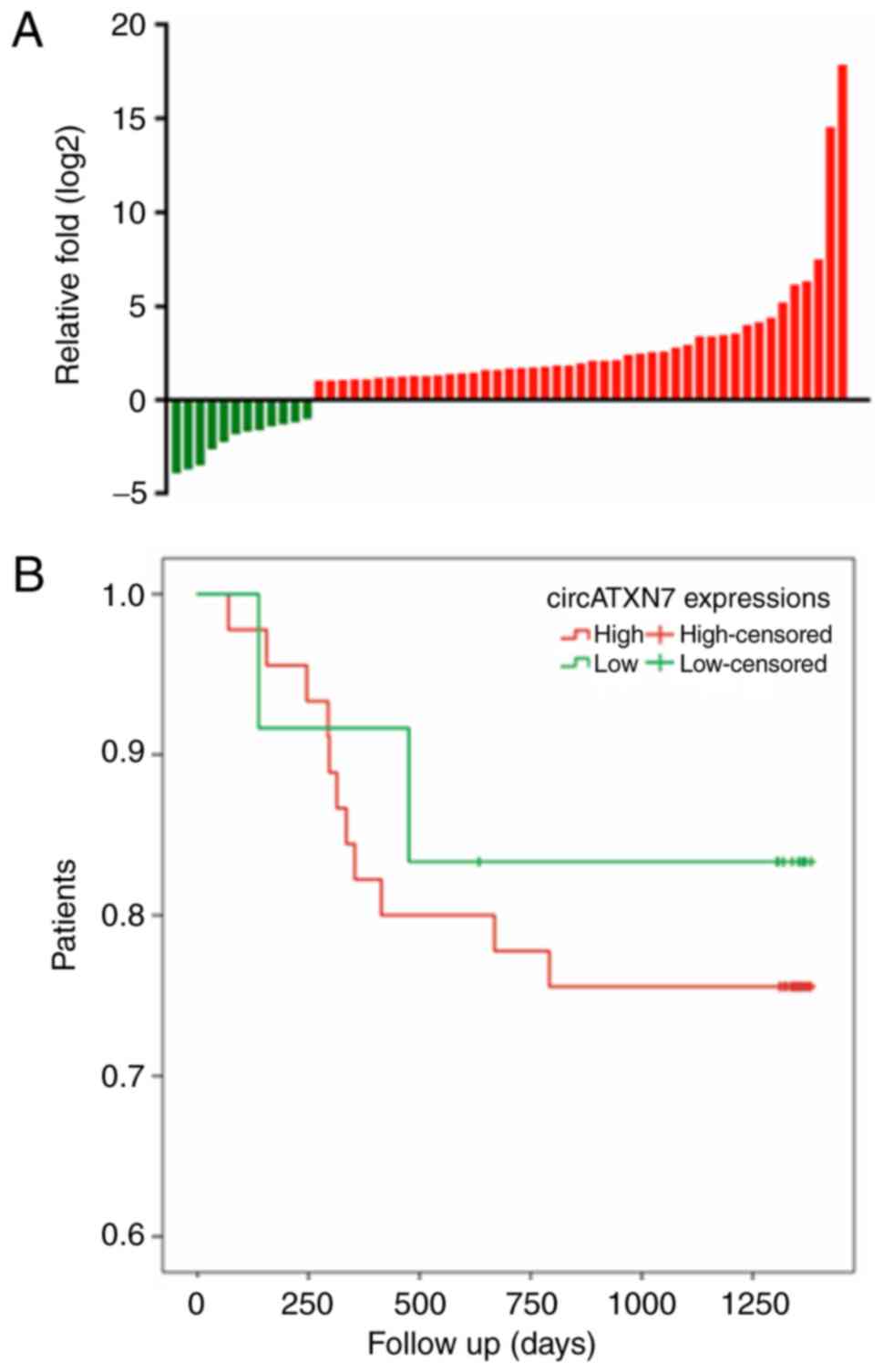
To investigate the expression of circATXN7 in NSCLC,
RT-qPCR analysis was performed in 57 pairs of NSCLC tissues and
matched adjacent non-tumor tissues. It was observed that circATXN7
was upregulated in 45 of the 57 NSCLC tissues as compared with its
expression in the matched non-tumor tissues (Fig. 2A). However, no significant
associations were detected between the expression level of
circATXN7 and the clinicopathological characteristics, including
age, sex, lymph node metastasis or TNM stage (Table I) (18). The prognostic value of circATXN7
expression was demonstrated by the Kaplan-Meier survival curves. It
was observed that the survival time of patients with high levels of
circATXN7 was shorter compared with that of patients with low
levels of circATXN7, although the association was not statistically
significant (P>0.05; Fig.
2B).
 | Table I.Associations between circATXN7 level
and clinicopathological characteristics of patients with non-small
cell lung cancer. |
Table I.
Associations between circATXN7 level
and clinicopathological characteristics of patients with non-small
cell lung cancer.
|
| circATNX7 |
|
|---|
|
|
|
|
|---|
| Characteristic | Low expression
(n=12) | High expression
(n=45) | P-value |
|---|
| Sex |
|
| 0.577 |
| Male | 8 | 26 |
|
|
Female | 4 | 19 |
|
| Age, years |
|
| 0.177 |
| ≥60 | 9 | 24 |
|
|
<60 | 3 | 21 |
|
| TNM stage |
|
| 0.058 |
| I–II | 7 | 13 |
|
|
III–IV | 5 | 32 |
|
| Lymph node
metastasis |
|
| 0.533 |
| No | 6 | 18 |
|
| Yes | 6 | 27 |
|
circATXN7 promotes the proliferation
and invasion abilities of NSCLC cells
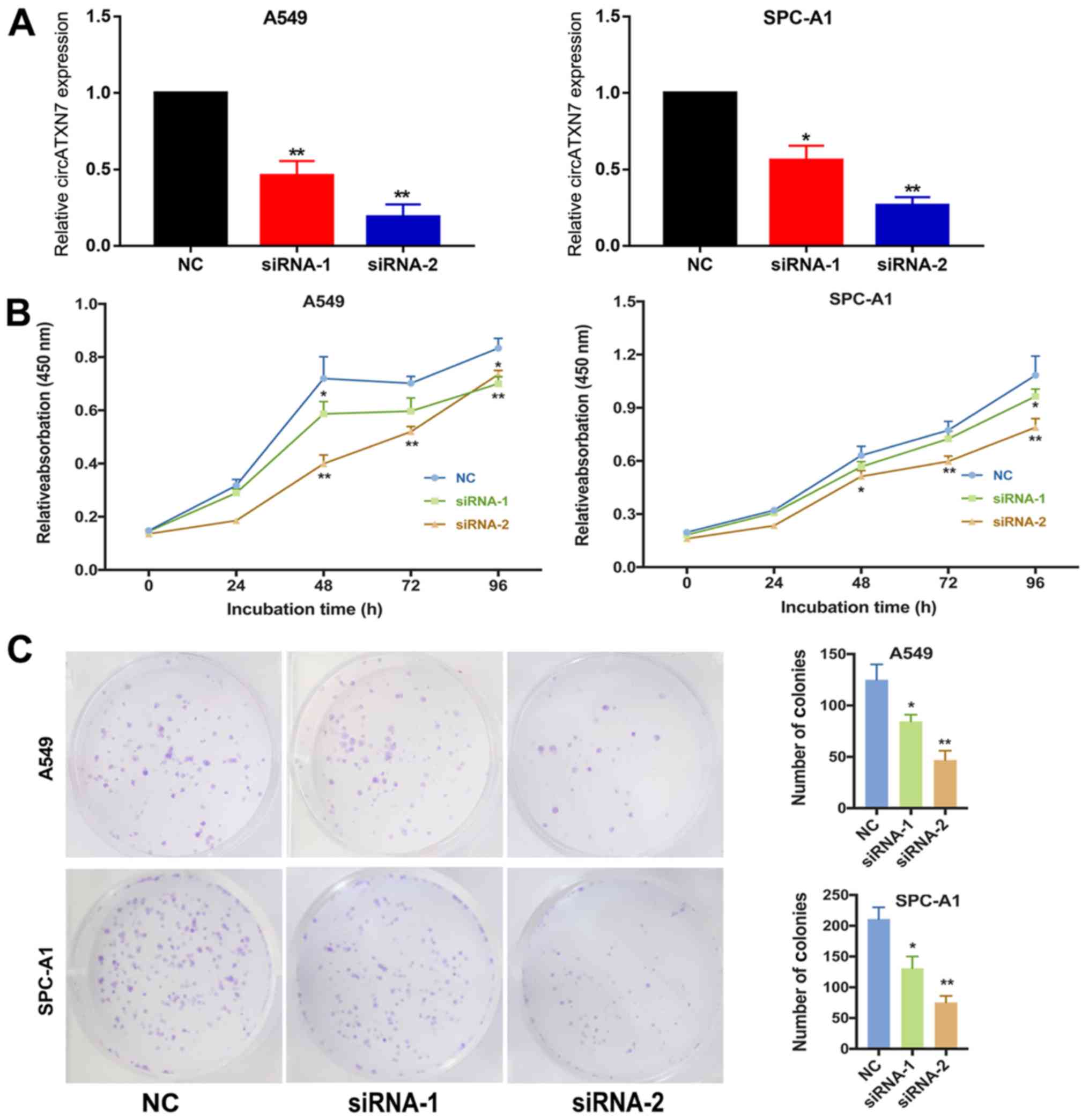
SPC-A1 and A549 were selected as the experimental
cell lines in the current study. Two siRNAs that specifically
target the junction of the covalently joined 3′ and 5′ ends were
designed to inhibit circATXN7 expression and investigate its
biological function in vitro, and the circATXN7 silencing in
A549 and SPC-A1 cells was successfully induced by transfection
(Fig. 3A). Next, the results of the
MTT assay indicated that the silencing of circATXN7 significantly
suppressed the proliferative ability compared with that in cells
transfected with negative control (NC) siRNA (Fig. 3B). Furthermore, the clonogenic assay
demonstrated that silencing circATXN7 significantly decreased the
number of clones in NSCLC cells (Fig.
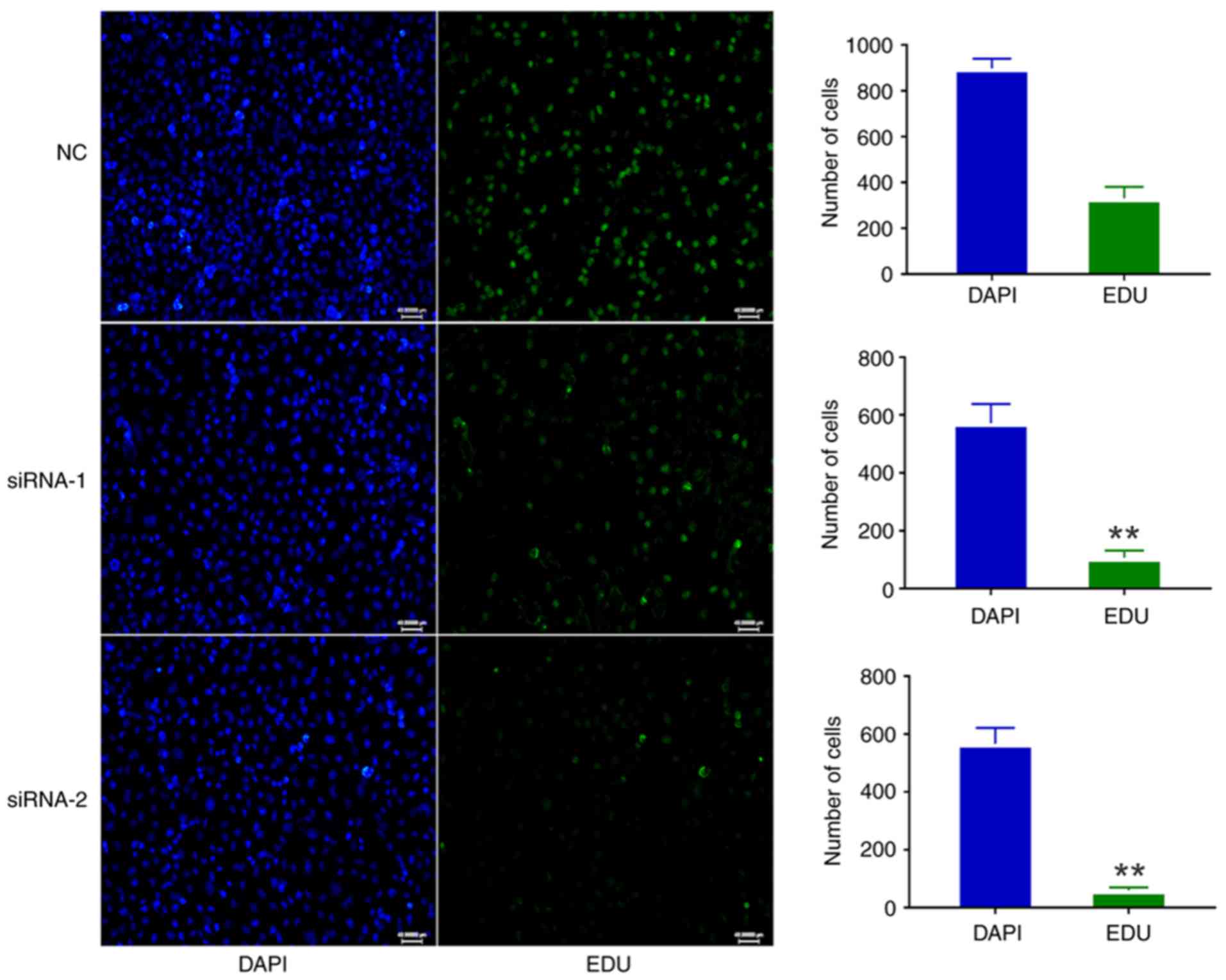
3C). The EdU assay also suggested that circATXN7 silencing
inhibited the proliferative ability of A549 cells (Fig. 4). Thus, these results revealed that
circATXN7 silencing suppressed the proliferation of NSCLC
cells.
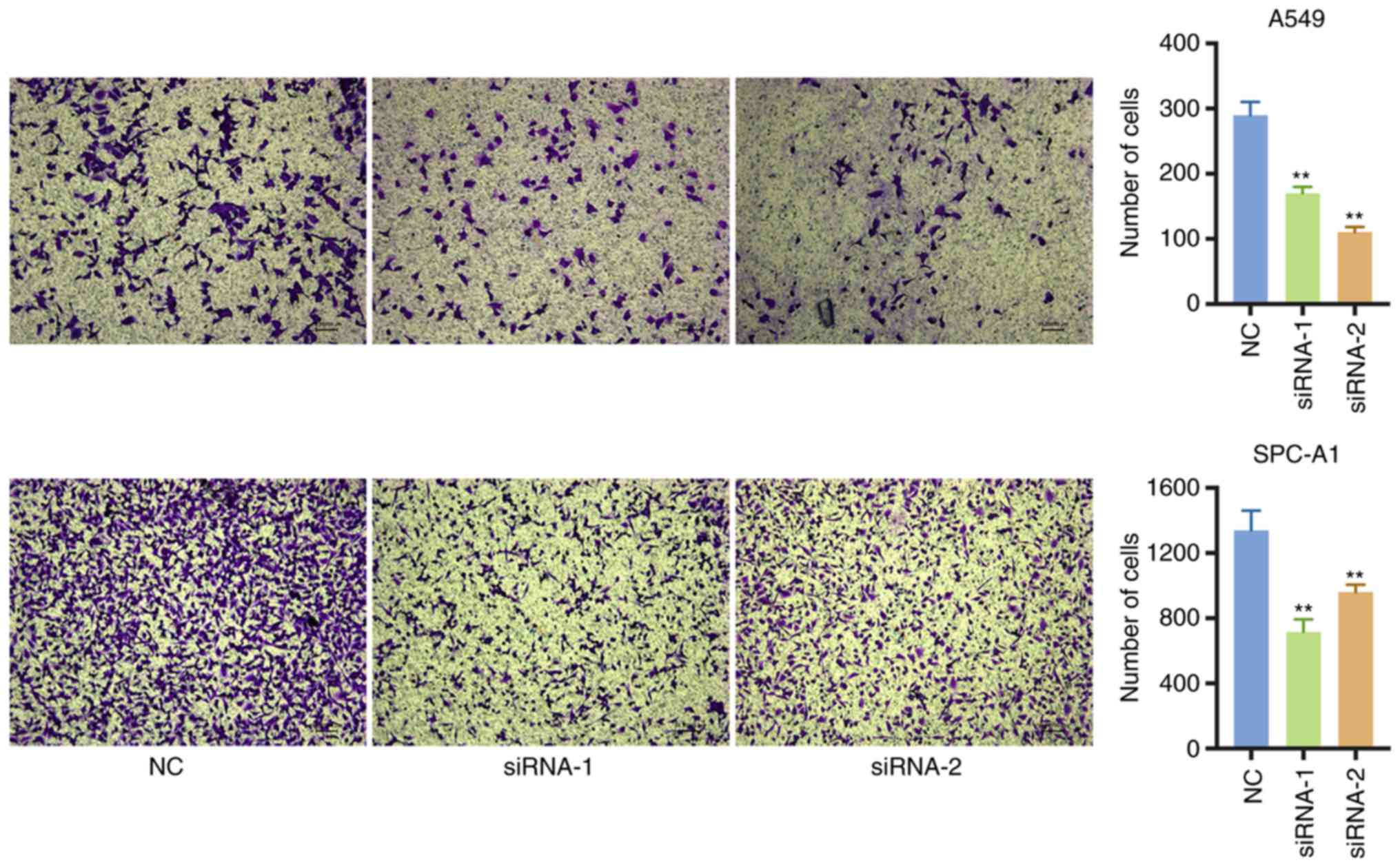
The effect of circATXN7 on the invasive ability of
NSCLC cells was then further analyzed. The Transwell assay
confirmed that the silencing of circATXN7 markedly inhibited the
invasive ability of A549 and SPC-A1 cells (Fig. 5). Taken together, the results
indicated that circATXN7 silencing was able to inhibit the
proliferation and invasion of lung cancer cells in
vitro.
Discussion
circRNAs are a type of non-protein coding RNAs that
are characterized by a covalently closed loop, and have been
detected for decades in viruses, plants and animals (10). To date, the role and mechanism of
circRNAs in the progression of malignancies remain to be identified
(19). circRNAs are characterized by
highly conserved sequences and have a specific covalently closed
circular construction (20). They
may serve considerable roles in the initiation and development of
cancer, and may be candidate biomarkers for the diagnosis of
diseases, such as cancer (21,22).
The tumorigenesis of NSCLC is a complex dynamic
biological process that involves multiple genes (23). However, a limited number of studies
have examined the association between circRNAs and NSCLC.
Previously, we demonstrated that a novel circular RNA, circPRKCI,
functions as a sponge for both miR-545 and miR-589 in NSCLC and
inhibits their suppression of the pro-tumorigenic transcription
factor E2F7 (14).
In the present study, a novel circRNA, namely
circATXN7, was identified in NSCLC. The potential association of
the expression of circATXN7 with clinical factors and its
prognostic value were investigated. Next, the potential function of
circATXN7 was investigated by siRNA-mediated silencing. The
findings of the current study indicated that the prognosis of
patients with upregulation of circATXN7 was poorer compared with
patients with low circATXN7 levels; however, the association was
not statistically significant. Furthermore, there was no marked
association between circATXN7 expression level and the majority of
clinicopathological characteristics, including the patient age,
cancer location, lymph node metastasis, tumor size, tumor
differentiation, T stage or TNM stage. Overall, the association
between circATXN7 expression level and the overall survival of
patients with NSCLC requires further research.
Silencing of circATXN7 by siRNA was subsequently
conducted in the current study to investigate its biological
functions in NSCLC cells. The results indicated that circATXN7
silencing inhibited the proliferation and invasion of NSCLC cells,
suggesting the oncogenic role of circATXN7 in NSCLC. Hsu and
Coca-Prados (24) detected circRNAs
in eukaryote cells using electron microscopy in 1979, and circRNAs
have since been hypothesized to be promising biomarkers in numerous
diseases, particularly in cancer, owing to sequence conservation
and biological stability (25,26).
circRNAs may harbor numerous miRNA binding sites, functioning as a
huge ‘sponges’ that can consume target miRNAs (27). circHIPK3, which is derived from exon
2 of the HIPK3 gene, was reported to sponge 9 miRNAs with 18
potential binding sites (28).
Therefore, further specific studies are required to analyze whether
circATXN7 serves as a ‘sponge’ for miRNAs.
In conclusion, the present study is the first to
identify that circATXN7 was upregulated in NSCLC tumor tissues.
Furthermore, the downregulation of circATXN7 inhibited the
proliferation and invasion abilities of NSCLC cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81702256), the
Natural Science Foundation of Beijing (grant no. 7182169) and
partly by the Postdoctoral Fellowship of Peking-Tsinghua Center for
Life Sciences (grant awarded to MQ; grant no. 4452-10148).
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MQ, JW and QH designed experiments. QH, MQ, SW, XL,
FY, CF and KZ performed the experiments. MQ and QH performed the
data analysis and wrote the manuscript. All authors discussed the
results and commented on the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All procedures in the current study involving human
participants were performed in accordance with the standards of the
Ethical Committee of Peking University People's Hospital, and with
the 1964 Helsinki Declaration and its later amendments or
comparable ethical standards. Informed consent was obtained from
all individual participants included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smith RA, Manassaram-Baptiste D, Brooks D,
Doroshenk M, Fedewa S, Saslow D, Brawley OW and Wender R: Cancer
screening in the United States, 2015: A review of current American
cancer society guidelines and current issues in cancer screening.
CA Cancer J Clin. 65:30–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ettinger DS, Wood DE, Akerley W, Bazhenova
LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D'Amico TA,
Demmy TL, et al: Non-small cell lung cancer, version 6.2015. J Natl
Compr Canc Netw. 13:515–524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 World health organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Antoni D and Mornex F: Chemoradiotherapy
of locally advanced nonsmall cell lung cancer: State of the art and
perspectives. Curr Opin Oncol. 28:104–109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Socinski MA, Obasaju C, Gandara D, Hirsch
FR, Bonomi P, Bunn P, Kim ES, Langer CJ, Natale RB, Novello S, et
al: Clinicopathologic features of advanced squamous NSCLC. J Thorac
Oncol. 11:1411–1422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan
X, Han S and Wu G: hsa_circ_0013958: A circular RNA and potential
novel biomarker for lung adenocarcinoma. FEBS J. 284:2170–2182.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu Y, Wang J, Qiu M and Xu L, Li M, Jiang
F, Yin R and Xu L: Upregulation of the long noncoding RNA TUG1
promotes proliferation and migration of esophageal squamous cell
carcinoma. Tumour Biol. 36:1643–1651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
13
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z,
Xu W, Zhang E, Wang J, Fang T, et al: The circular RNA circPRKCI
promotes tumor growth in lung adenocarcinoma. Cancer Res.
78:2839–2851. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding X, Zhang S, Li X, Feng C, Huang Q,
Wang S, Wang S, Xia W, Yang F, Yin R, et al: Profiling expression
of coding genes, long noncoding RNA, and circular RNA in lung
adenocarcinoma by ribosomal RNA-depleted RNA sequencing. FEBS Open
Bio. 8:544–555. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The eighth edition lung cancer stage classification. Chest.
151:193–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang SD, Yuan Y, Zhuang CW, Li BL, Gong
DJ, Wang SG, Zeng ZY and Cheng HZ: MicroRNA-98 and microRNA-214
post-transcriptionally regulate enhancer of zeste homolog 2 and
inhibit migration and invasion in human esophageal squamous cell
carcinoma. Mol Cancer. 11:512012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (Eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X, Sun Q, Chen C, Yin R, Huang X,
Wang X, Shi R, Xu L and Ren B: ZYG11A serves as an oncogene in
non-small cell lung cancer and influences CCNE1 expression.
Oncotarget. 7:8029–8042. 2016.PubMed/NCBI
|
|
20
|
Vannini I, Wise PM, Challagundla KB,
Plousiou M, Raffini M, Bandini E, Fanini F, Paliaga G, Crawford M,
Ferracin M, et al: Transcribed ultraconserved region 339 promotes
carcinogenesis by modulating tumor suppressor microRNAs. Nat
Commun. 8:18012017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Y, Li C, Tan C and Liu X: Circular
RNAs: A new frontier in the study of human diseases. J Med Genet.
53:359–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|