Introduction
Lung cancer occurs from uncontrolled cell growth in
the lung tissue, resulting in tumors (1). Non-small cell lung cancer (NSCLC) is
one of the most common types of lung tumor (2). The 5-year survival rate across all
stages of NSCLC is only 12% (3).
Unfortunately, the median survival time for NSCLC patients is <1
year following diagnosis (4). Thus,
improved outcomes for NSCLC are clearly required. The improvement
of clinical outcomes for NSCLC patients can be determined by a deep
investigation of the molecular mechanisms (5). Previous studies have shown that
abnormal expression of long non-coding RNAs (lncRNAs) is strongly
associated with the pathogenesis of NSCLC (6,7).
Differentially expressed lncRNAs (DE-lncRNAs), including HOX
transcript antisense RNA and metastasis-associated lung
adenocarcinoma transcript 1, serve important roles in the
progression of NSCLC via the epidermal growth factor
receptor-tyrosine kinase inhibitor resistance pathway (6). As a potential biomarker, the lncRNA
signature contributes to the survival prediction of NSCLC patients
(8). Ghadimi et al (9) showed that mRNAs, including
lung-specific X protein (LUNX) and carcinoembryonic antigen,
in the pleural fluid can serve as promising biomarkers for the
detection of NSCLC (9). The plasma
LUNX mRNA is another non-invasive specific biomarker for the
diagnosis and prognostic prediction of NSCLC (10). Apart from mRNAs, microRNAs
(miRNAs/miRs) are commonly used to identify novel NSCLC genes and
their associated networks. A previous study showed that miRNAs,
including miR-98-5p and miR-302e, can be used as
markers for predicting radio-sensitivity in NSCLC (11). The lncRNA-miRNA-mRNA regulatory
network, known as the competing endogenous RNA (ceRNA) network, is
an important tripartite axis in the regulation of the disease
process (12,13). Although the identification of
molecular factors contributes to therapeutic interventions in
NSCLC, the key lncRNAs and the possible regulatory mechanism of
ceRNAs in NSCLC remains unclear.
Recent technological advances in high-throughput
gene sequencing and expression profiling have allowed the analysis
of gene expression in NSCLC (14,15). As
a small molecular inhibitor, XAV939 promotes apoptosis in the tumor
cell line via telomere shortening (16). A previous study showed that XAV939
can inhibit the proliferation and migration of lung adenocarcinoma
cells (17). In the present study,
high-throughput sequencing was performed on XAV939-treated
NCI-H1299 cells (XAV939 group) and untreated controls (control
group), to explore the DE-lncRNAs and DEGs. Subsequently, a
protein-protein interaction (PPI) network was constructed for
DEG-encoded proteins. The associated functions of DEGs and
DE-lncRNAs were further explored. The miRNA-DEG associations were
subsequently explored, followed by investigation of the miRNA-DEG
regulatory network. Finally, the ceRNA network was constructed and
analyzed. The present study attempted to explore the potential
mechanism of NSCLC and provide information regarding novel genes as
potential targets for the treatment of NSCLC.
Materials and methods
Cell culture
The NSCLC cell line, NCI-H1299, was purchased from
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China) and maintained in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
1% penicillin-streptomycin (Sangon Biotech Co., Ltd., Shanghai,
China) in a humidified incubator with a 5% CO2
atmosphere at 37°C.
Cell Counting Kit-8 (CCK-8) assay
Following XAV939 (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) treatment, CCK-8 (Beyotime Institute of
Biotechnology, Shanghai, China) assay was used to determine the
viability of the NCI-H1299 cells. The NCI-H1299 cells were seeded
in a 96-well plate at a density of 5.0×103 per well overnight.
Subsequently, the cells were treated with low and high
concentrations of XAV939 (10 and 20 µM/ml, respectively) for 24 h
and NCI-H1299 cells without XAV939 were used as the control group,
with 5 repeats in each group. The doses and time point selected
were used as referenced by previous studies (17–19). In
addition, the NCI-H1299 cell morphology was observed following
treatemn with XAV939 under a light microscope (YYS-190E; Shanghai
Optical Instrument Co., Ltd., Shanghai, China). Following 22 h of
treatment, 10 µl CCK-8 solution (Beyotime Institute of
Biotechnology) was added to each well and incubated for 2 h.
Subsequently, the optical density (OD) of each well at 450 nm was
determined using the Infinite M100 PRO (Tecan Group Ltd.,
Mannedorf, Switzerland).
Apoptosis analysis
The NCI-H1299 cells were seeded in a 6-well plate at
a density of 1×105 cells/ml overnight. On the 2nd day, the
NCI-H1299 cells were treated with 10 µM XAV939 (Sigma-Aldrich) and
the NCI-H1299 cells without treatment were used as the control
group. Following treatment for 48 h, the NCI-H1299 cells from the
treatment and the control groups were washed with PBS and incubated
in 1 ml PBS containing 100 µl 1X binding buffer, 5 µl propidium
iodide (Sigma-Aldrich) and 5 µl FITC-Annexin-V (BD Biosciences, San
Jose, CA, USA) in the dark, at room temperature for 15 min.
Finally, 400 µl 1X binding buffer was added to each well and the
FACSCalibur flow cytometer (BD Biosciences) was used to analyze
apoptosis within 1 h and data were analyzed with FlowJo version 10
software (FlowJo LLC, Ashland, OR, USA).
Cell preparation for RNA-sequencing
(RNA-Seq)
The NCI-H1299 cells in the treatment group,
including three samples termed s1, s2 and s3, at confluence were
cultured in a 12-well plate supplemented with 10 µM XAV939 for 24 h
(XAV939 group), while those without XAV939 treatment were used as
controls (control group; including three samples termed k1, k2 and
k3). Following 24 h of culture, the six cell samples in the 12-well
plate were washed with PBS and the total RNA of samples were
extracted for RNA-seq.
RNA isolation and RNA-Seq
The six RNA samples from the two groups were
isolated using a TRIzol® kit (Takara Biotechnology Co.,
Ltd., Dalian, China). The total RNA was quantified with a NanoDrop
2000 (Thermo Fisher Scientific, Inc.). Subsequently, total RNA (3
µg/sample) was reverse-transcribed to construct a cDNA library
using a NEBNext® Ultra™ RNA Library Prep kit for
Illumina® (cat no. E7530L; New England Biolabs, Inc.,
Ipswich, MA, USA) according to the manufacturer's protocol. mRNAs
were enriched on a magnetic bead prior to shearing into fragments.
Following this, the cDNAs were synthesized and amplified with 15
cycles of the repair chain reaction. The cDNA clusters were
sequenced using the Illumina HiSeq 4000 platform (Illumina, Inc.,
San Diego, CA, USA) using the 150 paired end method (20).
Pretreatment of RNA-seq data
Quality control was performed with Trimmomatic tools
(v3.6) (21) as follows: i) Removal
of the barcode sequences, ii) elimination of the base at each end
with a quality <10, iii) exclusion of low-quality reads in which
bases with quality >20 accounted for <80% of the length and
iv) exclusion of reads with a length <50 nucelotides. Clean
reads from all the samples were obtained and aligned with the human
reference genome (GRCh38.p7) (22)
using TopHat software (v2.1.0) (23)
with the default parameters.
Gene expression levels analysis
Following pretreatment of the raw RNA-seq data, gene
expression levels were determined by counting reads in the genomic
region or the exon region. The read count was obtained based on
human genome annotation information in the GENCODE database (v25;
http://www.gencodegenes.org/human/releases.html)
using the featureCounts software (v1.6.2; http://subread.sourceforge.net/), followed by
normalization based on the reads per kilobase per million (RPKM)
mapped reads method. Genes with a RPKM value <0.1 in ≥3 samples
were defined as having low expression. Genes were classified as
lncRNA or mRNA based on the annotation information.
Identification of DEGs and
DE-lncRNAs
The DEGs and DE-lncRNAs between the XAV939 group and
control group were investigated using the quasi-likelihood F-tests
method of the edgeR package (24) in
R software (www.r-project.org), with the
thresholds of |log2 fold-change (FC)|>0.3 and P<0.05. The
bidirectional hierarchical clustering for DEGs was performed by
pheatmap software (v1.0.8; http://cran.r-project.org/web/packages/pheatmap/index.html).
Function and pathway enrichment
analysis
The Gene Ontology (GO) database was utilized to
identify potential biological processes associated with DEGs, while
the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was
used to reveal the pathways enriched in DEGs. GO functional
categories include molecular function (MF), biological process (BP)
and cellular component (CC) (25).
The function and pathway enrichment analyses were performed on DEGs
using the Database for Annotation, Visualization and Integrated
Discovery (DAVID) online tool (26).
The cut-off value for a significant function and pathway selection
was P<0.05.
Construction of PPI network
To examine the potential interactions of the DEGs,
the Search Tool for the Retrieval of Interacting Genes/Proteins
database (27) was used to establish
the PPI network under the condition of a required confidence
(combined score) of >0.4. Cytoscape software (version 3.2.1;
National Institute of General Medical Sciences, Seattle, WA, USA)
(28) was used to visualize the
network. A node in the PPI network represents the protein product
encoded by DEGs and the degree (Degree Centrality) (29) of a node indicates the number of
proteins interacting with this specific node. The top ten nodes
ranked by degree (degree >5) were considered as the hub
nodes.
lncRNAs target gene prediction
The correlations between DE-lncRNAs and DEGs were
calculated with the Pearson's correlation coefficient (30). The DEGs with Benjamini-Hochberg
corrected P-values of <0.05 (31)
and |ρ| (correlation coefficient) >0.9 were considered as
potential target genes of lncRNAs.
lncRNA function prediction
GO function and KEGG pathway enrichment analyses
were performed on the target genes of DE-lncRNAs to predict the
function of the DE-lncRNAs. Using the clusterProfiler package
(version 2.2.7; http://www.bioconductor.org/packages/3.1/bioc/html/clusterProfiler.html)
in R (32), ≥15 DE-lncRNA target
genes were found and selected for the subsequent analysis.
P<0.05 was set as the cut-off value for significant function and
pathway enrichment.
Functional similarity analysis of
DE-lncRNAs
The Resnik method is an information content-based
method (33), while the Wang method
is a graph-based method (34). In
the present study, the Resnik and Wang methods in the GOSemSim
software (version 2.8.0; http://www.bioconductor.org/packages/release/bioc/html/GOSemSim.html)
(35) were used to explore the
functional similarity among the lncRNAs. A sum of Wang and Resnik
methods score >1.2, generated by GOSemSim, represented a
functional similarity between 2 lncRNAs. The lncRNA functional
similarity network was constructed using Cytoscape.
miRNA prediction and ceRNA regulatory
network construction
DEG-associated miRNAs were predicted using Enrichr
software (2016 version; http://amp.pharm.mssm.edu/Enrichr) with P<0.05 as
the threshold (36). Subsequently,
the miRanda software (v.3.3a; parameters: -score 120, -energy −20)
was used to confirm the regulatory associations between miRNAs and
DE-lncRNAs.
Based on the miRNA-DEG, DE-lncRNA-miRNA and
DE-lncRNA-DEG interactions, the DEG-miRNA-DE-lncRNA network (ceRNA)
was explored further. Finally, the network topology analysis was
performed to obtain the key elements in the network.
Statistical analysis
All the OD values were presented as the mean ±
standard deviations (SD). The comparisons between the
XAV939-treatment group and control group were analyzed by one-way
analysis of variance, followed by the least significant difference
test. The SPSS 22.0 software (IBM Corp., Armonk, NY, USA) was used
for this analysis, where P<0.05 was considered to indicate a
statistically significant difference.
Results
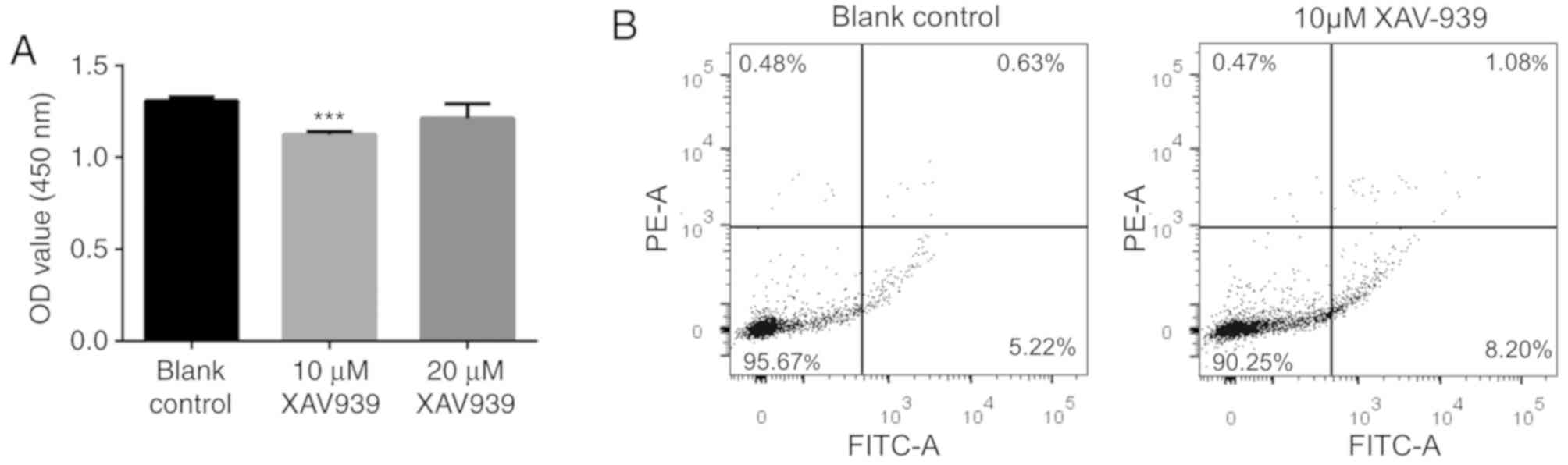
XAV939 decreases NCI-H1299 cell
proliferation, but increases apoptosis
Following treatment with XAV939, the size of
NCI-H1299 cells was identified to be slightly smaller under the
microscope. The CCK-8 assay was used to determine an appropriate
XAV939 concentration that may significantly reduce the cell
viability of NCI-H1299 cells compared with other XAV939
concentrations for downstream analysis. The result showed that 10
µM XAV939 could significantly inhibit the proliferation of
NCI-H1299 cells (P<0.001), but no significant differences were
identified between the 10- and 20-µM treatment groups (P>0.05),
or between the control and the 20-µM treatment group (P>0.05;
Fig. 1A). As a result, 10 µM XAV939,
which significantly reduced the cell viability of NCI-H1299 cells
compared with 20 µM XAV939, was used to treat the NCI-H1299 cells
in the following investigation. Following this, the apoptosis of
the NCI-H1299 cells after treatment with 10 µM XAV939 was also
determined and revealed to be significantly increased (P=0.004;
Fig. 1B).
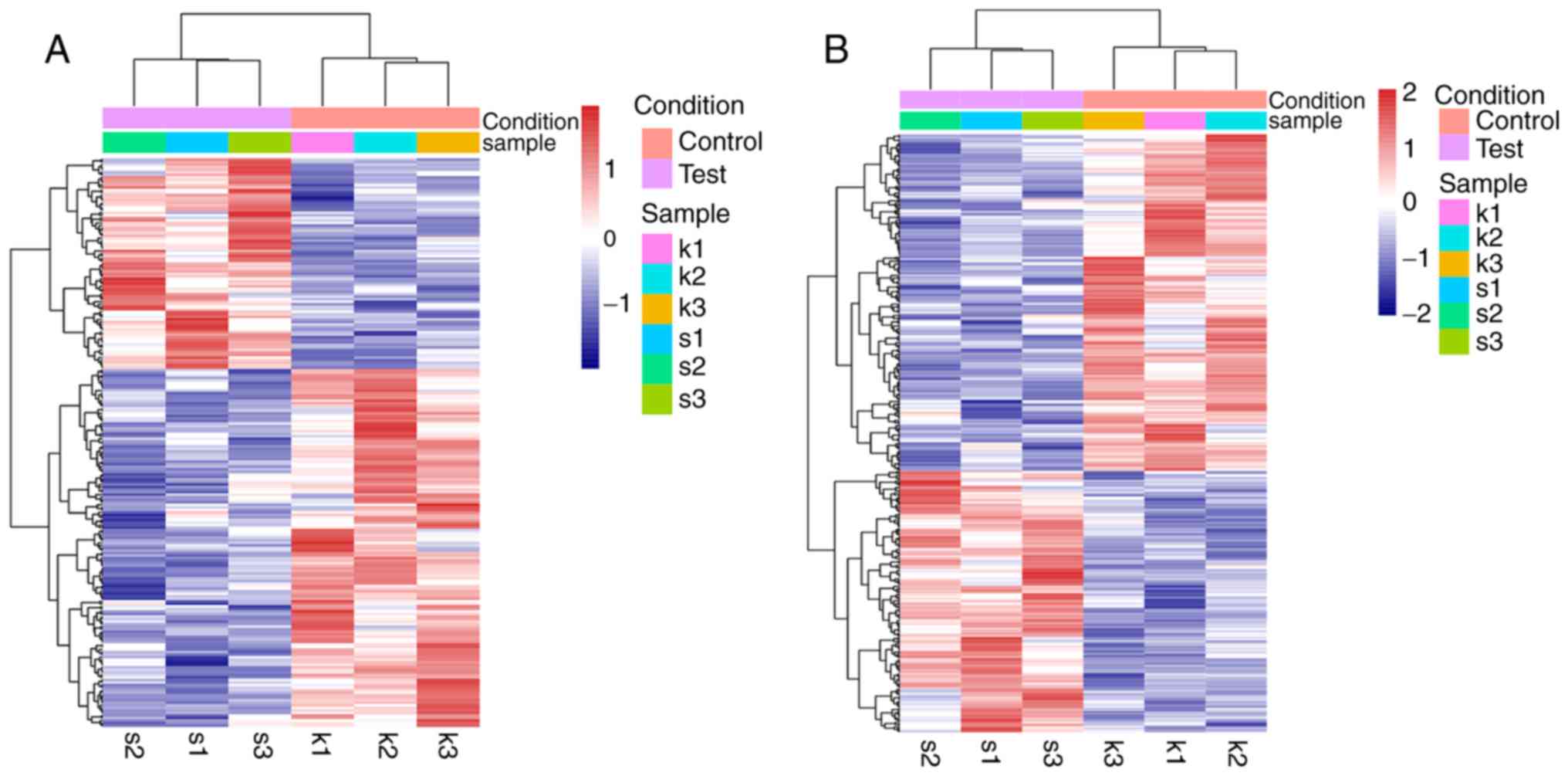
DEGs and DE-lncRNAs between XAV939 and
control groups
A total of 256 upregulated genes (including 173 DEGs
and 83 DE-lncRNAs) and 364 downregulated genes (including 223 DEGs
and 141 DE-lncRNAs) were revealed between the XAV939 treatment
group and the control group, using the threshold criteria
|log2FC|>0.3 and P<0.05. Heatmaps were generated
for these DEGs and DE-lncRNAs (Fig.
2).
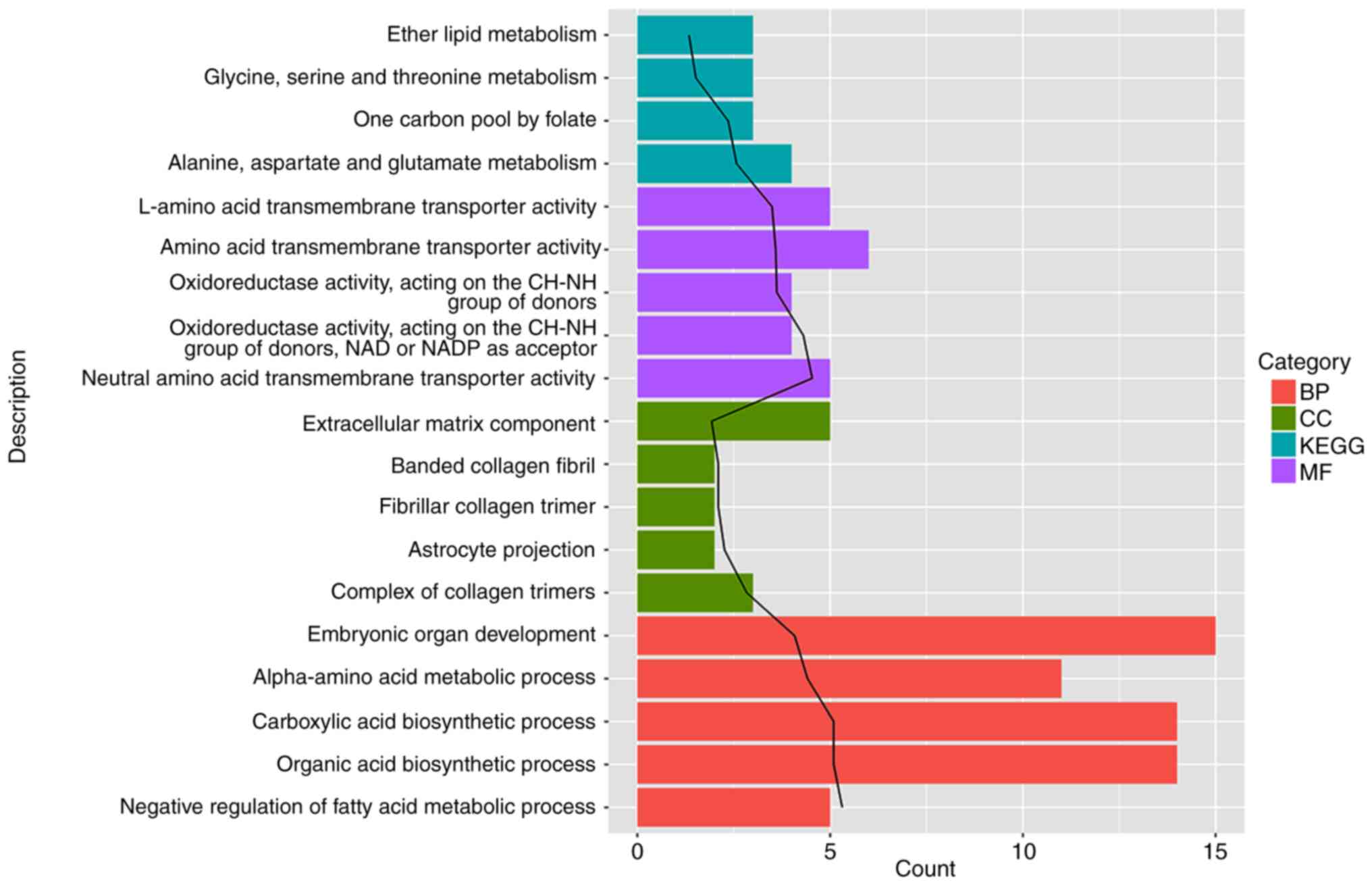
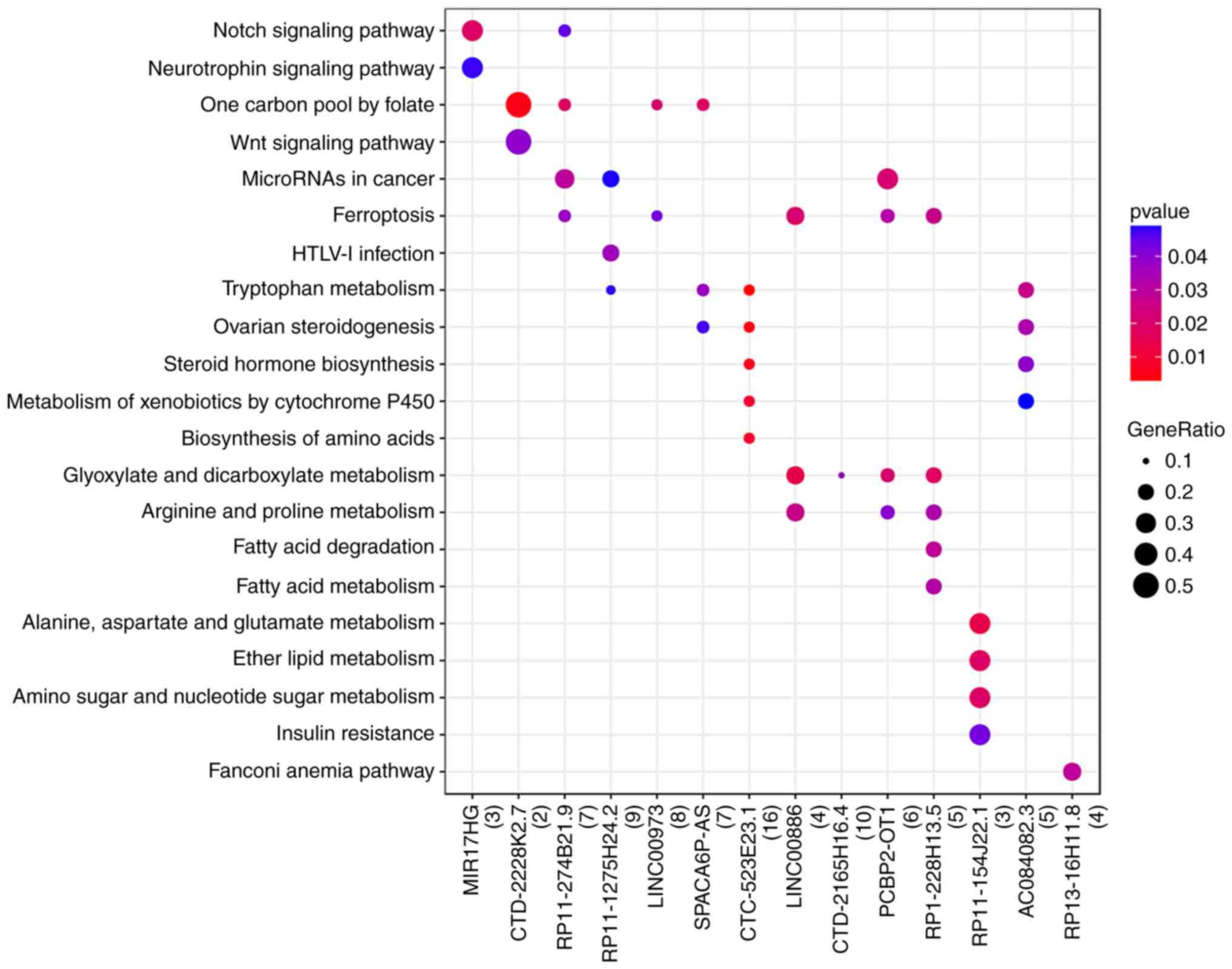
Enrichment analysis of DEGs
GO functions and KEGG pathways enriched by DEGs were
further investigated (Fig. 3;
Table I). The results showed that GO
DEGs were mainly associated with functions such as ‘embryonic organ
development’ (BP, GO:0048568; P=8.33×10-5), ‘extracellular matrix
component’ (CC, GO:0044420; P=1.19×10-2) and ‘amino acid
transmembrane transporter activity’ (MF, GO:0015171; P=2.59×10-4).
Meanwhile, the KEGG DEGs were mainly enriched in pathways such as
‘alanine, aspartate and glutamate metabolism’ (hsa00250;
P=2.67×10-3).
 | Table I.Top 5 GO functions and top 4 KEGG
pathways of differentially expression genes. |
Table I.
Top 5 GO functions and top 4 KEGG
pathways of differentially expression genes.
| Category | ID | Description | P-value | Count | Gene list |
|---|
| BP | GO:0045922 | Negative regulation
of fatty acid metabolic process |
4.86×10−6 | 5 | TRIB3, SIRT4,
ETFBKMT, ACADL, CNR1 |
|
| GO:0016053 | Organic acid
biosynthetic process |
8.09×10−6 | 14 | CYP1A1, ASNS,
PSAT1, MTHFD2, PHGDH, MTHFD1L, PYCR1, TRIB3, GPT2, DECR2, CBS,
LDHC, ACADL, HOGA1 |
|
| GO:0046394 | Carboxylic acid
biosynthetic process |
8.09×10−6 | 14 | CYP1A1, ASNS,
PSAT1, MTHFD2, PHGDH, MTHFD1L, PYCR1, TRIB3, GPT2, DECR2, CBS,
LDHC, ACADL, HOGA1 |
|
| GO:1901605 | α-amino acid
metabolic process |
3.85×10−5 | 11 | SLC7A5, ASNS,
PSAT1, PHGDH, PYCR1, GPT2, CBS, SIRT4, DDO, GFPT2, HOGA1 |
|
| GO:0048568 | Embryonic organ
development |
8.33×10−5 | 15 | DLX2, STRA6,
MTHFD1L, CEBPB, PRDM1, NES, HESX1, RNF207, RBPMS2, HLX, DLL1, IRX5,
SOX18, KCNQ4, HOXA2 |
| CC | GO:0098644 | Complex of collagen
trimers | 0.001451 | 3 | COL7A1, COL11A2,
TNXB |
|
| GO:0097449 | Astrocyte
projection | 0.005501 | 2 | ADGRG1,
GJB2 |
|
| GO:0005583 | Fibrillar collagen
trimer | 0.007948 | 2 | COL11A2,
TNXB |
|
| GO:0098643 | Banded collagen
fibril | 0.007948 | 2 | COL11A2,
TNXB |
|
| GO:0044420 | Extracellular
matrix component | 0.01185 | 5 | TNC, COL7A1,
COL11A2, SPARC, TNXB |
| MF | GO:0015175 | Neutral amino acid
transmembrane transporter activity |
2.91×10−5 | 5 | SLC7A11, SLC7A5,
SLC1A4, SLC6A9, SLC7A9 |
|
| GO:0016646 | Oxidoreductase
activity, acting on the CH-NH group of donors, NAD or NADP as
acceptor |
4.95×10−5 | 4 | MTHFD2, MTHFD1L,
PYCR1, ALDH1L2 |
|
| GO:0016645 | Oxidoreductase
activity, acting on the CH-NH group of donors | 0.000242 | 4 | MTHFD2, MTHFD1L,
PYCR1, ALDH1L2 |
|
| GO:0015171 | Amino acid
transmembrane transporter activity | 0.000259 | 6 | SLC7A11, SLC7A5,
SLC7A1, SLC1A4, SLC6A9, SLC7A9 |
|
| GO:0015179 | L-amino acid
transmembrane transporter activity | 0.000321 | 5 | SLC7A11, SLC7A5,
SLC1A4, SLC6A9, SLC7A9 |
| KEGG | hsa00250 | Alanine, aspartate
and glutamate metabolism | 0.002667 | 4 | ASNS, GPT2, DDO,
GFPT2 |
|
| hsa00670 | One carbon pool by
folate | 0.00442 | 3 | MTHFD2, MTHFD1L,
ALDH1L2 |
|
| hsa00260 | Glycine, serine and
threonine metabolism | 0.03032 | 3 | PSAT1, PHGDH,
CBS |
|
| hsa00565 | Ether lipid
metabolism | 0.045872 | 3 | JMJD7-PLA2G4B,
PLA2G7, PLA2G4B |
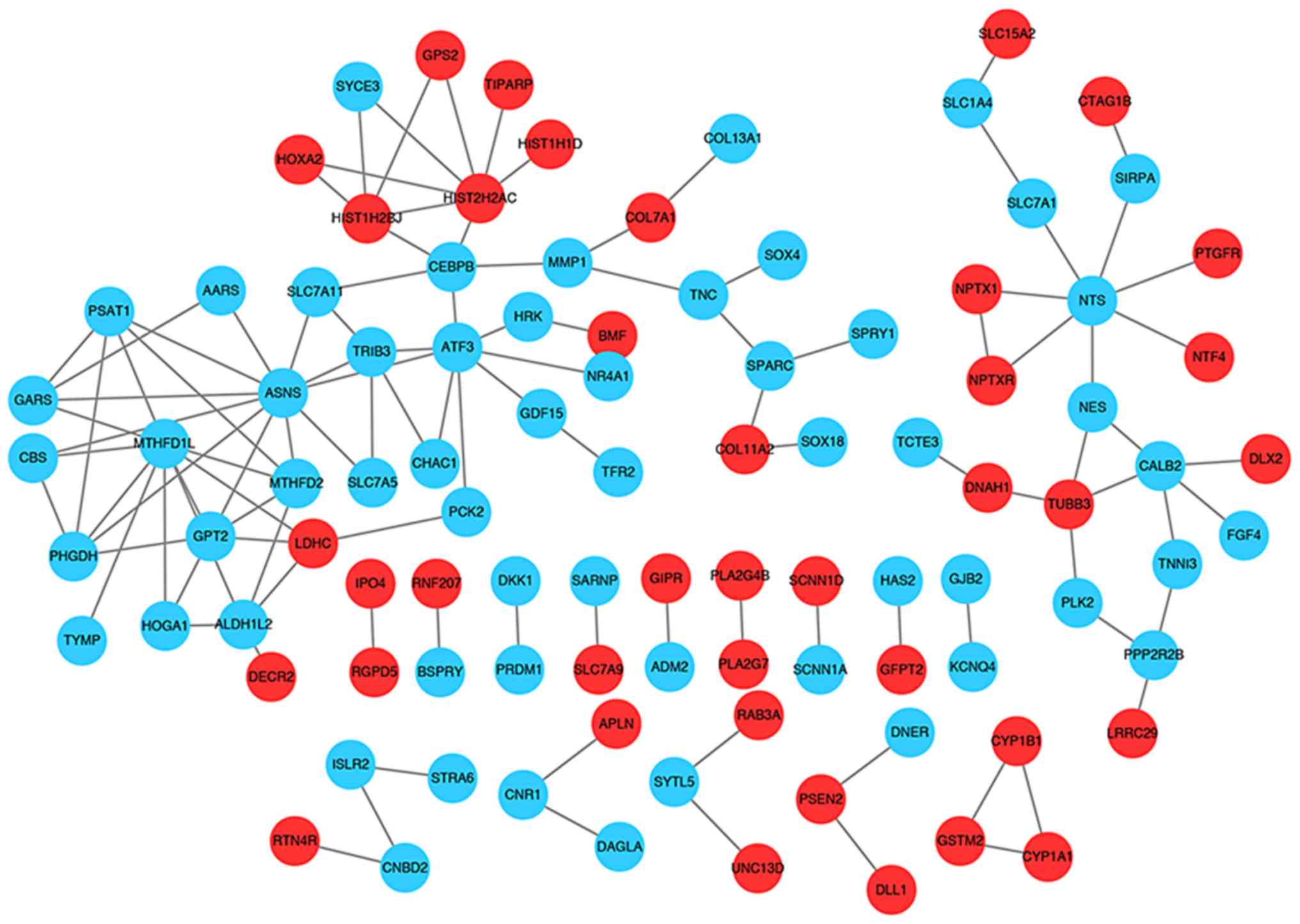
PPI network analysis
With a combined score of >0.4, a PPI network for
the present study was constructed with a total of 97 nodes and 112
interactions (Fig. 4). The top 10
nodes in the PPI network were asparagine synthetase
(glutamine-hydrolyzing) (degree=11), methylenetetrahydrofolate
dehydrogenase (NADP + dependent) 1 like (degree=10), activating
transcription factor 3 (degree=8), neurotensin (degree=7), histone
cluster 2 H2A family member C (degree=7), glutamic-pyruvic
transaminase 2 (degree=6), calbindin 2 (degree=5), tribbles
pseudokinase 3 (degree=5), CCAAT/enhancer binding protein β
(degree=5) and aldehyde dehydrogenase 1 family member L2
(degree=5).
DE-lncRNA-target gene interaction
Based on the Pearson's correlation coefficient
analysis, a total of 224 DE-lncRNAs had the predicted target genes.
Specifically, the top 10 lncRNAs were RP1-101A2.1,
RP11-584P21.2, CTD-2246P4.1, ST3 β-galactosile
α-2,3-sialyltransferase 6 (ST3GAL6)-antisense RNA 1 (AS1),
small integral membrane protein 2-AS1, CTD-2553L13.4,
AC018766.6, long intergenic non-protein coding RNA (LINC) 926
(LINC00926), RP11-43N16.4 and LINC00973.
DE-lncRNA function prediction and
comparison
The functional prediction for DE-lncRNAs was
obtained via GO and KEGG analyses of target genes of DE-lncRNAs.
The results showed that the target genes of DE-lncRNAs were mainly
enriched in pathways such as ‘ferroptosis’ [(DEG; solute carrier
family 7 member 11 (SLC7A11)] and ‘tryptophan metabolism’
(DEG; cytochrome p450 family 1 subfamily B member 1 (Fig. 5).
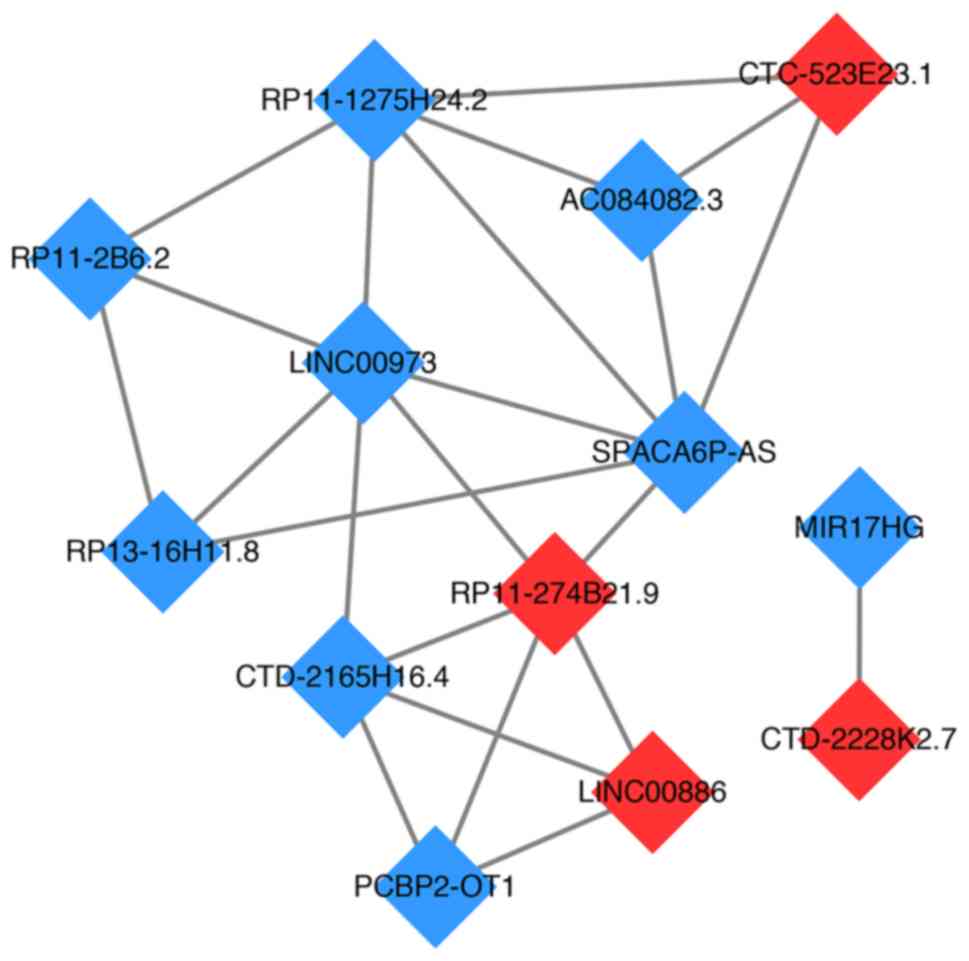
DE-lncRNA functional similarity
The DE-lncRNA functional similarity network was
constructed using Cytoscape software (Fig. 6). The results showed that there were
9 downregulated lncRNAs, including LINC00973, sperm
acrosome-associated 6 antisense RNA (SPACA6P-AS), AC084082.3,
RP11-1275H24.2, RP11-2B6.2, RP13-16H11.8, CTD-2165H16.4, poly
(rC) binding protein 2-overlapping transcript 1 and miR-17-92a-1
cluster host gene, and 4 upregulated lncRNAs, including
CTC-523E23.1, CTD-2228K2.7, RP11-274B21.9 and
LINC00886, in this network. Among these lncRNAs,
LINC00973 and SPACA6P-AS were 2 noteworthy DE-lncRNAs
that had the highest number of interactions.
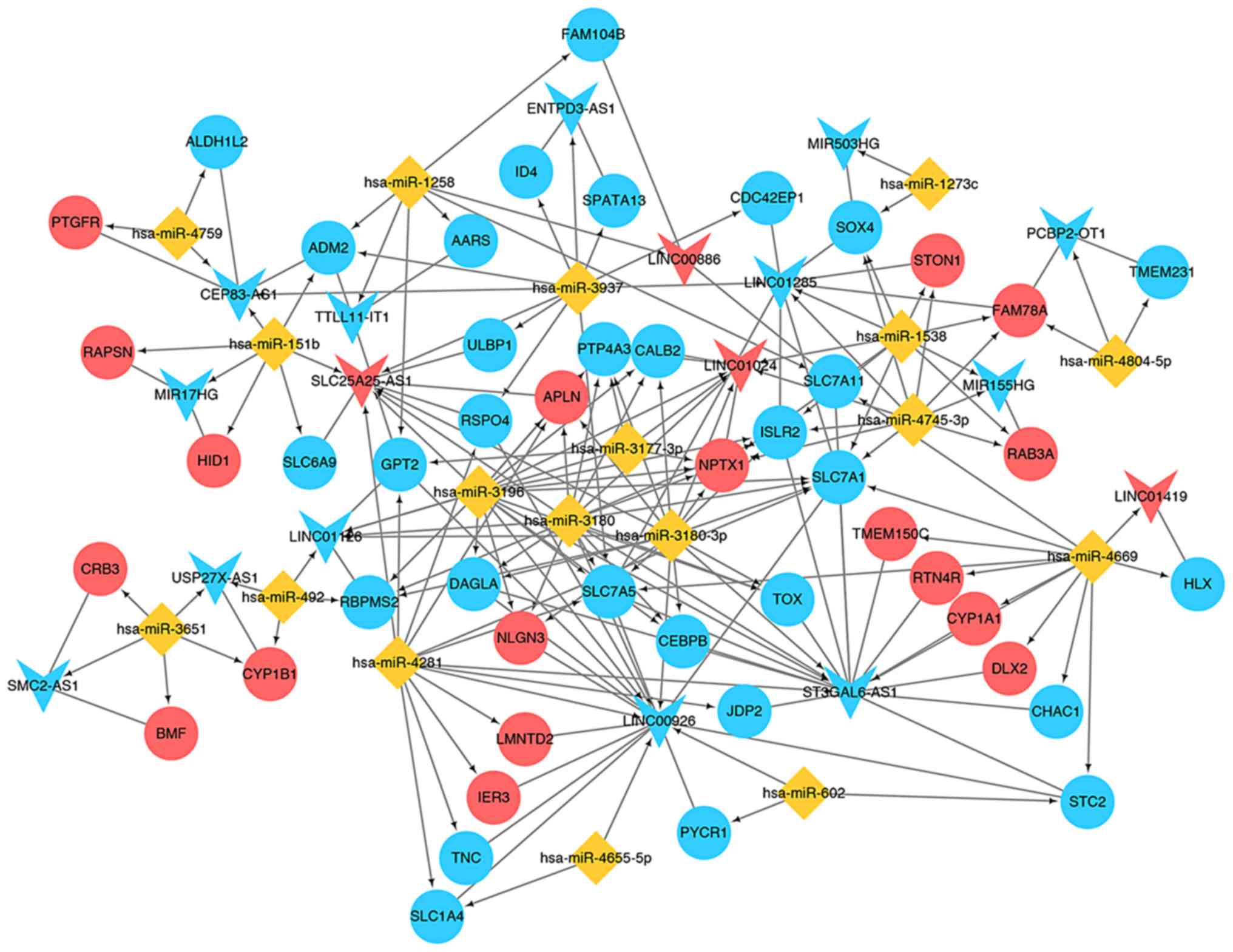
ceRNA regulatory network
investigation
Based on the miRNA-DEG interactions (Table II), the DE-lncRNA-miRNA interactions
and the DE-lncRNA-DEG interactions, the ceRNA network was further
explored (Fig. 7). There were 18
upregulated DEGs [including distal-less homeobox 2 (DLX2)], 30
downregulated DEGs [including SRY-box 4 (SOX4)], 4
upregulated lncRNAs (including LINC01419), 13 downregulated
lncRNAs [including MIR503 host gene (MIR503HG)] and 18
miRNAs (including hsa-miR-1273c) in this ceRNA network. The
results identified several miRNA-lncRNA-DEG associations, including
MIR503HG-miR1273c-SOX4 and ST3GAL6-AS1-miR4669-DLX2,
which were of note in the ceRNA network.
 | Table II.Top 10 predictive results of the
miRNA-mRNA interaction. |
Table II.
Top 10 predictive results of the
miRNA-mRNA interaction.
| miRNA | Target mRNA
number | P-value | Genes |
|---|
|
hsa-miR-4804-5p | 18 |
7.19×10−5 | CNTNAP3, GSTM2,
SLC15A2, LIMCH1, FAM78A, APLN, TMEM231, SLC7A5, GPR153, ADM2, STC2,
SIRP, SNCB, NPTXR, SOX7, LONRF2, AARS, STRA6 |
| hsa-miR-184 | 20 | 0.000206362 | CNTNAP3, POPDC2,
GSTM2, SLC15A2, LIMCH1, FAM78A, ISLR2, TMEM150C, APLN, TMEM231,
SLC7A5, SLC6A9, ADM2, STC2, SIRPA, SOX7, LONRF2, ZNF784, AARS,
STRA6, |
| hsa-miR-4669 | 35 | 0.001573186 | RTN4R, POPDC2,
DLX2, SPARC, MEGF10, SLC1A4, SLC7A11, SLC7A1, ALDH1L2, ADGRG1,
CCND2, KCNT2, FAM196B, HLX, STC2, HAS2, BMF, CHAC1, RSPO4, SLC15A2,
FAM78A, ADAM11, FAM86B1, TMEM150C, TMEM231, DYNAP, SLC7A5, PARP11,
GJB2, PTP4A3, SCNN1D, CYP1A1, KCNQ4, MAP6D1, PCDHB3 |
| hsa-miR-1273c | 10 | 0.005480298 | FAM156A, REP15,
PALM2, COL11A2, SIRPA, SLC1A4, BMF, CTAG1B, BEST3, SOX4 |
| hsa-miR-1538 | 20 | 0.012773605 | CEBPB, RAB3A,
DAGLA, FAM78A, STON1, ISLR2, SLC7A1, C1QTNF4, CCND2, GPR153,
SLC6A9, HLX, ADM2, SESN2, SIRPA, NPTX1, PI16, LONRF2, SOX4,
STRA6 |
|
hsa-miR-4745-3p | 20 | 0.012773605 | CEBPB, RAB3A,
DAGLA, FAM78A, STON1, ISLR2, SLC7A1, C1QTNF4, CCND2, GPR153,
SLC6A9, HLX, ADM2, SESN2, SIRPA, NPTX1, PI16, LONRF2, SOX4,
STRA6 |
| hsa-miR-3683 | 9 | 0.013763534 | ACADL, REP15,
PALM2, COL11A2, SIRPA, SLC1A4, BMF, SLC7A11, BEST |
|
hsa-miR-4655-5p | 11 | 0.021883755 | FAM78A, CNR1,
ADM2, SPATA13, CCDC170, KCNN1, SLC1A4, NPiTX1, UNC13D, ATF3,
TMEM231 |
| hsa-miR-3937 | 20 | 0.022144387 | DLX2, DAGLA,
LRRC29, GPT2, SLC1A4, DLL1, SLC7A5, PARP11, RNF207, ADM2, SPATA13,
TMEM217, ID4, HAS2, CDC42EP1, PI16, ULBP1, LONRF2, ATF3,
RSPO4 |
|
hsa-miR-4740-3p | 21 | 0.027471721 | DLX2, SLC15A2,
PALM2, COL11A2, PSEN2, ISLR2, SLC7A11, FAM104B, SLC7A1, FGF4,
SLC7A5, SLC6A9, ADM2, TMEM217, BMF, NPTX1, NPTXR, ULBP1, JDP2,
CBLN2, CRB3 |
Discussion
Worldwide, NSCLC is one of the most lethal cancer
affecting individuals (37).
Understanding the molecular mechanisms of NSCLC may contribute to
the therapeutic clinical outcomes in patients. In the present
study, a total of 396 DEGs and 224 DE-lncRNAs were identified in
the NSCLC NCI-H1299 cell line between the XAV939 treatment and
control groups. These lncRNAs included pathways such as the
‘ferroptosis’ pathway (DEG, SLC7A11). Furthermore, the
lncRNA-target interaction investigation revealed 10 lncRNAs that
were noteworthy, including ST3GAL6-AS1. Finally, the ceRNA
network identified several novel miRNA-lncRNA-mRNA regulatory
associations, including MIR503HG-miR1273c-SOX4.
XAV939 can inhibit the proliferation of carcinoma
cells by repressing specific gene expression, including that of
lactate dehydrogenase A (38). In
the present study, the ceRNA network analysis showed that
MIR503HG, which was inhibited by XAV939, was one of the most
significantly downregulated lncRNAs correlated with the
upregulation of miR1273c and downregulation of SOX4.
Importantly, lncRNAs have recently been reported in tumorigenesis
and serve a pivotal role in regulating cell cycle behavior
(39). A study by Muys et al
(40) showed that the lncRNA
MIR503HG impairs the migration and invasion capacities in a
choriocarcinoma tumor cell, indicating a potential role in human
reproduction and tumorigenesis. Knockdown of MIR503HG leads
to a strong downregulation of the target gene collagen type 1 α1
chain, indicating an important role of MIR503HG in diseases,
including systemic sclerosis (41).
As the lncRNAs target genes, mRNA expression is commonly involved
in the pathogenesis and prognosis of lung cancer (42). SOX4 is a critical
developmental transcription factor in vertebrates (43). A previous study indicated that a
SOX4 gene mutation is associated with lung carcinogenesis
and tumor metastasis (44). By
targeting SOX4, miR-212 functions as a tumor suppressor in
the metastasis of NSCLC (45). In
addition, Li et al (46)
reported that silencing SOX4 can inhibit the migration and
invasion of tumor cells in the lung. Actually, miRNAs can be used
as biomarkers for NSCLC (47). Lee
et al (48) indicated that
miR1273c, a member of the miR1273 family, serves an
inhibitory role in the progression of cancer cells. However, the
regulatory effect of miR1273c on NSCLC remains unclear. The
present study indicated that MIR503HG-miR1273c-SOX4 was one
of the notable interactions in the ceRNA network. Thus, we
hypothesize that the downregulation of lncRNA MIR503HG
induced by XAV939 may serve an important role in inhibiting the
progression of NSCLC by sponging miR-1273c and
downregulating SOX4 expression.
A previous study showed that XAV939 can be used as
an agent for lung cancer therapy (17). The inhibition of XAV939 in the
proliferation and migration of lung adenocarcinoma cells was
determined by modulating certain biological pathways, including
that of WNT (49). Ferroptosis, a
type of programmed cell death dependent on iron, inhibits tumor
growth and increases drug resistance (50,51). A
previous study revealed that the inhibition of ferroptosis
influences lung cancer progression (52). Although the association between the
ferroptosis pathway and lung cancer progression has already been
proved (53), the detailed mechanism
of ferroptosis on the development of NSCLC remains unclear. The
solute carrier family (SLF) is a group of membrane transport
proteins located in the cell membrane (54). As a member of the SLF family, SLC7A11
contributes to the temozolomide toxicity via the ferroptosis
pathway (55). A previous study
revealed that tumor suppression occurred by repression of
SLC7A11 in the ferroptosis pathway (56). More importantly, the upregulation of
SLC7A11 in transformed airway epithelial cells was able to induce
tumor formation in nude mice (57).
In the present study, the lncRNA function analysis revealed that
the ferroptosis pathway, which was associated with SLC7A11,
was one of the significant pathways affected. In addition, the
results revealed that SLC7A11 was downregulated in
XAV939-treated NCI-H1299 cells. Collectively, these results
indicate that the downregulation of SLC7A11 induced by
XAV939 may suppress the development of NSCLC via the ferroptosis
pathway.
However, there are limitations in the present study,
including the use of a single cell line and lack of a verification
test. Thus, a larger sample size using additional cell lines, with
a wide verification analysis is required in future investigations.
In addition, the dose and time point selected were used as
referenced by similar studies, and a lack of dose-dependent and
time-dependent experiments is therefore also a limitation of the
present study. The present study was preliminary and several
important bioinformatics results, including expression changes and
regulatory mechanisms discussed, require validation by biological
experiments in future studies.
In conclusion, the downregulation of the lncRNA
MIR503HG induced by XAV939 may serve an important role in
suppressing the progression of NSCLC via sponging miR1273c,
to downregulate its target SOX4. Furthermore, the
downregulation of SLC7A11 induced by XAV939 may inhibit
NSCLC development via participation in the ferroptosis pathway.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Study of the
Correlation Between ROCK1 and the Metastasis of Lung Cancer (grant
no. SCZSY201728).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HX, LX and HY were responsible for the conception
and design of the study, and drafted the manuscript. ZH and ZX
performed the data acquisition. HY and CA performed the data
analysis and interpretation. LX and HY performed the statistical
analysis. All authors have read and approved the manuscript.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
NSCLC
|
non-small cell lung cancer
|
|
DE-lncRNA
|
differentially-expressed lncRNA
|
|
PPI
|
protein-protein interaction
|
|
ceRNA
|
competing endogenous RNA
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
MF
|
molecular function
|
|
CC
|
cellular componenT
|
|
ST3GAL6
|
ST3 β-galactosile
α-2,3-sialyltransferase 6
|
|
AS1
|
antisense RNA 1
|
|
SLF
|
solute carrier family
|
References
|
1
|
Johnson BE: Divide and conquer to treat
lung cancer. N Engl J Med. 375:1892–1893. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schiller JH, Harrington D, Belani CP,
Langer C, Sandler A, Krook J, Zhu J and Johnson DH; Eastern
Cooperative Oncology Group, : Comparison of four chemotherapy
regimens for advanced non-small-cell lung cancer. N Engl J Med.
346:92–98. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Petty RD, Nicolson MC, Kerr KM,
Collie-Duguid E and Murray GI: Gene expression profiling in
non-small cell lung cancer: From molecular mechanisms to clinical
application. Clin Cancer Res. 10:3237–3248. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng N, Li X, Zhao C, Ren S, Chen X, Cai
W, Zhao M, Zhang Y, Li J, Wang Q and Zhou C: Microarray expression
profile of long non-coding RNAs in EGFR-TKIs resistance of human
non-small cell lung cancer. Oncol Rep. 33:833–839. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ghadimi K, Bahrami N, Fathi M, Farzanegan
B, Naji T, Emami M and Mohamadnia A: Diagnostic value of LunX mRNA
and CEA mRNA expression in pleural fluid of patients with non-small
cell lung cancer. Minerva Pneumol. 56:90–95. 2017.
|
|
10
|
Zhou HX, Yang MX, Wang Y, Cao WM, Lu KF,
Zong LY, Wu RQ and Zhang P: Plasma LUNX mRNA, a non-invasive
specific biomarker for diagnosis and prognostic prediction of
non-small cell lung cancer. Am J Cancer Res. 6:452–458.
2016.PubMed/NCBI
|
|
11
|
Xu C, Xu Y, Liao X, Liao R, Zhang L, Niu
K, Li T, Li D, Chen Z, Duan Y and Sun J: Plasma miRNAs in
predicting radiosensitivity in non-small cell lung cancer. Tumour
Biol. 37:11927–11936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu C and Arora P: Long noncoding
RNA-microRNA-mRNA: A novel tripartite axis in the regulation of
cardiac hypertrophy. Circ Cardiovasc Genet. 7:729–731. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye S, Yang L, Zhao X, Song W, Wang W and
Zheng S: Bioinformatics method to predict two regulation mechanism:
TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell
Biochem Biophys. 70:1849–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang ZH, Zheng R, Gao Y, Zhang Q and Zhang
H: Abnormal gene expression and gene fusion in lung adenocarcinoma
with high-throughput RNA sequencing. Cancer Gene Ther. 21:74–82.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leidinger P, Brefort T, Backes C, Krapp M,
Galata V, Beier M, Kohlhaas J, Huwer H, Meese E and Keller A:
High-throughput qRT-PCR validation of blood microRNAs in non-small
cell lung cancer. Oncotarget. 7:4611–4623. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tian X, Hou W, Bai S, Fan J, Tong H and
Bai Y: XAV939 promotes apoptosis in a neuroblastoma cell line via
telomere shortening. Oncol Rep. 32:1999–2006. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo W, Shen F, Xiao W, Chen J and Pan F:
Wnt inhibitor XAV939 suppresses the viability of small cell lung
cancer NCI-H446 cells and induces apoptosis. Oncol Lett.
14:6585–6591. 2017.PubMed/NCBI
|
|
18
|
Lin HH, Feng WC, Lu LC, Shao YY, Cheng AL
and Hsu CH: Abstract 2052: WNT/beta-catenin signaling inhibitors
improve the anti-proliferative effect of sorafenib against
hepatocellular carcinoma (HCC) cells. Cancer Res. 73:20522013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Su D and Song T: Programmed cell
death 4 inhibits proliferation and differentiation and induces
apoptosis of human mesenchymal stem cells through suppressing the
Wnt/β-catenin pathway. RSC Adv. 7:26566–26573. 2017. View Article : Google Scholar
|
|
20
|
Ran M, Chen B and Li Z, Wu M, Liu X, He C,
Zhang S and Li Z: Systematic identification of long non-coding RNAs
in immature and mature porcine testes. Biol Reprod. 94:772016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lun AT, Chen Y and Smyth GK: It's
DE-licious: A recipe for differential expression analyses of
RNA-seq experiments using quasi-likelihood methods in edgeR.
Methods Mol Biol. 1418:391–416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gene Ontology Consortium, . The gene
ontology in 2010: Extensions and refinements. Nucleic Acids Res.
38((Database Issue)): D331–D335. 2010.PubMed/NCBI
|
|
26
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35((Web Server Issue)): W169–W175. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Opsahl T, Agneessens F and Skvoretz J:
Node centrality in weighted networks: Generalizing degree and
shortest paths. Social Networks. 32:245–251. 2010. View Article : Google Scholar
|
|
30
|
Pearson K: Note on regression and
inheritance in the case of two parents. Proc R Soc Lond.
58:240–242. 1895. View Article : Google Scholar
|
|
31
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc. 57:289–300. 1995.
|
|
32
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Resnik P: Semantic similarity in a
taxonomy: An information-based measure and its application to
problems of ambiguity in natural language. J Artif Intell Res.
11:95–130. 1999. View Article : Google Scholar
|
|
34
|
Wang JZ, Du Z, Payattakool R, Yu PS and
Chen CF: A new method to measure the semantic similarity of GO
terms. Bioinformatics. 23:1274–1281. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu G, Li F, Qin Y, Bo X, Wu Y and Wang S:
GOSemSim: An R package for measuring semantic similarity among GO
terms and gene products. Bioinformatics. 26:976–978. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44((W1)):
W90–W97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhan Y, Zang H, Feng J, Lu J, Chen L and
Fan S: Long non-coding RNAs associated with non-small cell lung
cancer. Oncotarget. 8:69174–69184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zheng K, Wu G, Qin X, Wang Y, Xia S and
Meng X: Effects of XAV939 on proliferation and glycolysis of
hepatocellular carcinoma cells. J Mod Oncol. 13:2023–2026.
2016.
|
|
39
|
Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H,
Xi Z, Li Z and Liu Y: Role of HCP5-miR-139-RUNX1 feedback loop in
regulating malignant behavior of glioma cells. Mol Ther.
24:1806–1822. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Muys BR, Lorenzi JC, Zanette DL, Lima e
Bueno Rde B, de Araújo LF, Dinarte-Santos AR, Alves CP, Ramão A, de
Molfetta GA, Vidal DO and Silva WA Jr: Placenta-Enriched LincRNAs
MIR503HG and linc00629 decrease migration and invasion potential of
JEG-3 cell line. PLoS One. 11:e01515602016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pachera E, Assassi S, Cintora GS,
Frank-Bertoncelj M, Haunerdinger V, Dobrota R, Brock M, Vettori S,
Hellerbrand C, Feghali-Bostwick CA, et al: OP0284 long noncoding
RNA MIR503HG is a novel factor in the pathogenesis of systemic
sclerosis. Ann Rheum Dis. 74 (Suppl 2):S180.1–180. 2015. View Article : Google Scholar
|
|
42
|
Grimminger PP, Maus MK, Schneider PM,
Metzger R, Hölscher AH, Sugita H, Danenberg PV, Alakus H and
Brabender J: Glutathione S-transferase PI (GST-PI) mRNA expression
and DNA methylation is involved in the pathogenesis and prognosis
of NSCLC. Lung Cancer. 78:87–91. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Scharer CD, Mccabe CD, Aliseyed M, Berger
MF, Bulyk ML and Moreno CS: Genome-wide promoter analysis of the
SOX4 transcriptional network in prostate cancer cells. Cancer Res.
69:709–717. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen QL, Zheng WL, Yao WJ, Nie LW, Cheng
SH and Ma WL: Analysis of SOX4 gene mutation in non-small cell lung
cancer tissues. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 24:505–509.
2007.PubMed/NCBI
|
|
45
|
Tang T, Huan L, Zhang S, Zhou H, Gu L,
Chen X and Zhang L: MicroRNA-212 functions as a tumor-suppressor in
human non-small cell lung cancer by targeting SOX4. Oncol Rep.
38:2243–2250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li Y, Chen P, Zu L, Liu B, Wang M and Zhou
Q: MicroRNA-338-3p suppresses metastasis of lung cancer cells by
targeting the EMT regulator Sox4. Am J Cancer Res. 6:127–140.
2016.PubMed/NCBI
|
|
47
|
Foss KM, Sima C, Ugolini D, Neri M, Allen
KE and Weiss GJ: miR-1254 and miR-574-5p: Serum-based microRNA
biomarkers for early-stage non-small cell lung cancer. J Thorac
Oncol. 6:482–488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee CW, Kang MR, Yun J, Oh SJ and Kang JS:
Abstract 4387: Up-regulation of VHL by miR-1273C inhibits renal
cell carcinoma. Cancer Res. 74:43872014. View Article : Google Scholar
|
|
49
|
Li C, Zheng X, Han Y, Lv Y, Lan F and Zhao
J: XAV939 inhibits the proliferation and migration of lung
adenocarcinoma A549 cells through the WNT pathway. Oncol Lett.
15:8973–8982. 2018.PubMed/NCBI
|
|
50
|
Lachaier E, Louandre C, Ezzoukhry Z, Godin
C, Mazière JC, Chauffert B and Galmiche A: Ferroptosis, a new form
of cell death relevant to the medical treatment of cancer. Med Sci
(Paris). 30:779–783. 2014.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jiang Y, He Y, Liu S and Tao Y: Chromatin
remodeling factor lymphoid-specific helicase inhibits ferroptosis
through lipid metabolic genes in lung cancer progression. Chin J
Cancer. 36:822017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alvarez SW, Sviderskiy VO, Terzi EM,
Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K
and Possemato R: NFS1 undergoes positive selection in lung tumours
and protects cells from ferroptosis. Nature. 551:639–643. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
He L, Vasiliou K and Nebert DW: Analysis
and update of the human solute carrier (SLC) gene superfamily. Hum
Genomics. 3:195–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sehm T, Rauh M, Wiendieck K, Buchfelder M,
Eyüpoglu IY and Savaskan NE: Temozolomide toxicity operates in a
xCT/SLC7a11 dependent manner and is fostered by ferroptosis.
Oncotarget. 7:74630–74647. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jiang L: p53 promotes ferroptosis during
ROS stress to suppress tumorigenesis. Cancer Dis. 5:4652015.
View Article : Google Scholar
|
|
57
|
Ji XJ, Qian J, Rahman J, Harris B,
Hoeksema MD, Chen H, Eisenberg R and Young J: Abstract A10: SLC7A11
contributes to the pathogenesis of lung cancer. Mol Cancer Res.
14:A102016. View Article : Google Scholar
|