Introduction
Gliomas are the tumors with the highest morbidity
and mortality in adult central nervous system tumors (1). With the improvement of the medical
science level, the treatment methods for brain tumors (surgical
resection, chemotherapy, and radiotherapy) have greatly improved,
but the prognosis of glioma patients is still poor. At 5% is the
lowest five-year survival rate of all cancer patients, the median
survival time is approximately 14 months, even after receiving the
maximum safety resection plus adjuvant chemotherapy, the median
survival time can only be increased to approximately 15 months
(2,3). Due to abundant blood supply, gliomas
appear to be infiltrating growth, and it is difficult to excise the
gliomas completely. It is difficult to avoid postoperative
recurrence. Chemotherapy and radiotherapy are not highly specific
for glioma cells and have great toxic and side effects (4). Therefore, the search for new molecular
biomarkers and the development of safer and more effective
molecular targeted therapies are important tasks in the treatment
of gliomas.
lncRNA is an RNA with a base number of not less than
200 bp and does not encode protein, and regulates the expression of
genes at several levels such as chromatin modification,
transcription, and post-transcriptional modification (5). It has been found that many lncRNAs are
abnormally expressed during carcinogenesis. These lncRNAs may be
useful therapeutic targets for malignant tumors and molecular
markers for the diagnosis of prognosis (6). lncRNA ZEB1-AS1 is a newly discovered
lncRNA, and several studies have shown that lncRNA ZEB1-AS1 plays
an important role in the occurrence and development of many types
of tumors, including liver cancers (7), colorectal cancer (8), esophageal squamous cell carcinoma
(9), and lung cancer (10). Li et al (11) studied gastric cancer and found that
ZEB1-AS1 exerts carcinogenic effects by promoting the migration,
invasion and EMT process of gastric cancer cells. The expression of
ZEB1-AS1 is positively correlated with the prognosis of gastric
cancer. Wang et al (9)
investigated the expression of lncRNA ZEB1-AS1 in esophageal
squamous cell carcinoma and found that the expression of lncRNA
ZEB1-AS1 was increased in cancer tissues and was positively
correlated with tumor grade, depth of invasion, and lymph node
metastasis. However, the clinical significance, biological
function, and mechanism of action of lncRNA ZEB1-AS1 in gliomas
remain to be determined.
Lv et al (12)
found that ZEB1-AS1 is highly expressed in glioma tissues.
Therefore, the purpose of this study was to investigate the effects
of knockdown of lncRNA ZEB1-AS1 on proliferation, invasion, and
apoptosis of glioma U87 cells in order to treat gliomas and to
provide new therapeutic targets.
Materials and methods
Materials and reagents
U87MG glioblastoma cells of unknown origin (Shanghai
Zeye Biological Technology Co., Ltd., catalogue no.: AC319); lncRNA
ZEB1-AS1-specific SiRNA and non-specific scramble siRNA sequence
design (Shandong Weizheng Biotech Co., Ltd., Shandong, China);
fetal bovine serum, DMEM medium [Wuhan Puno (Sales Life Technology
Co., Ltd.), Wuhan, China]; TransScript Green Two-Step RT-qPCR
SuperMix kit (Beijing Quantum Biotechnology Co., Ltd., Beijing,
China); Lipofectamine® 3000 Transfection Reagent, TRIzol
Reagent, Thermo Scientific Revert Aid First Strand c DNA Synthesis
kit (Thermo Fisher Scientific (China) Co., Ltd., Shanghai, China;
qPCR Primer Sequences (Shanghai Sangon Bioengineering Co., Ltd.,
Shanghai, China); CCK8 kit (Shanghai Shengsheng Biotechnology Co.,
Ltd. Shanghai, China); and Transwell Cell (Beijing Yiming Fuxing
Biotechnology Co., Ltd. Beijing, China) were used in the study.
The study was approved by the Ethics Committee of
The Central Hospital of Wuhan, Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China).
Plasmid transfection
The U87MG glioblastoma cell line was cultured in
DMEM medium containing 20% fetal bovine serum at 37°C in a 5%
CO2 incubator. When the cell confluence reached 80%, the
cells were seeded in a 6-well plate at a concentration of
1x105/per well. When the cell confluence reached 60%, Si
RNA transfected with lncRNA ZEB1-AS1 was transfected into Si group,
NC group was transfected with non-specific scramble siRNA, blank
group was not transfected, and equal amount of complete medium was
added. The instructions for Lipofectamine 3000 transfection
reagents were strictly followed. After incubation in a 5%
CO2 incubator at 37°C, assays were performed when cell
confluency reached 80%.
RT-qPCR detection of lncZEB1-AS1
expression
TRIzol and chloroform reagents were used to extract
total RNA from each group. The concentration and purity of RNA were
measured by UV spectrophotometer (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The ratio of A260/A280 between 1.8 and 2.0 was
considered as qualified. The reverse transcription reaction system
was prepared strictly in accordance with the instructions,
incubated at 42°C for 15 min, and reverse transcribed with 1 µg of
total RNA at 85°C for 5 sec. The qPCR reaction was strictly
performed in accordance with the TransScript Green Two-Step RT-qPCR
SuperMix kit instructions. The amplification was performed using a
two-step method, pre-denaturing at 94°C for 30 sec, 94°C for 5 sec,
and 60°C for 30 sec for 40 cycles. Using GAPDH as an internal
reference, the sequences of each primer are shown in Table I.
 | Table I.qPCR primer sequences. |
Table I.
qPCR primer sequences.
| Gene | Upstream
sequence | Downstream
sequence |
|---|
| GAPDH |
5′-CCCATCACCATCTTCCAGGAG-3′ |
5′-GTTGTCATGGATGACCTTGGC-3′ |
| ZEB1-AS1 |
5′-AACCTTGTTGCTAGGGACCG-3′ |
5′-AGTCACTTCCCATCCCGGTT-3′ |
MTT test cell proliferation in each
group
When the cells grew to logarithmic phase, the cells
were digested and the cell suspension was prepared with RPMI-1640
containing 10% FBS. The cell density was adjusted to
2×104 ml and seeded in 96-well plates at
3×103/200 µl/well. The cell proliferation was measured
by MTT assay every 24 h until 120 h. Five tests were performed. Of
5 mg/ml MTT solution 10 µl was added to each well for 4 h.
Formanzan solution was added at 150 µl/well. The wells were mixed
for 10 min at room temperature and vortexed, the absorbance (OD)
was recorded at a wavelength of 490 nm using a microplate reader,
and the assay was repeated three times per well.
Transwell invasion experiment
After 12 h of culture in serum-free RPMI-1640
medium, cell suspension was prepared and counted to adjust the cell
density to 1×105/ml. Then, 100 µl cell suspension was
added to the Transwell upper chamber of 24-well plate, and 600 µl
DMEM medium with 20% fetal bovine serum was added to the lower
chamber, followed by incubation at 37°C, 5% CO2 for 24
h. The mixture was washed 3 times with PBS and the non-migrating
cells of the upper chamber on the polycarbonate membrane were
gently wiped off with a cotton swab. Then, 4% paraformaldehyde was
fixed on a polycarbonate membrane for 10 min, followed by crystal
violet staining for 10 min, prior to slowly washing 3 times with
PBS and counting under a microscope at a magnification of ×400.
Five fields were randomly selected for counting and average value
was recorded.
Flow cytometry detection of
apoptosis
After 48 h of transfection, the cells were first
trypsinized, and collected. Following digestion with trypsin, the
cells were re-precipitated (4°C) in 0.01 mol/l PBS, counted to
adjust the cell density to 1×105/ml, and 150 µl of
Annexin V-FITC was added to the cells to resuspend the cells, which
were then transferred to a flow test tube. Annexin V-FIT (5 µl) and
15 µl of propidium iodide was added to each tube and gently mixed,
and reacted at room temperature for 15 min in the dark. The samples
were detected by flow cytometry. The process was completed within 1
h, with each sample repeated 3 times. Apoptosis rate was expressed
as a percentage and data analysis was performed using flow software
Flowjo 7.5.
Statistical analysis
The statistical analysis was performed using the
SPSS 19.0 software system (IBM, SPSS, Chicago, IL, USA). Data were
expressed as the mean ± standard deviation (mean ± SD). The mean of
multiple groups was compared using a single factor variance
analysis and analysis of repeated measures. Multiple comparison was
performed using LSD test. P<0.05 was considered to indicate a
statistically significant difference.
Results
RT-qPCR detection of lncZEB1-AS1
expression
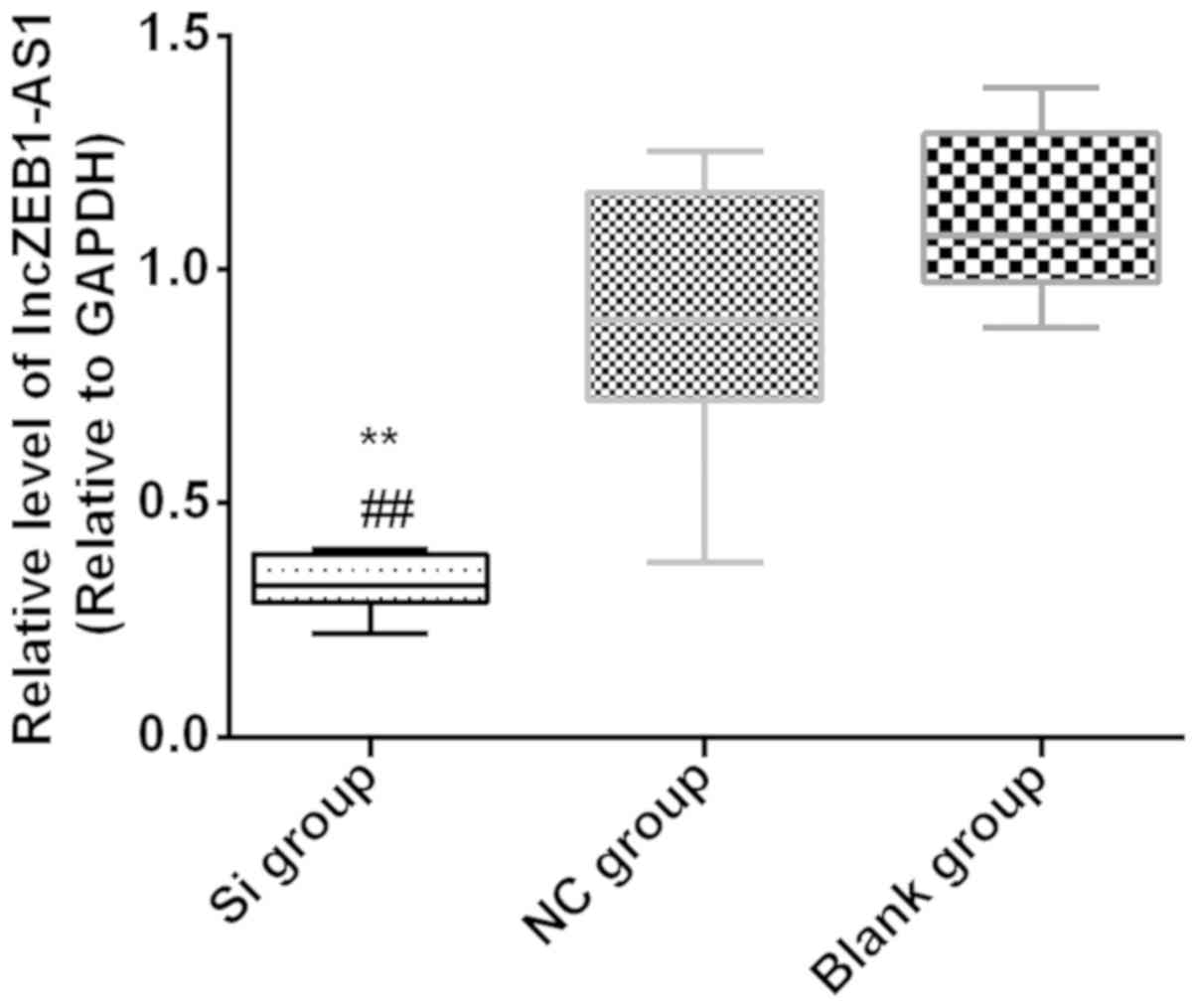
The expression of lncZEB1-AS1 was detected by
RT-qPCR 48 h after transfection. The expression of lncZEB1-AS1 in
the Si group (0.33±0.02) was significantly lower than that in the
NC group (0.91±0.10) and blank group (1.11±0.06) (P<0.01). There
was no statistical difference between the NC and blank groups
(P>0.05) (Fig. 1).
MTT detection of cell proliferation in
each group
The comparison between the three groups at the same
time point showed that there was no significant difference in OD490
among the three groups at 24 h (P>0.05). At 48 h, the Si group
was significantly lower than the NC group and the blank group
(P<0.05), there was no difference between the NC group and the
blank group (P>0.05). From 48 h, the three groups showed a
gradual upward trend, but at all time points, the Si group was
lower than the NC group and the blank group (P<0.01). There was
no statistical significance between the NC group and the blank
group (P<0.05); The variance analysis of repeated measures at
the same group at different time points showed that the OD values
of the blank group at 72, 96, and 120 h were significantly higher
than those at the previous time point (P<0.01). The OD values of
the NC groups at 72, 96 and 120 h were significantly higher than
those at the previous time point (P<0.01). The OD values of Si
group at 48 and 96 h were significantly higher than those at the
previous time point (P<0.01). Although there was an upward trend
between 72 and 48 h, the difference was not statistically
significant (P>0.05) (Fig.
2).
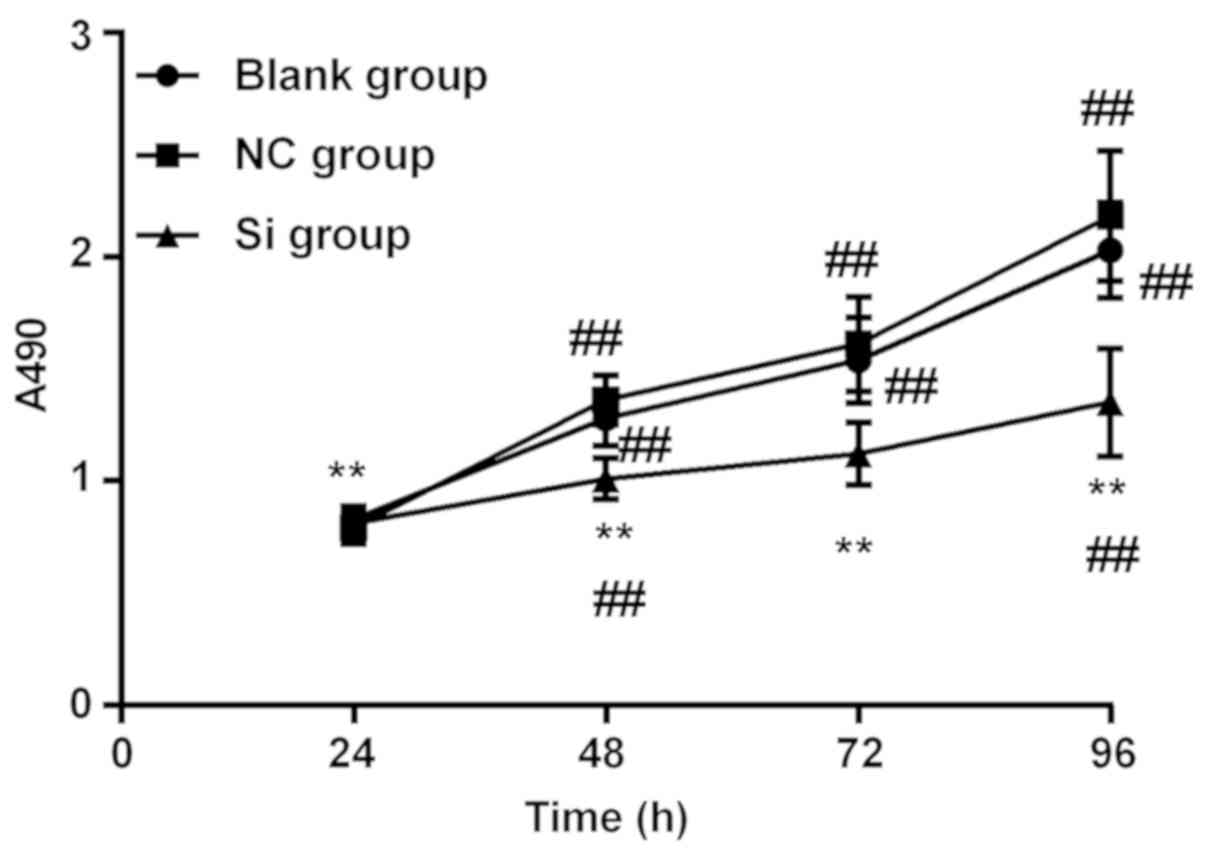 | Figure 2.MTT test for cell proliferation in
each group: the three groups were found at the same time point,
there was no statistically significant difference in OD490 in three
groups at 24 h (P>0.05), at 48 h, Si group was significantly
lower than the NC and blank groups (P<0.00), there was no
difference between the NC and blank groups (P>0.05). The three
groups showed a gradual upward trend from 48 h, but Si group was
always lower than the NC group and blank group at each time point
(P<0.01). There was no significant difference between the NC and
blank groups (P<0.05). The variance analysis of repeated
measurement at the same group at different time points showed that
the OD values of blank group at 72, 96, 120 h significantly
increased compared with the previous time point (P<0.01), OD
values of NC group at 72, 96, 120 h were significantly higher
compared with the previous time point (P<0.01). The OD values of
Si group at 48 and 96 h were significantly higher than those at the
previous time point (P<0.01). Although there was an upward trend
between 72 and 48 h, the difference was not statistically
significant (P>0.05). At the same time point, **P<0.01
compared with NC group and blank group. In the same group,
##P<0.01 compared with the previous time point. |
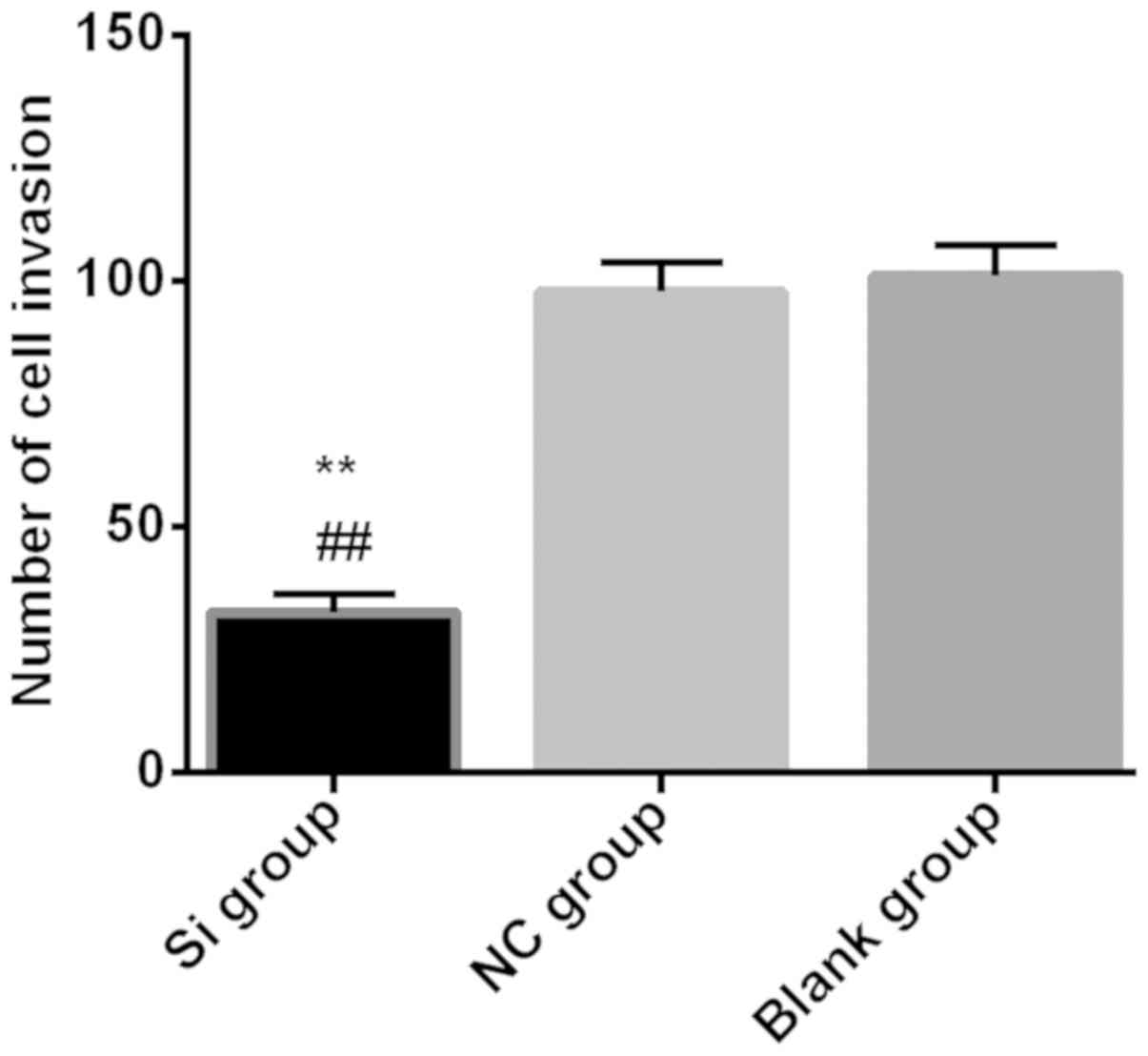
Transwell invasion experiment
Transwell invasion assay to detect the invasive
ability of each group found that the number of invading cells in
the Si group (32.51±3.71) was significantly lower than that in the
NC group (97.82±5.92) and the blank group (101.09±6.22)
(P<0.01), There was no statistical difference in the number of
invasive cells between NC group and blank group (P>0.05)
(Fig. 3).
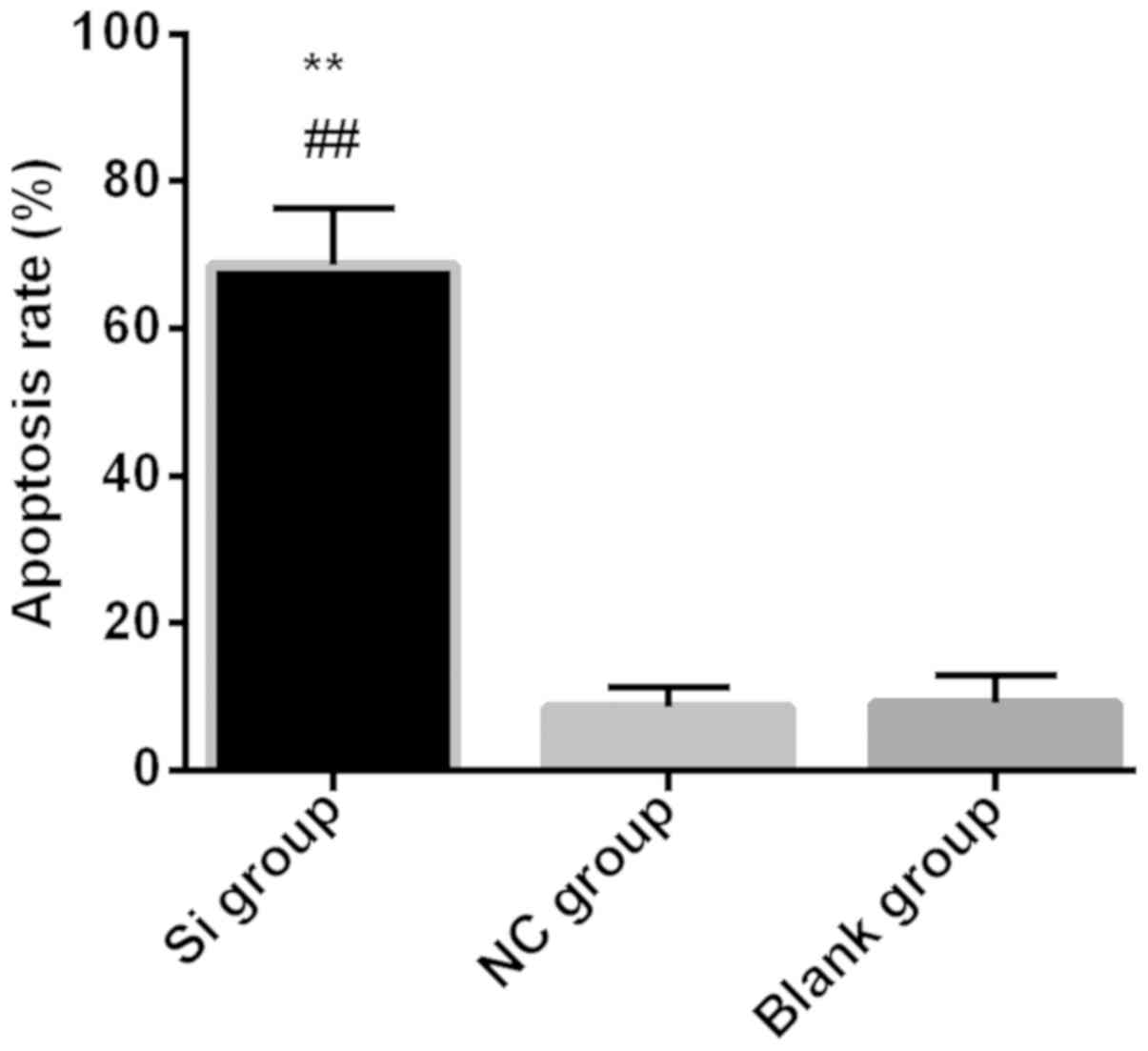
Flow cytometry detection of
apoptosis
Flow cytometry detected apoptosis in each group the
apoptosis rate in the Si group 68.54±7.71% was significantly higher
than that in the NC group 8.51±2.71% and in the blank group
9.16±3.71% (P<0.01). There was no statistical significance
between NC group and blank group (P>0.05) (Fig. 4).
Discussion
Gliomas are the most common type of malignant brain
tumors, accounting for approximately 46%. According to the time of
onset, they are divided into adult and child types. The prognosis
is extremely poor, which jeopardizes the patient's physical health
and social economic development, leading to great economic and
psychological burden to the family (13,14).
lncRNA ZEB1-AS1 is upregulated in a variety of tumors and is
closely related to clinical stage, prognosis and lymph node
metastasis, but the relationship between the mechanism of action of
glioma cells and clinical features has not yet been clarified
(15). This study explored the
effect of lncRNA ZEB1-AS1 on the proliferation, invasion and
apoptosis of U87MG glioblastoma cells and the role of lncRNA
ZEB1-AS1 in glioblastoma cells in order to provide
molecular-targeted therapy and theoretical basis for gliomas.
The expression of lncZEB1-AS1 was detected by
RT-qPCR and it was found that the expression of lncZEB1-AS1 in Si
group was significantly lower than that in NC group and blank group
after transfection, indicating that ZEB1-AS1 SiRNA was successfully
transfected into Si group; other subsequent results in cells were
comparable. MTT detection of cell proliferation in each group found
that at 24 h, OD490 of the three groups did not show statistical
difference, from 48 h, the three groups showed a gradual upward
trend, but at each time point Si group was lower than the NC and
blank groups, there was no statistical significance between the NC
and blank groups, indicating that transfection of ZEB1-AS1 SiRNA
inhibited the proliferation of U87MG cells. In other words,
lncZEB1-AS1 promote the proliferation of glioblastoma cells and
Transwell invasion assay detected the invasion ability of cells in
each group. The number of invading cells in the Si group was
significantly lower than that in the NC and blank groups. There was
no statistical difference in the number of invading cells between
the two groups, indicating that the transfection of ZEB1-AS1 SiRNA
inhibited the invasion of U87MG cells, i.e., lncZEB1-AS1 could
promote the invasion of brain glioma cells. Apoptosis was detected
by flow cytometry and it was found that the apoptosis rate of Si
group was significantly higher than that of the NC and blank
groups, indicating that ZEB1-AS1 SiRNA promoted apoptosis of U87MG
cells after transfection. Lnc ZEB1-AS1 can inhibit apoptosis of
glioma cells. Lv et al (12)
found that lncZEB1-AS1 is highly expressed in glioma tissues and is
closely related to the clinical stage and prognosis of glioma.
lncZEB1-AS1 is an independent prognostic factor for glioma
patients. lncRNA ZEB1-AS1 inhibits proliferation and invasion and
promotes apoptosis in U87MG glioblastoma cells. The possible reason
is that lncRNA ZEB1-AS1 is derived from the promoter region of
ZEB1, and lncRNA ZEB1-AS1 can induce H3K4me3 modification of ZEB1
promoter region and activate ZEB1 transcription. Thus, ZEB1 exerts
the role of oncogenes by regulating its downstream targets
(16,17). The miR-101/ZEB1 axis can promote the
hypoxia-induced epithelial-mesenchymal transition in malignant
gliomas (18). ZEB1-AS1 can also
regulate the proliferation and migration of osteosarcoma cells
through miR-200 (19).
The present findings showed that lncRNA ZEB1-AS1 can
promote the proliferation and invasion of U87MG glioblastoma cells
and inhibit apoptosis, indicating that lncRNA ZEB1-AS1 plays an
oncogenic role, and if there is an additional grouping of normal
brain glial cells, with RT-qPCR detection of lncZEB1-AS1
expression, lncRNA ZEB1-AS1 level in the normal brain glial cells
was significantly lower than glioma U87 cells, we can further
verify this conclusion. In this study, only one type of glioma cell
was selected for testing. It does not fully represent the actual
progress of glioma. It should be tested in other glioma cell lines.
Moreover, the cell test is in vitro, it does not represent
the true situation in the body, so it should also be used to
further verify the glioma cells in the rat subcutaneous
tumorigenesis test (20).
In summary, inhibition of lncRNA ZEB1-AS1 can
inhibit proliferation and invasion of U87MG glioblastoma cells and
promote apoptosis. lncRNA ZEB1-AS1 is expected to become a new
target for the treatment of glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WZ drafted the manuscript. WZ and LX were mainly
devoted to collecting and interpreting the data. WZ helped with
flow cytometry detection. LX performed Transwell invasion
experiment. Both authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Central Hospital of Wuhan, Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Agarwal S, Sane R, Oberoi R, Ohlfest JR
and Elmquist WF: Delivery of molecularly targeted therapy to
malignant glioma, a disease of the whole brain. Expert Rev Mol Med.
13:e172011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Delgado-López PD and Corrales-García EM:
Survival in glioblastoma: A review on the impact of treatment
modalities. Clin Transl Oncol. 18:1062–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yan W, Zhang W and Jiang T: Oncogene
addiction in gliomas: Implications for molecular targeted therapy.
J Exp Clin Cancer Res. 30:582011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jia L, Tian Y, Chen Y and Zhang G: The
silencing of LncRNA-H19 decreases chemoresistance of human glioma
cells to temozolomide by suppressing epithelial-mesenchymal
transition via the Wnt/β-catenin pathway. Onco Targets Ther.
11:313–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Q, Du X, Yang M, Xiao S, Cao J, Song
J and Wang L: LncRNA ZEB1-AS1 contributes to STAT3 activation by
associating with IL-11 in B-lymphoblastic leukemia. Biotechnol
Lett. 39:1801–1810. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin J, Zhan Y, Liu Y, Chen Z, Liang J, Li
W, He A, Zhou L, Mei H, Wang F, et al: Increased expression of
ZEB1-AS1 correlates with higher histopathological grade and
promotes tumorigenesis in bladder cancer. Oncotarget.
8:24202–24212. 2017.PubMed/NCBI
|
|
7
|
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z,
Deng X, Chen H, Shen B, Peng C, et al: Upregulation of long
noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor
prognosis in hepatocellular carcinoma. Oncogene. 35:1575–1584.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong H, Wen H, Zhu X, Lian Y, Yang X, Qian
Z and Zhu J: High expression of long non-coding RNA ZEB1-AS1
promotes colorectal cancer cell proliferation partially by
suppressing p15 expression. Tumour Biol. 39:10104283177053362017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang YL, Bai Y, Yao WJ, Guo L and Wang ZM:
Expression of long non-coding RNA ZEB1-AS1 in esophageal squamous
cell carcinoma and its correlation with tumor progression and
patient survival. Int J Clin Exp Pathol. 8:11871–11876.
2015.PubMed/NCBI
|
|
10
|
Rong L, Zhao R and Lu J: Highly expressed
long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer
progression via Wnt/β-catenin signaling. Biochem Biophys Res
Commun. 484:586–591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Wen X, Wang L, Sun X, Ma H, Fu Z and
Li L: LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric
cancer. Surg Oncol. 26:527–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lv QL, Hu L, Chen SH, Sun B, Fu ML, Qin
CZ, Qu Q, Wang GH, He CJ and Zhou HH: A long noncoding RNA ZEB1-AS1
promotes tumorigenesis and predicts poor prognosis in glioma. Int J
Mol Sci. 17:172016. View Article : Google Scholar
|
|
13
|
Fu J and Cui Y: Long noncoding RNA
ZEB1-AS1 expression predicts progression and poor prognosis of
colorectal cancer. Int J Biol Markers. 32:e428–e433. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bonavia R, Inda MM, Vandenberg S, Cheng
SY, Nagane M, Hadwiger P, Tan P, Sah DW, Cavenee WK and Furnari FB:
EGFRvIII promotes glioma angiogenesis and growth through the NF-κB,
interleukin-8 pathway. Oncogene. 31:4054–4066. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, Li Z, Leng K, Xu Y, Ji D, Huang L,
Cui Y and Jiang X: ZEB1-AS1: A crucial cancer-related long
non-coding RNA. Cell Prolif. 51:e124232018. View Article : Google Scholar
|
|
16
|
Su W, Xu M, Chen X, Chen N, Gong J, Nie L,
Li L, Li X, Zhang M and Zhou Q: Long noncoding RNA ZEB1-AS1
epigenetically regulates the expressions of ZEB1 and downstream
molecules in prostate cancer. Mol Cancer. 16:1422017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang C, Zan J, Yue B, Liu C, He C and Yan
D: Long non-coding ribonucleic acid zinc finger antisense 1
promotes the progression of colonic cancer by modulating ZEB1
expression. J Gastroenterol Hepatol. 32:1204–1211. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang S, Wang W, Liu G, Xie S, Li Q, Li Y
and Lin Z: Long non-coding RNA HOTTIP promotes hypoxia-induced
epithelial-mesenchymal transition of malignant glioma by regulating
the miR-101/ZEB1 axis. Biomed Pharmacother. 95:711–720. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu C, Pan C, Cai Y and Wang H: Interplay
between long noncoding RNA ZEB1-AS1 and miR-200s regulates
osteosarcoma cell proliferation and migration. J Cell Biochem.
118:2250–2260. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lenting K, Verhaak R, Ter Laan M,
Wesseling P and Leenders W: Glioma: Experimental models and
reality. Acta Neuropathol. 133:263–282. 2017. View Article : Google Scholar : PubMed/NCBI
|