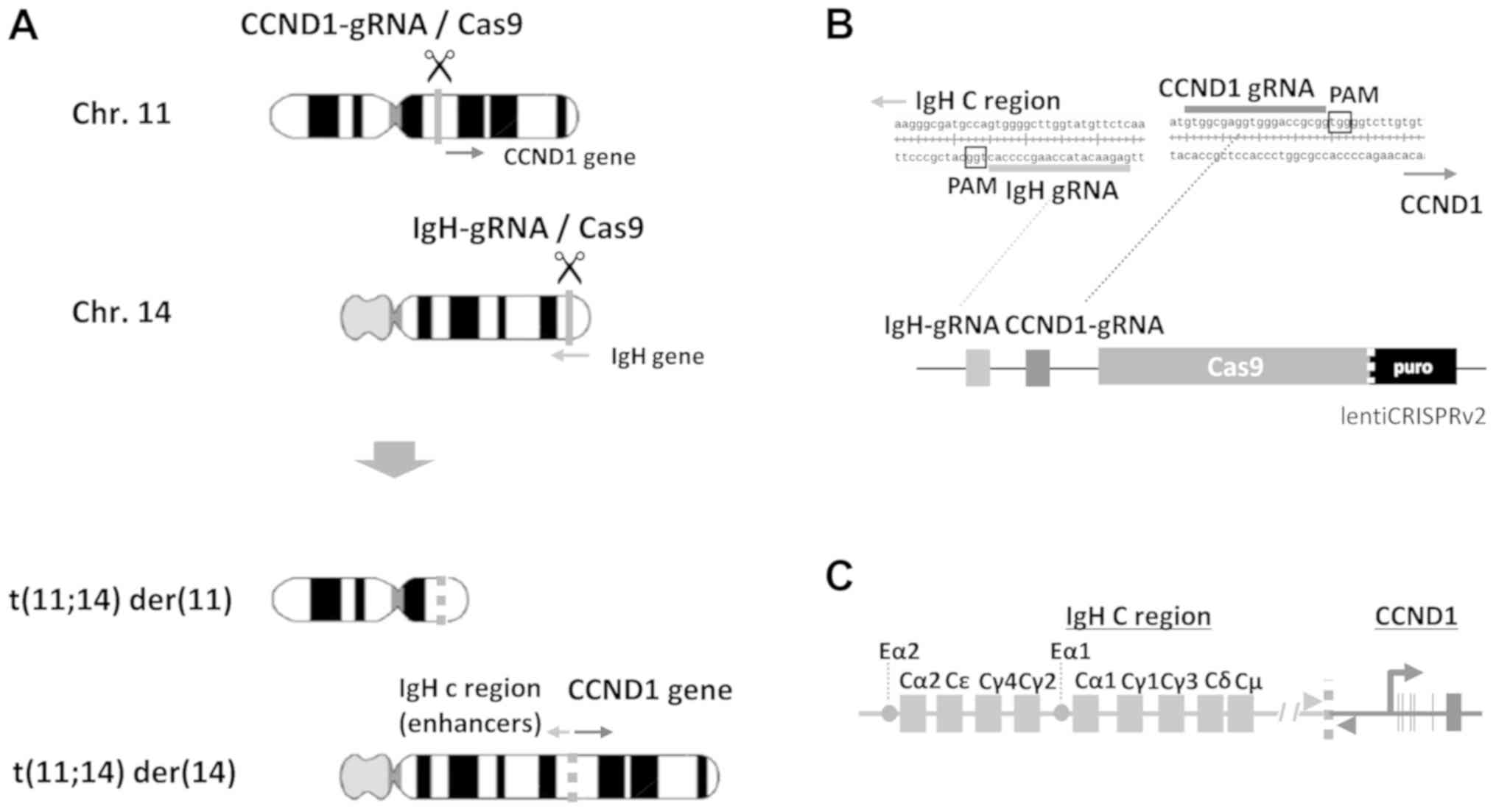
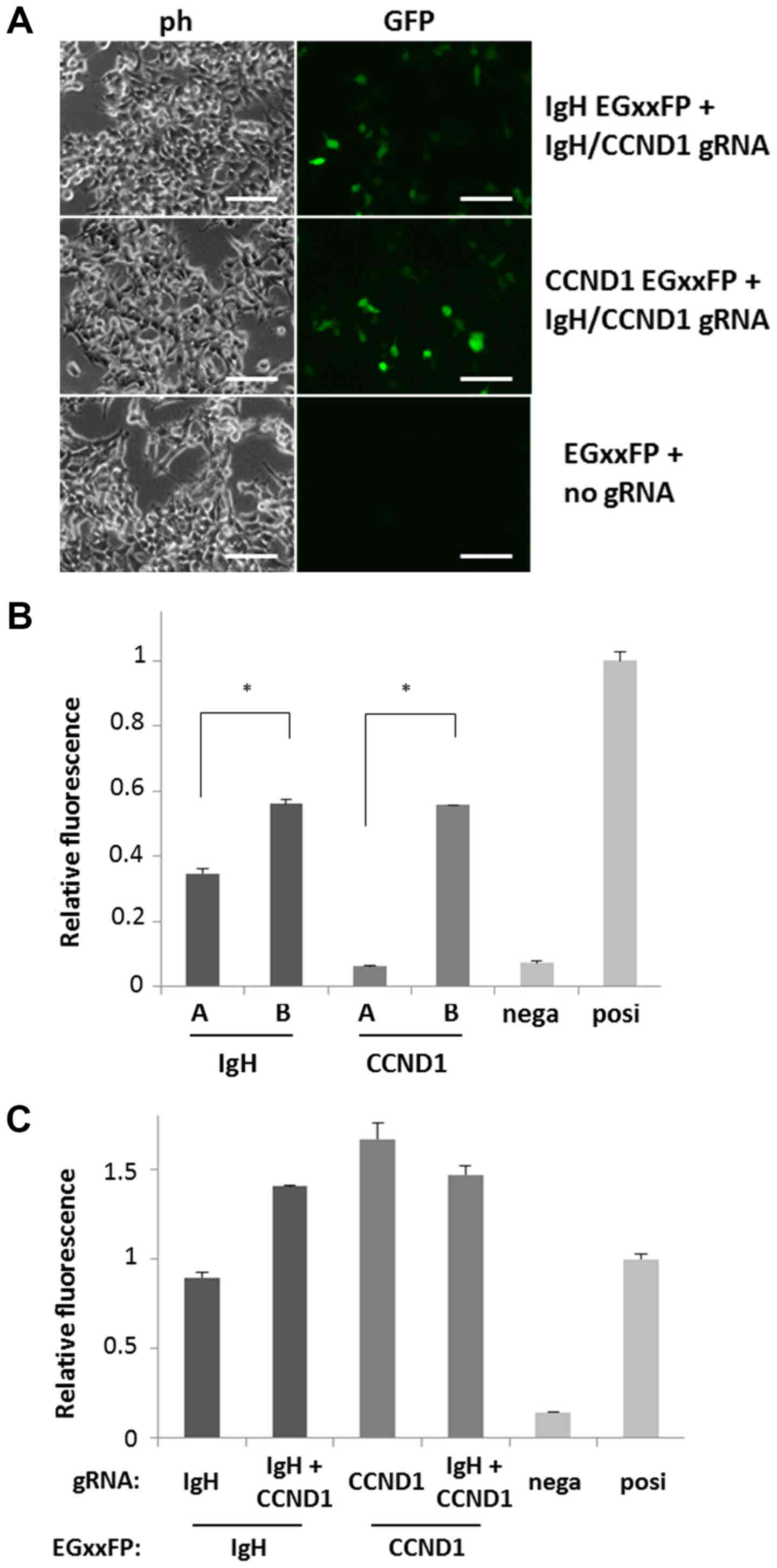
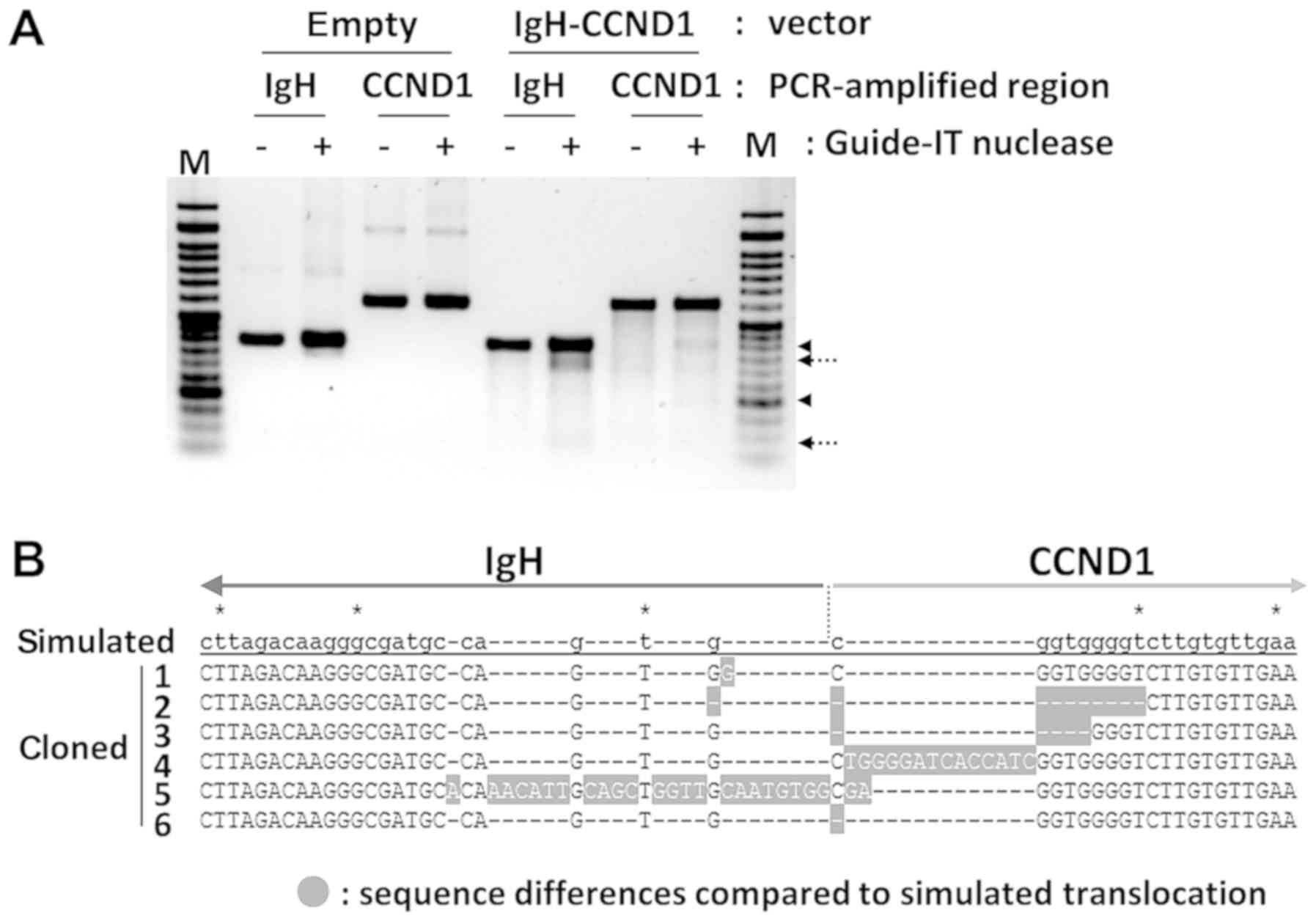
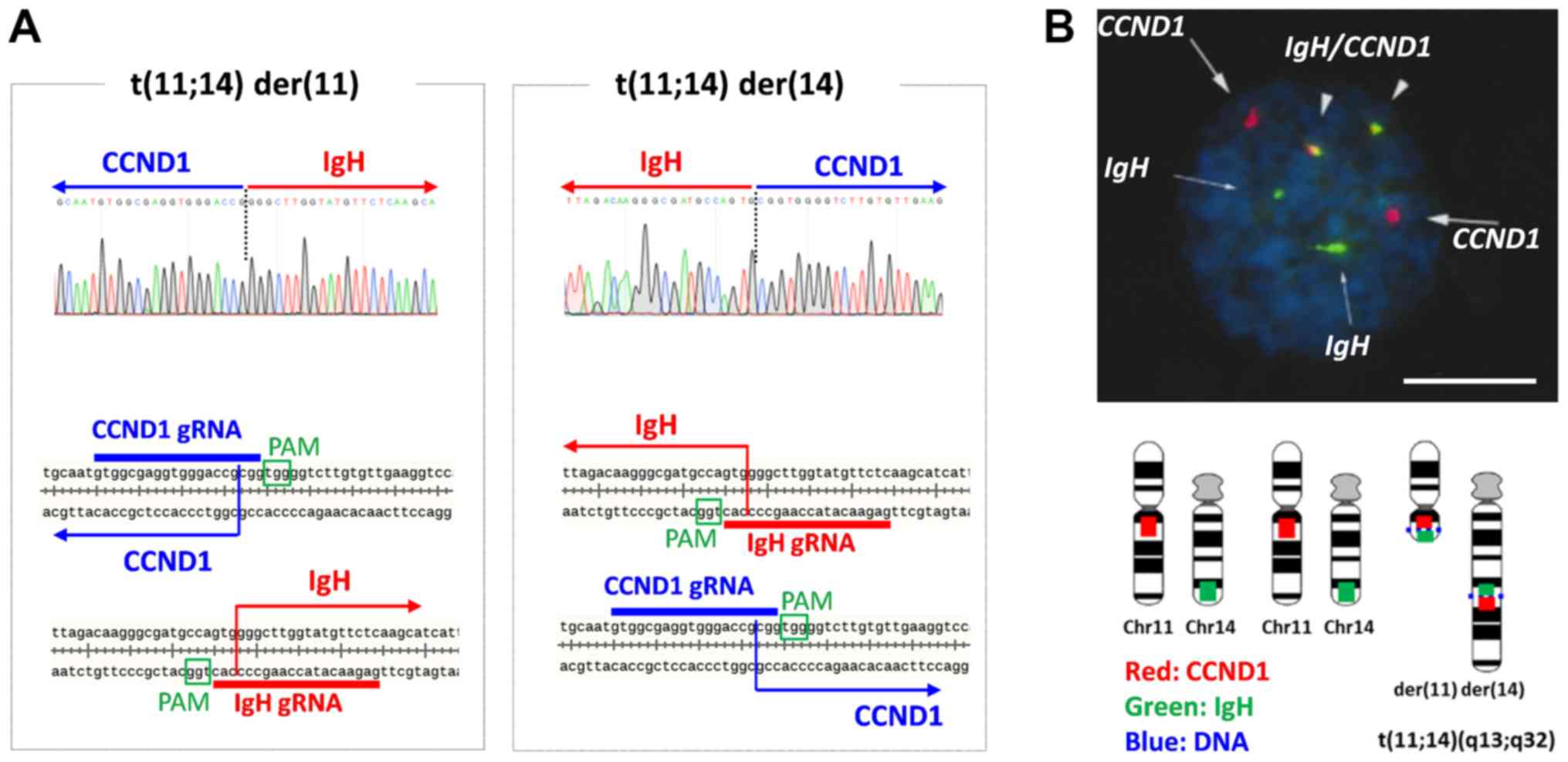
|
1
|
Rabbitts TH: Chromosomal translocations in
human cancer. Nature. 372:143–149. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nambiar M, Kari V and Raghavan SC:
Chromosomal translocations in cancer. Biochim Biophys Acta.
1786:139–152. 2008.PubMed/NCBI
|
|
3
|
Lieber MR: Mechanisms of human lymphoid
chromosomal translocations. Nat Rev Cancer. 16:387–398. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chesi M, Bergsagel PL, Brents LA, Smith
CM, Gerhard DS and Kuehl WM: Dysregulation of cyclin D1 by
translocation into an IgH gamma switch region in two multiple
myeloma cell lines. Blood. 88:674–681. 1996.PubMed/NCBI
|
|
5
|
Chesi M, Nardini E, Brents LA, Schrock E,
Ried T, Kuehl WM and Bergsagel PL: Frequent translocation
t(4;14)(p16.3;q32.3) in multiple myeloma is associated with
increased expression and activating mutations of fibroblast growth
factor receptor 3. Nat Genet. 16:260–264. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chesi M, Bergsagel PL, Shonukan OO,
Martelli ML, Brents LA, Chen T, Schrock E, Ried T and Kuehl WM:
Frequent dysregulation of the c-maf proto-oncogene at 16q23 by
translocation to an Ig locus in multiple myeloma. Blood.
91:4457–4463. 1998.PubMed/NCBI
|
|
7
|
Inaba T, Matsushime H, Valentine M,
Roussel MF, Sherr CJ and Look AT: Genomic organization, chromosomal
localization, and independent expression of human cyclin D genes.
Genomics. 13:565–574. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Szepetowski P, Perucca-Lostanlen D and
Gaudray P: Mapping genes according to their amplification status in
tumor cells: Contribution to the map of 11q13. Genomics.
16:745–750. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sherr CJ: Mammalian G1 cyclins. Cell.
73:1059–1065. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fonseca R, Blood EA, Oken MM, Kyle RA,
Dewald GW, Bailey RJ, Van Wier SA, Henderson KJ, Hoyer JD,
Harrington D, et al: Myeloma and the t(11;14)(q13;q32); evidence
for a biologically defined unique subset of patients. Blood.
99:3735–3741. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fenton JA, Pratt G, Rothwell DG, Rawstron
AC and Morgan GJ: Translocation t(11;14) in multiple myeloma:
Analysis of translocation breakpoints on der(11) and der(14)
chromosomes suggests complex molecular mechanisms of recombination.
Genes Chromosomes Cancer. 39:151–155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Walker BA, Wardell CP, Johnson DC, Kaiser
MF, Begum DB, Dahir NB, Ross FM, Davies FE, Gonzalez D and Morgan
GJ: Characterization of IGH locus breakpoints in multiple myeloma
indicates a subset of translocations appear to occur in pregerminal
center B cells. Blood. 121:3413–3419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Z, Zan H, Pone EJ, Mai T and Casali P:
Immunoglobulin class-switch DNA recombination: Induction, targeting
and beyond. Nat Rev Immunol. 12:517–531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Casellas R, Basu U, Yewdell WT, Chaudhuri
J, Robbiani DF and Di Noia JM: Mutations, kataegis and
translocations in B cells: Understanding AID promiscuous activity.
Nat Rev Immunol. 16:164–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nambiar M and Raghavan SC: How does DNA
break during chromosomal translocations? Nucleic Acids Res.
39:5813–5825. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Van der Oost J: Molecular biology. New
tool for genome surgery. Science. 339:768–770. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gaj T, Gersbach CA and Barbas CF III: ZFN,
TALEN, and CRISPR/Cas-based methods for genome engineering. Trends
Biotechnol. 31:397–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA and Zhang F:
Multiplex genome engineering using CRISPR/Cas systems. Science.
339:819–823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mali P, Yang L, Esvelt KM, Aach J, Guell
M, DiCarlo JE, Norville JE and Church GM: RNA-guided human genome
engineering via Cas9. Science. 339:823–826. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moreno-Mateos MA, Vejnar CE, Beaudoin JD,
Fernandez JP, Mis EK, Khokha MK and Giraldez AJ: CRISPRscan:
Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in
vivo. Nat Methods. 12:982–988. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choi PS and Meyerson M: Targeted genomic
rearrangements using CRISPR/Cas technology. Nat Commun. 5:37282014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Breese EH, Buechele C, Dawson C, Cleary ML
and Porteus MH: Use of genome engineering to create patient
specific MLL translocations in primary human hematopoietic stem and
progenitor cells. PLoS One. 10:e01366442015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang J, Zhang L, Zhou X, Chen X, Huang G,
Li F, Wang R, Wu N, Yan Y, Tong C, et al: Induction of
site-specific chromosomal translocations in embryonic stem cells by
CRISPR/Cas9. Sci Rep. 6:219182016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lekomtsev S, Aligianni S, Lapao A and
Bürckstummer T: Efficient generation and reversion of chromosomal
translocations using CRISPR/Cas technology. BMC Genomics.
17:7392016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reimer J, Knoss S, Labuhn M, Charpentier
EM, Gohring G, Schlegelberger B, Klusmann JH and Heckl D:
CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate
leukemia in human hematopoietic progenitor cells in vivo.
Haematologica. 102:1558–1566. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vanoli F, Tomishima M, Feng W, Lamribet K,
Babin L, Brunet E and Jasin M: CRISPR-Cas9-guided oncogenic
chromosomal translocations with conditional fusion protein
expression in human mesenchymal cells. Proc Natl Acad Sci USA.
114:3696–3701. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mashiko D, Fujihara Y, Satouh Y, Miyata H,
Isotani A and Ikawa M: Generation of mutant mice by pronuclear
injection of circular plasmid expressing Cas9 and single guided
RNA. Sci Rep. 3:33552013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cepko C: Large-scale preparation and
concentration of retrovirus stocks. Curr Protoc Mol Biol. Chapter
9: Unit 0.12. 1997.doi: 10.1002/0471142727.mb0912s37.
|
|
30
|
Doench JG, Fusi N, Sullender M, Hegde M,
Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et
al: Optimized sgRNA design to maximize activity and minimize
off-target effects of CRISPR-Cas9. Nat Biotechnol. 34:184–191.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ronchetti D, Finelli P, Richelda R,
Baldini L, Rocchi M, Viggiano L, Cuneo A, Bogni S, Fabris S,
Lombardi L, et al: Molecular analysis of 11q13 breakpoints in
multiple myeloma. Blood. 93:1330–1337. 1999.PubMed/NCBI
|
|
33
|
Jhunjhunwala S, van Zelm MC, Peak MM,
Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA and Murre
C: The 3D structure of the immunoglobulin heavy-chain locus:
Implications for long-range genomic interactions. Cell.
133:265–279. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zwijsen RM, Klompmaker R, Wientjens EB,
Kristel PM, van der Burg B and Michalides RJ: cyclin D1 triggers
autonomous growth of breast cancer cells by governing cell cycle
exit. Mol Cell Biol. 16:2554–2560. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Janssen JW, Vaandrager JW, Heuser T, Jauch
A, Kluin PM, Geelen E, Bergsagel PL, Kuehl WM, Drexler HG, Otsuki
T, et al: Concurrent activation of a novel putative transforming
gene, myeov, and cyclin D1 in a subset of multiple myeloma cell
lines with t(11;14)(q13;q32). Blood. 95:2691–2698. 2000.PubMed/NCBI
|
|
36
|
Spraggon L, Martelotto LG, Hmeljak J,
Hitchman TD, Wang J, Wang L, Slotkin EK, Fan PD, Reis-Filho JS and
Ladanyi M: Generation of conditional oncogenic chromosomal
translocations using CRISPR-Cas9 genomic editing and
homology-directed repair. J Pathol. 242:102–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gondo Y, Nakamura K, Nakao K, Sasaoka T,
Ito K, Kimura M and Katsuki M: Gene replacement of the p53 gene
with the lacZ gene in mouse embryonic stem cells and mice by using
two steps of homologous recombination. Biochem Biophys Res Commun.
202:830–837. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Haapaniemi E, Botla S, Persson J,
Schmierer B and Taipale J: CRISPR-Cas9 genome editing induces a
p53-mediated DNA damage response. Nat Med. 24:927–930. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ihry RJ, Worringer KA, Salick MR, Frias E,
Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, et al: p53
inhibits CRISPR-Cas9 engineering in human pluripotent stem cells.
Nat Med. 24:939–946. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y,
Liu H, La Russa M, Xie M, Ding S and Qi LS: Small molecules enhance
CRISPR genome editing in pluripotent stem cells. Cell Stem Cell.
16:142–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Song J, Yang D, Xu J, Zhu T, Chen YE and
Zhang J: RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in
efficiency. Nat Commun. 7:105482016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li G, Zhang X, Zhong C, Mo J, Quan R, Yang
J, Liu D, Li Z, Yang H and Wu Z: Small molecules enhance
CRISPR/Cas9-mediated homology-directed genome editing in primary
cells. Sci Rep. 7:89432017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kawamura F, Inaki M, Katafuchi A, Abe Y,
Tsuyama N, Kurosu Y, Yanagi A, Higuchi M, Muto S, Yamaura T, et al:
Establishment of induced pluripotent stem cells from normal B cells
and inducing AID expression in their differentiation into
hematopoietic progenitor cells. Sci Rep. 7:16592017. View Article : Google Scholar : PubMed/NCBI
|