Introduction
Ovarian cancer (OC) affects one in every 70 females
and is one of the most severe gynecologic malignancies, with a
5-year survival rate of <50% if diagnosed at a late stage in the
USA between 2011 and 2015 (1). There
are no notable symptoms of early OC and no effective screening
tools are available (2). The
majority of patients with OC are generally asymptomatic until tumor
progression and metastasis occur, and >66% of tumors are already
in an advanced stage upon diagnosis (3). Once clinically diagnosed as high-grade
epithelial OC, treatment outcomes are poor, even following surgery
and appropriate chemotherapy (4).
Therefore, clarification of the key molecular events that are
associated with the development of OC may provide potential
molecular targets for the clinical treatment of advanced epithelial
OC.
Cluster of differentiation 44 (CD44), a leukocyte
differentiation antigenic determinant, is a multi-structural and
multi-functional cell surface adhesion molecule that is involved in
the homing, activation, movement, and cell-cell and cell-matrix
interactions of lymphocytes (5,6). This
molecule is widespread and has a number of subtypes, which can be
divided into standard CD44s and variant CD44s (CD44v1-v10), which
are involved in the regulation of numerous physiological and
pathological processes (7). The key
role of CD44 is to provide defense against inflammatory processes
by cellular transmigration and cell signaling. Saegusa et al
(8) demonstrated that CD44 can
inhibition the spread and metastasis of local tumors. Additionally,
a number of studies indicated that CD44 is a specific cell surface
marker of cancer stem cells in certain solid tumors, including OC
(9–14). Notably, studies suggested that
variant CD44s are associated with the occurrence and development of
various malignancies, including pancreatic (15), breast (16) and colorectal cancer (17). Among these variants, CD44v6 is the
most associated with tumor invasion and metastasis (18,19).
CD44v6 has been demonstrated to be a beneficial
prognostic factor of a variety of cancer types, including those of
the stomach (20), head and neck
(21), prostate (22) and lung (23). Additionally, investigations
identified that CD44v6 may serve a vital role in tumor progression
and pervasion (24–27). It has been reported that CD44v6 may
participate in the metastasis of OC by mediating OC cell adhesion
and migration (19). In OC, changes
occurring in the extracellular environment are crucial for
tumorigenesis and progression, as well as for intraperitoneal
transmission (28). The
extracellular matrix molecules versican and hyaluronic acid
interact with CD44 and serve a pivotal role in OC metastasis
(29). Certain studies demonstrated
that CD44v6 is only upregulated in tumor tissues during OC cell
adhesion and metastasis, indicating that CD44v6, as an adhesion
molecule, may participate in OC cell adhesion and metastasis
(30–32).
Nuclear factor-κB (NF-κB), a transcription factor,
is widely identified in mammalian cells and is a key factor in
tumor anti-apoptotic mechanisms (33). This molecule can regulate the cell
cycle and facilitate cell proliferation by activating the
transcription of cyclin D1 (34).
Additionally, NF-κB is involved in the regulation of tumorigenesis,
invasion and metastasis (35). NF-κB
inhibits apoptosis via the regulation of the transcription of
anti-apoptotic genes, including B-cell lymphoma (Bcl)-2, Bcl-extra
large and X-linked inhibited of apoptosis (36,37).
Furthermore, NF-κB can accelerate tumor invasion and metastasis by
regulating matrix metalloproteinase-9 and intercellular adhesion
molecule-1 expression, and by degrading the extracellular matrix
(38,39). Activated or abnormally expressed
NF-κB can be detected in numerous malignancy types, including
leukemia, breast cancer, liver cancer and thyroid cancer (40). Sasaki et al (41) investigated gastric cancer tissues and
identified that NF-κB expression is associated with tumor size,
lymph node invasion, invasion depth and peritoneal metastasis.
Activated NF-κB promotes the transcription of apoptotic genes,
resulting in the proliferation of tumor cells. NF-κB has been
demonstrated to be a pro-survival and pro-inflammatory
transcription factor that can promote cancer development (42). Therefore, the specific inhibition of
NF-κB is considered a potential therapeutic target. Studies
reported that the use of NF-κB inhibitors, including Eriocalyxin B,
may prevent the recurrence of OC (43–45).
The present study hypothesized that CD44v6 may
affect the proliferation, migration and invasion of OC cells via
the NF-κB pathway. To the best of our knowledge, no previous
studies investigated the association between these two molecules in
the molecular mechanism of OC development. Therefore, the present
study detected CD44v6 and NF-κB expression in human ovarian
tissues. CD44v6 gene knockout was performed in OC cells to detect
the expression levels of CD44v6 and NF-κB, and examine the
associations between the expression of CD44v6, and cellular
proliferation, migration and invasion. The nuclear translocation of
NF-κB was further detected by immunofluorescence to investigate
molecules that may cause the development of OC.
Materials and methods
Tissue specimens
A total of 46 fresh surgical specimens, including 24
malignant and 22 benign ovarian neoplasms, were collected between
June 2016 and February 2017. Patients with multiple tumors were
excluded. All patients were female, the age range of the patients
was 30–60 years and the mean age was 50±2.34 years. The Medical
Ethics Committee of Medical College of Wuhan University (Wuhan,
China) approved the present study and written informed consent was
obtained from all participants. All tissue specimens were from
primary tumors and were histologically verified by pathologists who
were blind to the study at the Department of Pathology, Renmin
Hospital of Wuhan University (Wuhan, China) to confirm the
diagnosis, histological type and tumor grade (46).
Cells and cell culture
The human serous OC cell line A2780, obtained from
the American Type Culture Collection (Manassas, VA, USA), was
cultured in RPMI-1640 medium (catalog no. SH30022; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (FBS; catalog no. 141215; Hangzhou HuaAn
Biotechnology Co., Ltd., Hangzhou, China), and 100 U/ml penicillin
and streptomycin in a 5% CO2 atmosphere at 37°C.
Small interfering RNA (siRNA)
knockdown of CD44v6
A2780 cells were transiently transfected with siRNA
obtained from Shanghai Integrated Biotech Solutions Co., Ltd.
(Shanghai, China). This included siRNA against CD44v6
(5′-GCCAACATTCATTCAATAC-3′) and negative control siRNA
(5′-GCCTTACTTACAACAATAC-3′). siRNA (100 nM) was transfected using
Lipofectamine® 2000 (catalog no. 11668019; Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. Following transfection, the cells were
incubated at 37°C in a CO2 incubator for 24 or 48 h
prior to being harvested for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). Stably transfected cells were
maintained from wells with only one green fluorescent clone.
Immunohistochemistry
The obtained ovarian tissue was fixed in 4%
paraformaldehyde for 10 h at room temperature. The tissue sections
were then cut to a thickness of 0.4 cm. Paraffin slices were placed
in an oven at 65°C on a baking sheet for 2 h, dewaxed in water and
washed three times with PBS for 5 min each time. The slices were
placed in EDTA buffer (catalog no. AS1016; Wuhan Boster Biological
Technology, Ltd., Wuhan, China) for microwave antigen retrieval at
medium power until boiling, then at low power until boiling at 10
min intervals. Slides were washed with PBS three times following
cooling for 5 min each time. Slides were placed in 3% hydrogen
peroxide solution and then incubated for 10 min at room temperature
in the dark. Slides were washed with PBS three times for 5 min each
time, dried and blocked with 5% bovine serum albumin (BSA; Roche
Diagnostics, Basel, Switzerland; catalog no. 10735078001) for 20
min at room temperature. The BSA solution was removed and ~50 µl
diluted primary antibodies against CD44v6 (catalog no. 8242; 1:50;
CST Biological Reagents Co., Ltd., Shanghai, China) and NF-κB
(catalog no. ab78960; 1:200; Abcam, Cambridge, UK) were added to
each section overnight at 4°C. The slides were washed three times
with PBS for 5 min each time. Following the removal of PBS
solution, 100 µl horseradish peroxidase-labeled goat anti-rabbit
IgG secondary antibody (catalog no. AS-1107; 1:200; Aspen
Biotechnology Co., Ltd. Wuhan, China) was added to each section and
the sections were incubated at 37°C for 50 min. The slides were
washed with PBS three times for 5 min each time. The PBS solution
was then removed, 3,3′-diaminobenzidine (50–100 µl) was added to
each slice and color development was monitored using a light
microscope (magnification, ×400). Following color development, the
slide was washed with water and counterstained with hematoxylin for
2 min at room temperature. Hydrochloric acid alcohol (1%) was used
for differentiation for ~1 sec and the slide was then washed with
water. The slide was dehydrated using an ethanol gradient (75, 90
and 100% each for 10 min), cleared in xylene and sealed with a
neutral gum seal.
RNA extraction and complementary DNA
(cDNA) synthesis
For the transfected/control cells, the medium was
carefully removed from the cell culture dish with a pipette and 1
ml pre-cooled PBS solution at 4°C was added. The residual medium
was gently removed, PBS solution was added by pipette and 1 ml
TRIzol® (catalog no. 15596-026; Invitrogen; Thermo
Fisher Scientific, Inc.) was added. The solution was repeatedly
pipetted around the bottom of the flask to ensure the cells were
fully covered by TRIzol. Chloroform (250 µl; catalog no. 10006818;
Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) was added
and mixed well, and then the solution was placed on ice for 10 min.
The mixture was centrifuged at 4°C and 10,000 × g for 10 min. The
supernatant (400 µl) was carefully transferred to a 1.5-ml
Eppendorf tube and an equal volume of 4°C pre-cooled isopropanol
(catalog no. 80109218; Sinopharm Chemical Reagent Co., Ltd.) was
added. The solution was inverted to mix and then incubated at −20°C
for 15 min. The solution was centrifuged at 4°C and 10,000 × g for
10 min. The liquid was carefully removed and 1 ml 75% ethanol,
pre-cooled at 4°C, was added. The RNA was precipitated three times,
and the RNA pellet was washed and centrifuged at 4°C and 10,000 × g
for 5 min. The RNA was dried for 10 min, and then the ethanol was
volatilized thoroughly, and 10 µl RNase-free water was added to
fully dissolve the RNA.
For human tissue samples, ~100 mg of tissue was
placed on ice using a sterile tool and thoroughly ground in 1 ml of
pre-cooled TRIzol. The homogenate was carefully poured into a
1.5-ml Eppendorf tube and chloroform (250 µl) was added. The
mixture was mixed well and incubated on ice for 5 min. The
aforementioned protocol was then performed for the remaining steps
of RNA extraction.
cDNA synthesis was performed using a PrimeScript™ RT
reagent kit with gDNA Eraser (catalog no. RR047A; Takara Bio, Inc.,
Otsu, Japan), and the following reaction solution was placed on
ice: 2.0 µl 5X gDNA Eraser buffer, 1.0 µl gDNA Eraser and 10.0 µl
RNA, all from the PrimeScript RT reagent kit. The mixture was then
placed at 42°C for 2 min and incubated at 4°C for 5 min. The
following reaction solution was added to the Eppendorf tube
containing RNA and placed on ice: 1.0 µl PrimeScript RT Enzyme mix
I, 1.0 µl RT Primer mix, 4.0 µl 5X PrimeScript Buffer 2 and 4.0 µl
RNase-free dH2O, all from the kit. The reaction solution
was then placed in a PCR machine at 37°C for 15 min, 85°C for 5 sec
and temporally kept at 4°C for 10 min. The reaction fixture was
finally stored at −20°C.
RT-qPCR
PCR was performed using a Step One™ Real-Time PCR
system (Thermo Fisher Scientific, Inc.) with the following
conditions: 2 min at 50°C, followed by 40 cycles at 95°C for 10
min, 95°C for 15 sec and 60°C for 1 min. Each 10-µl reaction
contained 5.0 µl SYBR® Premix Ex Taq (Takara Bio, Inc.),
0.2 µl forward primer (final concentration, 0.2 µM), 0.2 µl reverse
primer (final concentration, 0.2 µM), ROX reference dye, 1.0 µl
cDNA and 3.4 µl ddH2O. The threshold cycle value
(Cq) was recorded for all samples for both the target
gene and the reference gene GAPDH. Melt curve analysis was
performed for each run. The relative gene expression of the target
gene was calculated as ΔCq and was determined by
subtracting the Cq of the reference gene from the
Cq of the target gene. Differential expression of the
target gene in the control, negative control and CD44v6 siRNA
groups was calculated as ∆∆Cq (47), as determined by subtracting the
ΔCq of the CD44v6 siRNA group from the ΔCq of
the matched control group. Primer sequences are presented in
Table I.
 | Table I.Primers used for polymerase chain
reaction. |
Table I.
Primers used for polymerase chain
reaction.
| Primer | Sequence | Amplification
length, bp |
|---|
| GAPDH | Forward,
5′-GGTCGGAGTCAACGGATTTG-3′ | 218 |
|
| Reverse,
5′-GGAAGATGGTGATGGGATTTC-3′ |
|
| CD44v6 | Forward,
5′-GCCTTTGATGGACCAATTACC-3′ | 266 |
|
| Reverse,
5′-TCATTCCTATTGGTAGCAGGGA-3′ |
|
| NF-κB | Forward,
5′-CGCATCCAGACCAACAACA-3′ | 208 |
|
| Reverse,
5′-TGCCAGAGTTTCGGTTCAC-3′ |
|
Colony formation assay
Monolayer-cultured A2780 cells in the logarithmic
growth phase were digested with 0.25% trypsin and the suspension
was pipetted until the cells were fully dispersed. Subsequently,
the cells were suspended in RPMI-1640 medium with 10% FBS. The
cells were then plated at a density of 300 cells/plate in 35-mm
plates, which were then incubated at 37°C in an atmosphere
containing 5% CO2 for ~14 days, with fresh RPMI-1640
medium with 10% FBS provided every 3 days. When macroscopic
colonies appeared in the medium, the culture was terminated.
Subsequently, 4% formaldehyde was added for 20 min at room
temperature to fix the cells. Following two washes with PBS (pH
7.4), the cells were stained with Wright-Giemsa dye complex for 5
min at room temperature. Colonies were counted using an inverted
light microscope (magnification, ×400). Colonies with >50 cells
were counted. The cloning rate was calculated as follows: Clone
formation rate=cloning/inoculation cells ×100%.
Cell invasion and migration
assays
Matrigel was diluted (1:5) and coated on the upper
surface of a polycarbonate membrane (12-mm diameter and 8-mm pore
size) in a Transwell filter (EMD Millipore, Billerica, MA, USA).
The transfected A2780 cells were cultured to logarithmic growth
phase. The cells were digested, washed with PBS and serum-free
RPMI-1640 medium, and then suspended in serum-free RPMI-1640
medium. Each group of cells (4×105) was added to the
upper chamber and 500 µl RPMI-1640 containing 10% FBS was added to
the lower chamber. Following incubation in a 5% CO2
incubator at 37°C for 16 h, the cells were fixed with 4%
paraformaldehyde at room temperature for 20 min and stained with
crystal violet at room temperature for 30 min to determine how many
cells had invaded the lower surface of the filter. The invaded
cells were counted by light microscope (magnification, ×400) and
the mean number of cells in at least three fields per well was
calculated. The experiment was repeated three times. The migration
assay was similar, except that Matrigel was not added to the upper
membrane.
Immunofluorescence
Firstly, the transfected A2780 cells were cultured
on glass slides coated with poly-L-lysine and washed on the slides
in PBS three times. The cells were then fixed in 4%
paraformaldehyde for 30 min at room temperature and washed two
times with PBS. The cells were then permeabilized with 0.5% Triton
X-100 and 0.25% Tween 20 in TBS for 15 min, and incubated with
Image iT FX Signal Enhancer (Invitrogen; Thermo Fisher Scientific,
Inc.) for 30 min at room temperature in a humid chamber. Following
blocking with 5% BSA for 20 min at room temperature, the cells were
washed. Primary antibodies against CD44v6 (catalog no. 8242; 1:500;
CST Biological Reagents Co., Ltd.) and NF-κB (catalog no. ab78960;
1:1,000; Abcam) were added and the cells were incubated overnight
at 4°C. Subsequently, Alexa Fluor-conjugated secondary antibodies
(CY3 labeled goat anti-rabbit, catalog no. AS-1109; 1:50; Aspen
Biotechnology Co., Ltd.) were added and the cells were incubated
for 1 h at room temperature prior to being washed with PBS and 1%
Tween 20. The cells were then incubated with DAPI in the dark for 5
min at room temperature. Excess DAPI was rinsed off with PBS and 1%
Tween 20, and the slides were blotted dry with water-absorbent
paper. The slides were then sealed with anti-fluorescence quencher
and images were obtained using a fluorescent microscope
(magnification, ×400).
Statistical analysis
SPSS 20.0 software (IBM Corp., Armonk, NY, USA) was
used to analyze the data. Data are presented as the mean ± standard
deviation and are representative of three individual experiments.
An unpaired Student's t-test was used to compare two groups and
three or more groups were analyzed by one-way analysis of variance
(ANOVA) followed by a post hoc Bonferroni test. mRNA expression
levels of CD44v6 and NF-κB in the tissue samples were compared
using an unpaired Student's t-test. CD44v6 and NF-κB mRNA
expression levels in different cells were analyzed by ANOVA
followed by a post hoc Bonferroni test. The number of migrating and
invading cells, and the number of colonies were compared by one-way
ANOVA followed by a post-hoc Bonferroni correction test. P<0.05
was considered to indicate a statistically significant
difference.
Results
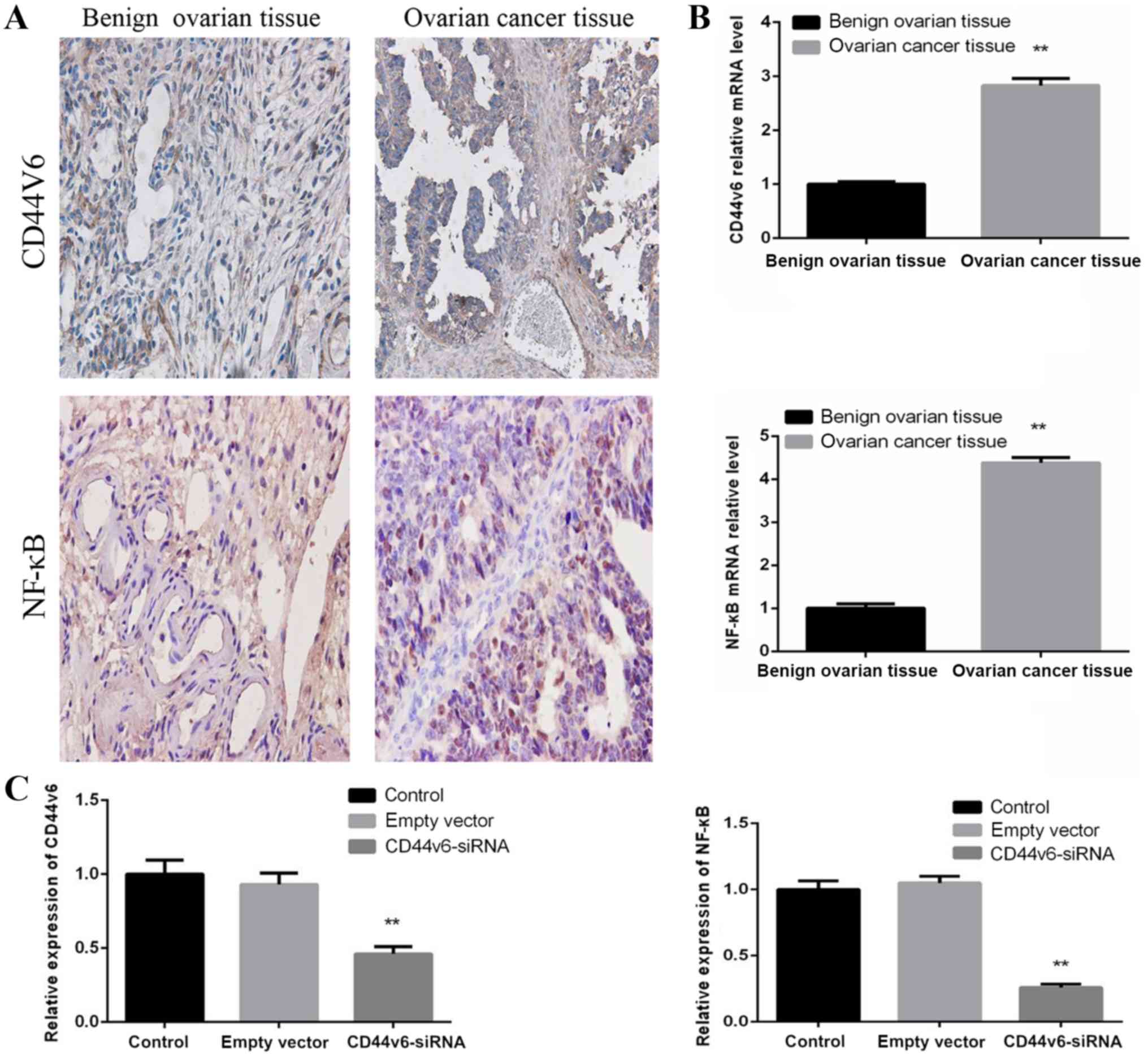
Expression of CD44v6 and NF-κB in OC
and normal ovarian tissue samples
To determine whether CD44v6 and NF-κB are abnormally
expressed in OC tissues, OC tissue samples and benign ovarian
tissue samples were collected. Immunohistochemical analysis
supported the observation that OC tissues exhibit increased levels
of CD44v6 and NF-κB proteins, compared with normal ovarian tissue
(Fig. 1A). The CD44v6 and NF-κB mRNA
levels were detected by RT-qPCR, which revealed that CD44v6 and
NF-κB mRNA levels in OC tissues were significantly increased,
compared with normal ovarian tissues (P<0.01; Fig. 1B). These data demonstrated that the
expression of CD44v6 and NF-κB is upregulated in OC tissue,
compared with normal ovarian tissue.
CD44v6-siRNA decreases NF-κB mRNA
expression in OC cells
To further investigate whether NF-κB is affected by
CD44v6 expression in OC cells, the A2780 OC cell line was used to
generate a normal control group, an empty vector control group and
a CD44v6-siRNA transfection group. The mRNA levels of CD44v6 and
NF-κB were then detected in each group. The results demonstrated
that there was no significant difference in the mRNA levels of
CD44v6 and NF-κB between the normal control group and the empty
vector control group. However, compared with the normal control
group, the siRNA-transfected group exhibited significantly reduced
expression levels of CD44v6 and NF-κB (P<0.01; Fig. 1C). This observation indicates that
NF-κB mRNA expression levels are affected by CD44v6 expression.
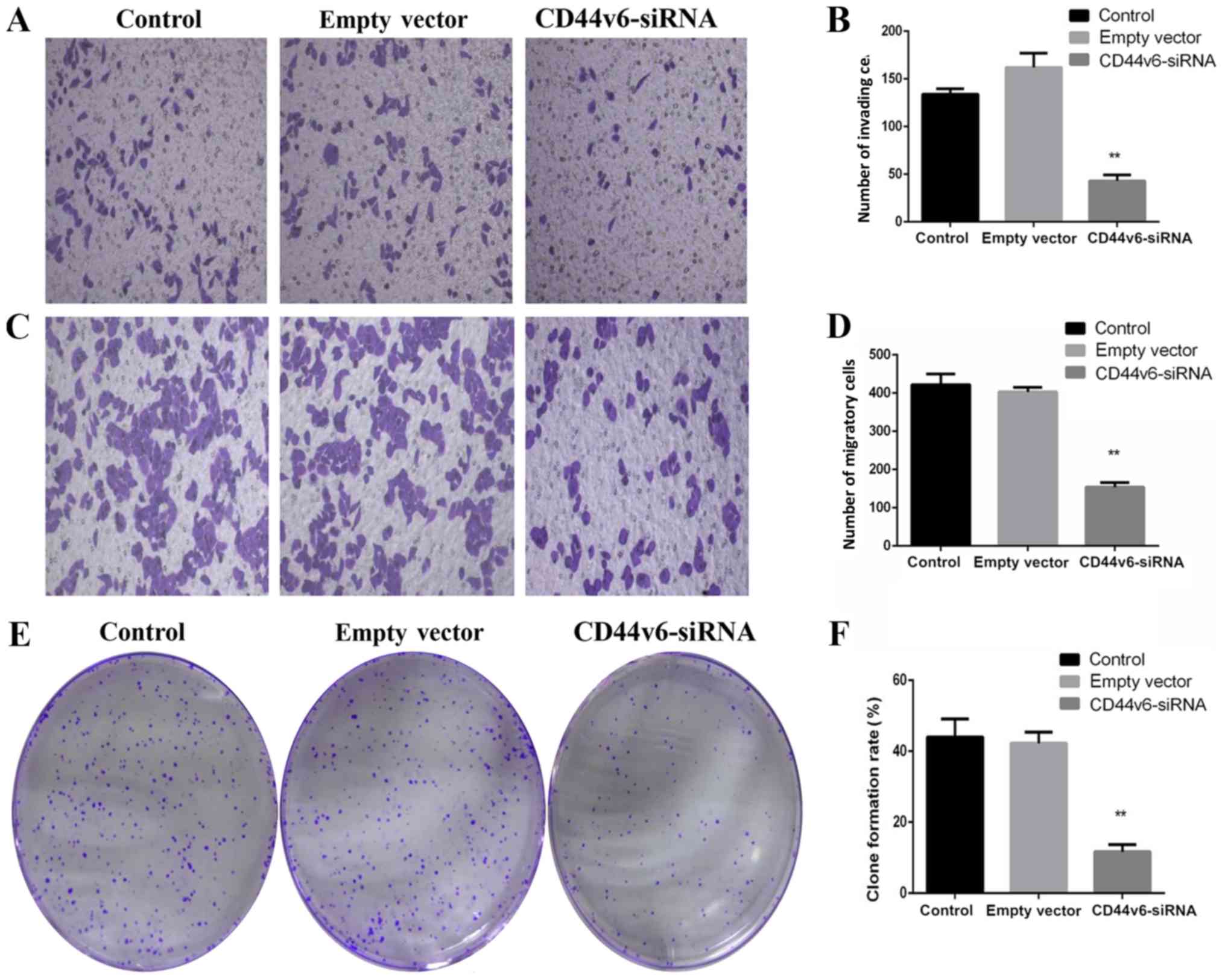
CD44v6-siRNA impairs the
proliferation, migration and invasion of OC cells
To determine whether knockdown of CD44v6 affects OC
cell proliferation, migration and invasion, OC cell Transwell and
colony formation experiments were performed. The results
demonstrated the effect of silencing CD44v6 on the proliferation
and invasion of A2780 cells. As presented in Fig. 2, in vitro Transwell invasion
assays revealed that following the silencing of CD44v6, the number
of invading cells per field in the CD44v6 siRNA group (43.25±3.02)
was significantly reduced, compared with that in the control group
(134.39±9.69; P<0.01; Fig. 2A and
B). The transwell migration assays demonstrated that the number
of migratory cells per field in the CD44v6 siRNA group
(154.45±21.75) markedly decreased compared with the control group
(421.67±48.95; P<0.01; Fig. 2C and
D). Similarly, the colony formation ability in the CD44v6-siRNA
group was significantly reduced, compared with the control group
(P<0.01; Fig. 2E and F).
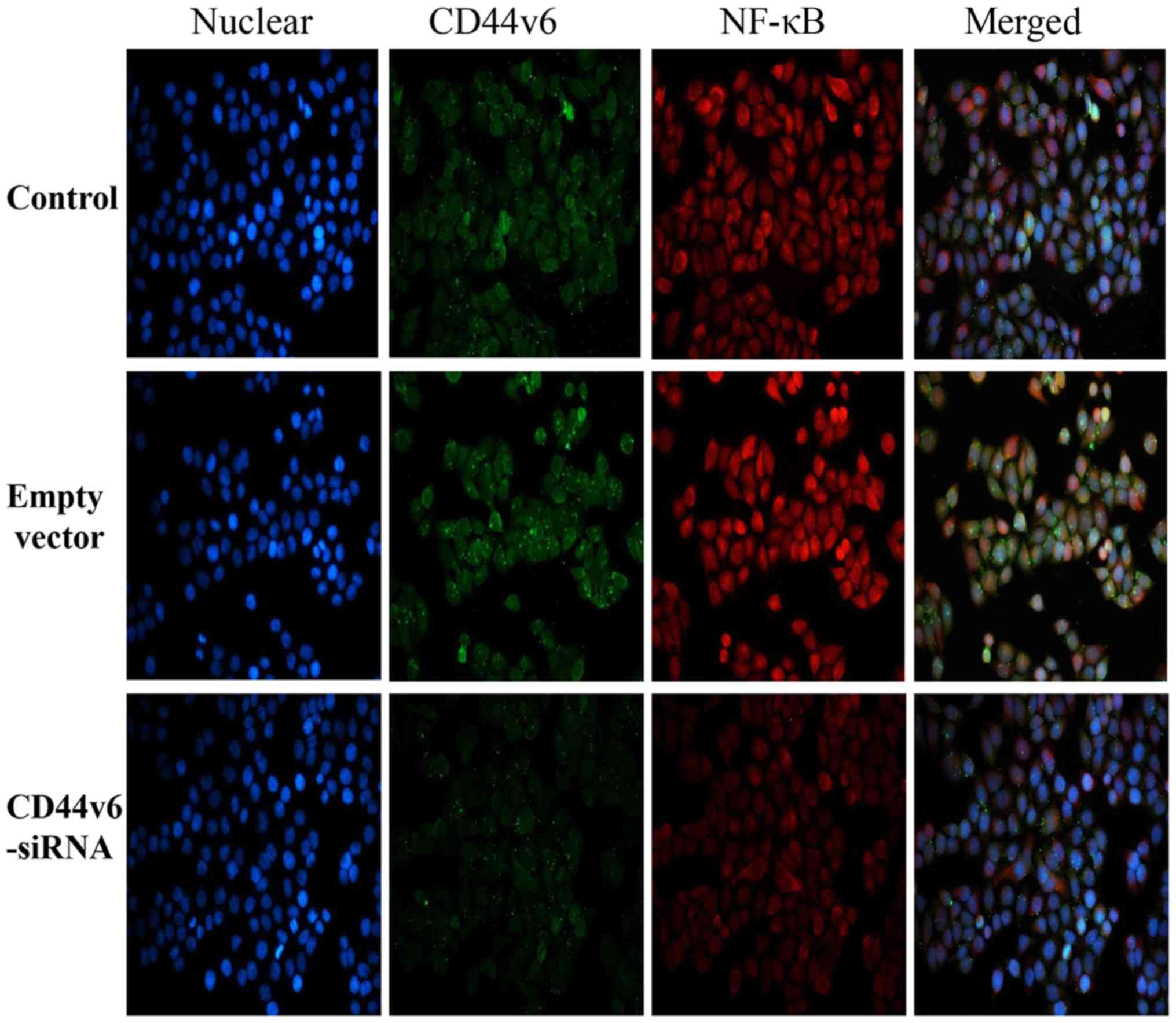
CD44v6-siRNA impairs the NF-κB pathway
in OC cells
The aforementioned PCR results demonstrated that
NF-κB mRNA expression was significantly reduced following knockdown
of CD44v6. To investigate whether silencing CD44v6 affects the
expression of NF-κB and its nuclear translocation, the expression
of NF-κB in each group was detected by immunofluorescence. As
demonstrated in Fig. 3, the
fluorescence intensity of NF-κB in the CD44v6-siRNA group was
markedly reduced, compared with the control group, which indicates
that the NF-κB pathway may affect CD44v6 expression.
Discussion
The present study investigated the effects of NF-κB
pathway inhibition by CD44v6 knockdown in OC cells. It was
identified that CD44v6-siRNA OC cells exhibit decreased invasion
ability and disrupted NF-κB activation.
In addition to the present study, previous studies
also examined the expression of CD44v6 in the ovary to investigate
the association between CD44v6 and the development of OC (27,48,49). The
results from the present study are in agreement with a study by
Tjhay et al (49), which
demonstrated that CD44v6-positive OC cells possess an increased
capacity for migration and invasion. Additionally, Zhang et
al (31) reported that the
expression of CD44v6 mRNA was significantly elevated in clinical
samples of benign ovarian tumor and OC, and no significant
differences were revealed in CD44v6 expression between normal
ovarian tissue and benign ovarian tumor at the protein level.
However, the authors identified that expression of CD44v6 in OC was
significantly increased, compared with that in normal ovaries and
benign ovarian tumors, which is in agreement with the present
results.
Using western blot analysis, Tjhay et al
(49) compared the expression of
epithelial-mesenchymal transition (EMT) regulatory proteins,
including E-cadherin, N-cadherin, fibronectin and vimentin, in
fluorescence-activated cell sorting (FACS)-sorted CD44v6-positive
cells with that in FACS-sorted CD44v6-negative cells. Compared with
FACS-sorted CD44v6-negative cells, E-cadherin expression was
downregulated in FACS-sorted CD44v6-positive cells, and N-cadherin,
fibronectin, and vimentin expression was increased in
CD44v6-positive cells. In contrast to immortalized normal cells,
prostate cancer cells with CD44v6 suppression demonstrated
downregulated EMT markers and reduced tumorigenic potential
(50). These observations indicate
that a subpopulation of CD44v6-positive cells may regulate the
metastatic ability of OC cells by influencing EMT. Additionally, a
previous study demonstrated that CD44v6-positive OC cells serve a
pivotal role in disseminated metastatic tumors of the pelvic
peritoneum and exhibit potential as metastatic initiators in OC
mouse models (51). CD44v6-positive
cancer cells possess a significant effect on the survival of
patients with OC (52). CD44v6 has
been demonstrated to promote OC metastasis by mediating ovarian
tumor cell invasion into the peritoneum (49). Furthermore, siRNA knockdown of CD44v6
expression has been demonstrated to decrease the ability of SKOV3
cells to adhere and migrate, indicating that CD44v6 may be involved
in mediating tumor cell adhesion and migration during metastasis
(53), an observation that is
consistent with the present results. Overall, this indicates that
CD44v6 expression is associated with the progression, metastasis
and recurrence of epithelial OC.
NF-κB generally exists as a dimer, most commonly
with the p50 and p65 subunits. In a non-stimulated state, NF-κB
binds to inhibitor of κB (IκB) in a non-activated form in the
cytoplasm. When cells receive an external stimulus, IκB is
phosphorylated and degraded, allowing NF-κB to translocate to the
nucleus, resulting in NF-κB activation. The nuclear localization
signal on NF-κB is then exposed in response to various cytokines,
growth factors, apoptosis-associated factors and other target genes
(54). Combined with the basic
sequence, this results in increased nuclear transcription, which is
involved in a number of human pathophysiological processes,
including the inflammatory and immune responses (55,56).
Numerous studies demonstrated that NF-κB regulates apoptosis
through the following pathways: i) Direct regulation of apoptotic
genes; ii) regulation of the S phase of the cell cycle, which
interferes with the cellular response to apoptotic signals; and
iii) interaction with certain cellular apoptosis proteins (54,56,57).
NF-κB controls a number of features of cancer cells by modulating
the transcriptional activation of genes involved in cell
proliferation, angiogenesis, metastasis and apoptosis arrest
(57).
NF-κB is one of the most significant molecules
involved in inflammation and innate immunity and has been
demonstrated to be an important endogenous tumor promoter (58). NF-κB serves a supervisory role in
regulating transformation and inflammation in the context of cancer
and inflammatory cells (59). NF-κB
activation in cancer cells is responsible for the progression of
inflammation-associated cancer; conversely, inhibiting NF-κB
activation can inhibit tumor growth (55). The present study observed that NF-κB
mRNA levels were reduced when CD44v6 was knocked down. Furthermore,
immunofluorescence demonstrated that NF-κB expression was also
inhibited. These results indicated that CD44v6 may participate in
the proliferation and metastasis of OC via activation of the NF-κB
pathway.
This hypothesis is supported by a number of other
studies. Kawana et al (60)
first demonstrated a direct association between CD44 and Toll-like
receptors (TLRs) by detecting the activation of NF-κB, which is the
principal signal transducer of TLR signaling in CD44-deficient bone
marrow macrophages. The results indicated that the cytoplasmic
domain of CD44 serves a regulatory role in TLR signaling and
results in NF-κB activation by zymosan or lipopolysaccharide.
Bourguignon et al (61)
revealed that the downregulation of CD44 expression by treatment
with CD44 siRNA not only effectively blocks CD44s association with
TLR2, TLR4 and myeloid differentiation primary response 88 (MyD88)
in MDA-MB-231 cells treated with low-molecular-weight hyaluronan
(LMW-HA), but also significantly inhibits LMW-HA-mediated signaling
and function, including NF-κB-mediated transcriptional activation,
interleukin (IL)-1β/IL-8 gene expression and protein production.
These results strongly indicate that CD44 and TLR2/4-associated
MyD88 serve important roles in regulating NF-κB p65-specific
transcriptional activation, in addition to cytokine/chemokine gene
expression and protein production in MDA-MB-231 cells in response
to LMW-HA treatment. Therefore, it is plausible that
LMW-HA-mediated NF-κB p65 signaling and cytokine/chemokine
production are functionally coupled in a CD44-dependent manner in
breast tumor cells.
Therefore, it is reasonable to suggest that the
mechanism by which CD44v6 causes tumor cell proliferation and
metastasis is associated with its ability to induce NF-κB
activation. This characteristic may serve an important role in the
potential use of chemotherapeutic drugs that target this critical
molecular mechanism for the treatment of OC.
In conclusion, the biological and molecular
heterogeneity of OC is a highly promising research area, and
studies may provide novel insights into the diagnosis and treatment
of OC as well as breakthroughs in the treatment and prognosis of
advanced ovarian epithelial cancer. CD44v6 may promote the
proliferation, invasion and migration of OC cells by activating the
NF-κB pathway. CD44v6 in cancer cells may represent a potential
molecular therapeutic target to prevent the initiation of
proliferation and metastasis of OC cells. However, which specific
molecular signaling pathways downstream of CD44v6 affect the NF-κB
pathway and which upstream molecules regulate the expression of
CD44v6 requires further research. Observations from future studies
may provide more effective targets for the treatment of OC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Department of Hubei Province Science and Technology
Support Program Project (grant no. 2015BCA313), the Independent
Research Project of Wuhan University (grant no. 413000117) and the
Special Fund for Clinical Medicine Research of Chinese Medical
Association (grant no. 17020310700).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
YC and XY conceived the study. YW designed the
experiments and performed the majority of them. XY contributed to
the acquisition of data, data analysis and interpretation of the
data. LZ contributed to the cell experiments. YW and SX performed
cell culture and collected clinical samples. YW wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The Medical Ethics Committee of Medical College of
Wuhan University (Wuhan, China) approved this study and written
informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Towne SD Jr: Socioeconomic, Geospatial,
and Geopolitical disparities in access to health care in the US
2011-2015. Int J Environ Res Public Health. 14:5732017. View Article : Google Scholar
|
|
2
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yap TA, Carden CP and Kaye SB: Beyond
chemotherapy: Targeted therapies in ovarian cancer. Nat Rev Cancer.
9:167–181. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kujawa KA and Lisowska KM: Ovarian
cancer-from biology to clinic. Postepy Hig Med Dosw (Online).
69:1275–1290. 2015.(In Polish). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Screaton GR, Bell MV, Bell JI and Jackson
DG: The identification of a new alternative exon with highly
restricted tissue expression in transcripts encoding the mouse
Pgp-1 (CD44) homing receptor. Comparison of all 10 variable exons
between mouse, human, and rat. J Biol Chem. 268:12235–12238.
1993.PubMed/NCBI
|
|
6
|
Jackson DG, Buckley J and Bell JI:
Multiple variants of the human lymphocyte homing receptor CD44
generated by insertions at a single site in the extracellular
domain. J Biol Chem. 267:4732–4739. 1992.PubMed/NCBI
|
|
7
|
Prochazka L, Tesarik R and Turanek J:
Regulation of alternative splicing of CD44 in cancer. Cell Signal.
26:2234–2239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saegusa M, Machida D, Hashimura M and
Okayasu I: CD44 expression in benign, premalignant, and malignant
ovarian neoplasms: Relation to tumour development and progression.
J Pathol. 189:326–337. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang S, Balch C, Chan MW, Lai HC, Matei
D, Schilder JM, Yan PS, Huang TH and Nephew KP: Identification and
characterization of ovarian cancer-initiating cells from primary
human tumors. Cancer Res. 68:4311–4320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yae T, Tsuchihashi K, Ishimoto T, Motohara
T, Yoshikawa M, Yoshida GJ, Wada T, Masuko T, Mogushi K, Tanaka H,
et al: Alternative splicing of CD44 mRNA by ESRP1 enhances lung
colonization of metastatic cancer cell. Nat Commun. 3:8832012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zoller M: CD44: Can a cancer-initiating
cell profit from an abundantly expressed molecule? Nat Rev Cancer.
11:254–267. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dallas MR, Liu G, Chen WC, Thomas SN,
Wirtz D, Huso DL and Konstantopoulos K: Divergent roles of CD44 and
carcinoembryonic antigen in colon cancer metastasis. FASEB J.
26:2648–2656. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishimoto T, Nagano O, Yae T, Tamada M,
Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, et al:
CD44 variant regulates redox status in cancer cells by stabilizing
the xCT subunit of system xc(−) and thereby promotes tumor growth.
Cancer Cell. 19:387–400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagano O, Okazaki S and Saya H: Redox
regulation in stem-like cancer cells by CD44 variant isoforms.
Oncogene. 32:5191–5198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li XP, Zhang XW, Zheng LZ and Guo WJ:
Expression of CD44 in pancreatic cancer and its significance. Int J
Clin Exp Pathol. 8:6724–6731. 2015.PubMed/NCBI
|
|
16
|
Ouhtit A, Rizeq B, Saleh HA, Rahman MM and
Zayed H: Novel CD44-downstream signaling pathways mediating breast
tumor invasion. Int J Biol Sci. 14:1782–1790. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abbasian M, Mousavi E, Arab-Bafrani Z and
Sahebkar A: The most reliable surface marker for the identification
of colorectal cancer stem-like cells: A systematic review and
meta-analysis. J Cell Physiol. 234:8192–8202. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gunthert U, Hofmann M, Rudy W, Reber S,
Zöller M, Haussmann I, Matzku S, Wenzel A, Ponta H and Herrlich P:
A new variant of glycoprotein CD44 confers metastatic potential to
rat carcinoma cells. Cell. 65:13–24. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liotta LA, Delisi C, Saidel G and
Kleinerman J: Micrometastases formation: A probabilistic model.
Cancer Lett. 3:203–208. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okayama H, Kumamoto K, Saitou K, Hayase S,
Kofunato Y, Sato Y, Miyamoto K, Nakamura I, Ohki S, Sekikawa K and
Takenoshita S: CD44v6, MMP-7 and nuclear Cdx2 are significant
biomarkers for prediction of lymph node metastasis in primary
gastric cancer. Oncol Rep. 22:745–755. 2009.PubMed/NCBI
|
|
21
|
Kawano T, Nakamura Y, Yanoma S, Kubota A,
Furukawa M, Miyagi Y and Tsukuda M: Expression of E-cadherin, and
CD44s and CD44v6 and its association with prognosis in head and
neck cancer. Auris Nasus Larynx. 31:35–41. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gu H, Shang P and Zhou C: Expression of
CD44v6 and E-cadherin in prostate carcinoma and metastasis of
prostate carcinoma. Zhonghua Nan Ke Xue. 1032–34. (38)2004.(In
Chinese). PubMed/NCBI
|
|
23
|
Afify AM, Tate S, Durbin-Johnson B, Rocke
DM and Konia T: Expression of CD44s and CD44v6 in lung cancer and
their correlation with prognostic factors. Int J Biol Markers.
26:50–57. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stickeler E, Vogl FD, Denkinger T, Mobus
VJ, Kreienberg R and Runnebaum IB: Soluble CD44 splice variants and
pelvic lymph node metastasis in ovarian cancer patients. Int J Mol
Med. 6:595–601. 2000.PubMed/NCBI
|
|
25
|
Sun Y, Shen Z and Ji X: Study on the
relationship between CD44v6, p53 gene mutation and ovarian
carcinoma metastasis. Zhonghua Fu Chan Ke Za Zhi. 35:225–228.
2000.(In Chinese). PubMed/NCBI
|
|
26
|
Bar JK, Grelewski P, Popiela A, Noga L and
Rabczynski J: Type IV collagen and CD44v6 expression in benign,
malignant primary and metastatic ovarian tumors: Correlation with
Ki-67 and p53 immunoreactivity. Gynecol Oncol. 95:23–31. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hong SC, Song JY, Lee JK, Lee NW, Kim SH,
Yeom BW and Lee KW: Significance of CD44v6 expression in
gynecologic malignancies. J Obstet Gynaecol Res. 32:379–386. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liotta LA, Delisi C, Saidel G and
Kleinerman J: Micrometastases formation: A probabilistic model.
Cancer Lett. 3:203–208. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ricciardelli C, Lokman NA, Ween MP and
Oehler MK: Women in Cancer Thematic Review: Ovarian
cancer-peritoneal cell interactions promote extracellular matrix
processing. Endocr Relat Cancer. 23:T155–T168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ween MP, Oehler MK and Ricciardelli C:
Role of versican, hyaluronan and CD44 in ovarian cancer metastasis.
Int J Mol Sci. 12:1009–1029. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang HF, Hu P and Fang SQ: Understanding
the role of CD44V6 in ovarian cancer. Oncol Lett. 14:1989–1992.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuhn S, Koch M, Nubel T, Ladwein M,
Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L,
Franke WW, et al: A complex of EpCAM, claudin-7, CD44 variant
isoforms, and tetraspanins promotes colorectal cancer progression.
Mol Cancer Res. 5:553–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Afify A, Purnell P and Nguyen L: Role of
CD44s and CD44v6 on human breast cancer cell adhesion, migration,
and invasion. Exp Mol Pathol. 86:95–100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sokolova O and Naumann M: NF-kB signaling
in gastric cancer. Toxins (Basel). 9(pii): E1192017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Olivier S, Robe P and Bours V: Can
NF-kappaB be a target for novel and efficient anti-cancer agents?
Biochem Pharmacol. 72:1054–1068. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie DH, Tang XD, Xia SJ, Tan JM, Wang XH
and Cai Y: Expression of NF-kappa B in human bladder cancer and its
clinical significance. Ai Zheng. 21:663–667. 2002.(In Chinese).
PubMed/NCBI
|
|
37
|
Pommier Y, Sordet O, Antony S, Hayward RL
and Kohn KW: Apoptosis defects and chemotherapy resistance:
Molecular interaction maps and networks. Oncogene. 23:2934–2949.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Takada Y, Kobayashi Y and Aggarwal BB:
Evodiamine abolishes constitutive and inducible NF-kappaB
activation by inhibiting IkappaBalpha kinase activation, thereby
suppressing NF-kappaB-regulated antiapoptotic and metastatic gene
expression, up-regulating apoptosis, and inhibiting invasion. J
Biol Chem. 280:17203–17212. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Felx M, Guyot MC, Isler M, Turcotte RE,
Doyon J, Khatib AM, Leclerc S, Moreau A and Moldovan F:
Endothelin-1 (ET-1) promotes MMP-2 and MMP-9 induction involving
the transcription factor NF-kappaB in human osteosarcoma. Clin Sci
(Lond). 110:645–654. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen Y, Li R, Wang R and Liu Z: The
significance of nuclear factor kappa Bp65 (NF kappa Bp65)
expression on the vascular endothelial cells of rectum
adenocarcinoma of human. Hua Xi Yi Ke Da Xue Xue Bao. 32:196–199.
2001.(In Chinese). PubMed/NCBI
|
|
41
|
Sasaki N, Morisaki T, Hashizume K, Yao T,
Tsuneyoshi M, Noshiro H, Nakamura K, Yamanaka T, Uchiyama A, Tanaka
M and Katano M: Nuclear factor-kappaB p65 (RelA) transcription
factor is constitutively activated in human gastric carcinoma
tissue. Clin Cancer Res. 7:4136–4142. 2001.PubMed/NCBI
|
|
42
|
Hodge JC, Bub J, Kaul S, Kajdacsy-Balla A
and Lindholm PF: Requirememt of Rho A activity for increased
nuclear factor kappa B activity and PC23 human prostate cancer cell
invasion. Cancer Res. 63:1359–1364. 2003.PubMed/NCBI
|
|
43
|
Jana A, Krett NL, Guzman G, Khalid A,
Ozden O, Staudacher JJ, Bauer J, Baik SH, Carroll T, Yazici C and
Jung B: NFkB is essential for activin-induced colorectal cancer
migration via upregulation of PI3K-MDM2 pathway. Oncotarget.
8:37377–37393. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ning Y, Xu M, Cao X, Chen X and Luo X:
Inactivation of AKT, ERK and NF-κB by genistein derivative,
7-difluoromethoxyl-5,4′-di-n-octylygenistein, reduces ovarian
carcinoma oncogenicity. Oncol Rep. 38:949–958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang S, Leng T, Zhang Q, Zhao Q, Nie X
and Yang L: Sanguinarine inhibits epithelial ovarian cancer
development via regulating long non-coding RNA CASC2-EIF4A3 axis
and/or inhibiting NF-κB signaling or PI3K/AKT/mTOR pathway. Biomed
Pharmacother. 102:302–308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lu Z and Chen J: Introduction of WHO
classification of tumours of female reproductive organs, fourth
edition. Zhonghua Bing Li Xue Za Zhi. 43:649–650. 2014.(In
Chinese). PubMed/NCBI
|
|
47
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang J, Xiao L, Luo CH, Zhou H, Zeng L,
Zhong J, Tang Y, Zhao XH, Zhao M and Zhang Y: CD44v6 promotes
β-catenin and TGF-β expression, inducing aggression in ovarian
cancer cells. Mol Med Rep. 11:3505–3510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tjhay F, Motohara T, Tayama S, Narantuya
D, Fujimoto K, Guo J, Sakaguchi I, Honda R, Tashiro H and Katabuchi
H: CD44 variant 6 is correlated with peritoneal dissemination and
poor prognosis in patients with advanced epithelial ovarian cancer.
Cancer Sci. 106:1421–1428. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ni J, Cozzi PJ, Hao JL, Beretov J, Chang
L, Duan W, Shigdar S, Delprado WJ, Graham PH, Bucci J, et al: CD44
variant 6 is associated with prostate cancer metastasis and
chemo-/radioresistance. Prostate. 74:602–617. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang HF, Hu P and Fang SQ: Understanding
the role of CD44V6 in ovarian cancer. Oncol Lett. 14:1989–1992.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shi J, Zhou Z, Di W and Li N: Correlation
of CD44v6 expression with ovarian cancer progression and
recurrence. BMC Cancer. 13:1822013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Motohara T, Fujimoto K, Tayama S,
Narantuya D, Sakaguchi I, Tashiro H and Katabuchi H: CD44 Variant 6
as a predictive biomarker for distant metastasis in patients with
epithelial ovarian cancer. Obstet Gynecol. 127:1003–1011. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pramanik KC, Makena MR, Bhowmick K and
Pandey MK: Advancement of NF-κB signaling pathway: A novel target
in pancreatic cancer. Int J Mol Sci. 19(pii): E38902018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
DiDonato JA, Mercurio F and Karin M: NF-κB
and the link between inflammation and cancer. Immunol Rev.
246:379–400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang S, Robinson JB, Deguzman A, Bucana
CD and Fidler IJ: Blockade of nuclear factor-kappaB signaling
inhibits angiogenesis and tumorigenicity of human ovarian cancer
cells by suppressing expression of vascular endothelial growth
factor and interleukin 8. Cancer Res. 60:5334–5339. 2000.PubMed/NCBI
|
|
57
|
Malinen M, Niskanen EA, Kaikkonen MU and
Palvimo JJ: Crosstalk between androgen and pro-inflammatory
signaling remodels androgen receptor and NF-κB cistrome to
reprogram the prostate cancer cell transcriptome. Nucleic Acids
Res. 45:619–630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Luo JL, Maeda S, Hsu LC, Yagita H and
Karin M: Inhibition of NF-kappaB in cancer cells converts
inflammation-induced tumor growth mediated by TNFalpha to
TRAIL-mediated tumor regression. Cancer Cell. 6:297–305. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Perkins ND: The diverse and complex roles
of NF-κB subunits in cancer. Nat Rev Cancer. 12:121–132. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kawana H, Karaki H, Higashi M, Miyazaki M,
Hilberg F, Kitagawa M and Harigaya K: CD44 suppresses TLR-mediated
inflammation. J Immunol. 180:4235–4245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bourguignon LY, Wong G, Earle CA and Xia
W: Interaction of low molecular weight hyaluronan with CD44 and
toll-like receptors promotes the actin filament-associated protein
110-actin binding and MyD88-NFκB signaling leading to
proinflammatory cytokine/chemokine production and breast tumor
invasion. Cytoskeleton (Hoboken). 68:671–693. 2011. View Article : Google Scholar : PubMed/NCBI
|