Introduction
Lung cancer is the leading cause of
cancer-associated mortality, and non-small cell lung cancer (NSCLC)
accounts for >80% of all lung cancer cases (1). Despite advances in chemotherapy and
targeted therapy, the overall 5-year survival rate of lung cancer
remains poor (<15%), which is mainly attributed to resistance
and metastasis (2,3). Epidermal growth factor receptor (EGFR)
mutations have been reported in 15% of NSCLC cases in Western
countries, and in up to 40% of cases in Asian countries (4). Osimertinib (AZD9291) is a
third-generation EGFR-tyrosine kinase inhibitor (TKI) that was
approved by the Food and Drug Administration (FDA) in 2015 for the
treatment of EGFR T790M-positive NSCLC patients (5). Inevitably, these patients eventually
acquire resistance to AZD9291. Recent clinical data indicated that
the emergence of C797S mutations has been identified in a subset of
tresistant patients (6). However,
other mechanisms contributing to the development of AZD9291
resistance in the majority of cases remain unknown.
Placenta-specific 8 (PLAC8) is a relatively small
protein containing an evolutionarily-conserved cysteine-rich
domain, which is expressed in human oocytes and preimplantation
embryos, regulating placental and embryonic development (7,8).
Increasing evidence has demonstrated that PLAC8 is involved in the
regulation of various cellular processes, and serves an important
role in tumor progression and resistance (9–11). In
colon cancer, elevated PLAC8 levels led to the development of
epithelial-mesenchymal transition features and increased
invasiveness, while knockdown of endogenous PLAC8 resulted in
smaller tumors and reduced local invasion (12). Additionally, silencing of PLAC8 in
renal cell carcinoma cells significantly increased their
sensitivity to cisplatin (13).
However, to the best of our knowledge, the functional role of PLAC8
in AZD9291 resistance in NSCLC has never been investigated.
The aldehyde dehydrogenase (ALDH) family is a group
of enzymes that oxidize aldehyde into carboxylic acid and are
involved in modulating early stem cell differentiation (14,15).
ALDH 1 family member A1 (ALDH1A1), a member of the ALDH family, has
been demonstrated to be a marker for cancer stem cells (CSCs) in
numerous types of tumors (16,17). In
lung CSCs, ALDH1A1 is upregulated and positively correlated with
the stage and grade of lung cancer patients (18). Furthermore, ALDH1A1 was reported to
be overexpressed in cisplatin-resistant lung cancer cell lines, and
ALDH1A1 silencing significantly increased apoptosis and drug
sensitivity (19). Taken together,
these previous studies suggested that ALDH1A1 may contribute to
AZD9291 resistance.
In the present study, the expression levels of PLAC8
in AZD9291-resistant and AZD9291-sensitive NSCLC cell lines were
analyzed. To the best of our knowledge, it was demonstrated for the
first time that the PLAC8 levels were significantly upregulated in
the resistant cells, indicating a potential regulatory role of
PLAC8 in AZD9291 resistance in NSCLC. Subsequently, PLAC8
overexpression was induced in the AZD9291-sensitive NSCLC cells,
and drug sensitivity and biological functions were assessed.
Finally, the potential association between PLAC8 expression and
AZD9291 resistance in NSCLC was preliminarily studied.
Materials and methods
Cell lines and culture
The AZD9291-resistant NSCLC cell lines PC9/AZD9291
and HCC827/AZD9291, as well as the respective sensitive cell lines
PC9 and HCC827, were kindly provided by Dr Tianxiang Chen (Shanghai
Chest Hospital, Shanghai, China). These four cell lines were
cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Invitrogen; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.). For resistance maintenance, 1 µmol/l AZD9291 (Selleck
Chemicals, Houston, TX, USA) was also added to the culture of the
two resistant cell lines. All cells were cultured at 37°C in a
humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR assay was performed to detect the relative
mRNA expression levels of PLAC8 and ALDH1A1. Briefly, cells
(6×106 cells/well) were incubated in a 6-well plate for
24 h, and then the total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The concentration of the total RNA was
measured with a Nano-100 micro-spectrophotometer (Hangzhou Allsheng
Instruments Co., Ltd., Hangzhou, China). Next, ReverTra ACE (Toyobo
Life Science, Osaka, Japan) was applied to reverse transcribe the
total mRNA into single-stranded cDNA. The qPCR assay was
subsequently performed using a SYBR Premix Ex Taq™ II
kit (Takara Biotechnology Co., Ltd., Dalian, China) in a final
volume of 10 µl, containing 0.5 µl cDNA, 0.5 µl of each primer, 5
µl SYBR Green and 3.5 µl deionized water. The primers used were as
follows: PLAC8 forward, 5′-GTGTGACTGTTTCAGCGACTG-3′, and reverse,
5′-CTGCAACTTGACACCCAAGG-3′; ALDH1A1 forward,
5′-GCACGCCAGACTTACCTGTC-3′, and reverse,
5′-CCTCCTCAGTTGCAGGATTAAAG-3′; GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′, and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. Relative mRNA expression was
normalized to GAPDH expression, which served as the internal
control. The reactions were performed in a 96-well optical plate at
95°C for 5 min, followed by 42 cycles of 95°C for 5 sec and 60°C
for 1 min. Relative quantification of gene expression was performed
by the 2−ΔΔCq method (20).
Western blot analysis
Western blot assay was performed to evaluate the
relative protein expression levels of PLAC8 and ALDH1A1. Briefly,
cells (6×106 cells/well) were incubated for 24 h in a
6-well plate and the lysates were prepared with
radioimmunoprecipitation assay buffer (containing 50 mM Tris-HCl,
pH 8.0, 150 mM sodium chloride, 1.0% IGEPAL CA-630, 0.5% sodium
deoxycholate and 0.1% sodium dodecyl sulfate; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany), followed by centrifugation (13,800 × g;
10 min; 4°C). Total protein concentration was determined using a
Bradford assay. Next, 20 µg protein was separated by SDS-PAGE (15%
gel) and subsequently transferred to polyvinylidene fluoride
membranes. The membranes were incubated with the corresponding
primary antibodies at 4°C overnight, followed by incubation with
horseradish peroxidase-conjugated secondary antibodies for 1 h at
room temperature. The primary antibodies, including PLAC8 (cat. no.
13885; 1:1,000), ALDH1A1 (cat. no. 54135; 1:1,000) and GAPDH (cat.
no. 5174; 1:1,000), and the secondary antibody (cat. no. 7074;
1:2,000) used in the assay were obtained from Cell Signaling
Technology, Inc. (Danvers, MA, USA). GAPDH was used as a loading
control to measure the expression of each protein. Bands were
visualized using Pierce™ ECL Western Blotting Substrate
(Pierce; Thermo Fisher Scientific, Inc.). Gray values of the blot
bands were then analyzed using ImageJ software (National Institutes
of Health, Bethesda, MD, USA), and the relative gray value was used
to represent the relative protein expression.
Lentivirus production and
infection
Full-length human PLAC8 was amplified and cloned
into the PLVX-mCherry-N1 plasmid (Clontech Laboratories, Inc.,
Mountainview, CA, USA). 293T cells were subsequently transfected
with lentiviruses containing PLAC8-PLVX and packaging plasmids
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), following the manufacturer's protocols. The
supernatant medium containing lentivirus was collected by
centrifugation (5,000 × g; 60 min; 4°C) to remove cellular
contaminants. The resulting lentiviruses were used to infect the
sensitive cell lines PC9 and HCC827, and then successfully infected
cells were selected using 2 µg/ml puromycin (Sigma-Aldrich; Merck
KGaA). Positive clones were screened and verified by RT-qPCR
analysis. Cells overexpressing PLAC8 were termed PC9-PLAC8 and
HCC827-PLAC8, while control cells were termed PC9-CTL and
HCC827-CTL.
Cell proliferation
Cells were digested and seeded at a density of 3,000
cells/well in 96-well plates. Then cells were cultured for 1, 2, 3,
4, 5, 6 and 7 days, separately, and 10 µl
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich; Merck KGaA) was added to the appropriate wells and
incubated for another 3 h. Subsequent to discarding the
supernatant, 100 µl dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck
KGaA) was added to the cells, and the absorbance was detected at
492 nm. Cell proliferation was evaluated using the following
equation: Proliferation rate=A(sample)/A(control).
Cell viability
Cells were seeded in 96-well plates (5,000
cells/well) and cultured overnight. The transfected cells were
treated with 0, 0.01, 0.05, 0.1, 0.5, 1 and 10 µM AZD9291
respectively the following morning and incubated for 72 h.
Subsequently, 10 µl MTT was added to each well and incubated for a
further 3 h. The supernatant was then removed and 100 µl DMSO was
added to each well. Finally, the absorbance was measured at 492 nm,
and the cell viability was determined as follows: Cell viability
(%)=A(sample)/A(control) ×100%. The half-maximal inhibitory
concentration (IC50) was also calculated with SPSS
software (version 16.0; SPSS, Inc., Chicago, IL, USA). Resistance
index was then calculated as follows: Resistance
index=IC50(cells overexpressing PLAC8)/IC50
(corresponding control cells).
Colony formation assay
Cells were seeded in 6-well plates (400 cells/well)
and incubated for 10–14 days, until visible colonies were observed.
The medium was replaced every 3 days during the culture period.
Subsequently, the plates were washed with PBS, and the colonies
were fixed with methanol for 20 min, stained with 1% crystal violet
for 30 min and counted. The number of colonies containing >50
cells was determined.
Migration assay
The migration assay was performed using 24-well
transwell inserts (8-µm pore size; EMD Millipore, Billerica, MA,
USA), according to manufacturer's protocol. Briefly,
2×104 cells/well suspended in serum-free RPMI-1640
medium were seeded in the upper chamber, and RPMI-1640 containing
10% FBS was added to the lower chamber. Following incubation for 24
h, cells on the upper surface of the membrane were removed. Cells
remaining on the bottom surface were fixed with methanol for 20 min
and then stained with a 1% crystal violet solution for 0.5 h.
Finally, the stained cells were photographed and counted in at
least five randomly selected fields.
Statistical analysis
The data are presented as the mean ± standard
deviation, and an unpaired Student's t-test was used to analyze the
statistical significance of the data. Analysis was performed with
SPSS software and GraphPad Prism software (version 5; GraphPad
Software, Inc., La Jolla, CA, USA). Each experiment was performed
at least three times. A statistically significant difference was
denoted by a value of P<0.05.
Results
PLAC8 expression is upregulated in
AZD9291-resistant NSCLC cell lines
Increasing evidence has demonstrated that PLAC8 is
associated with various cellular processes, and is an important
regulator of tumor progression and resistance (10). In the present study, the PLAC8 levels
in AZD9291-resistant and AZD9291-sensitive NSCLC cell lines were
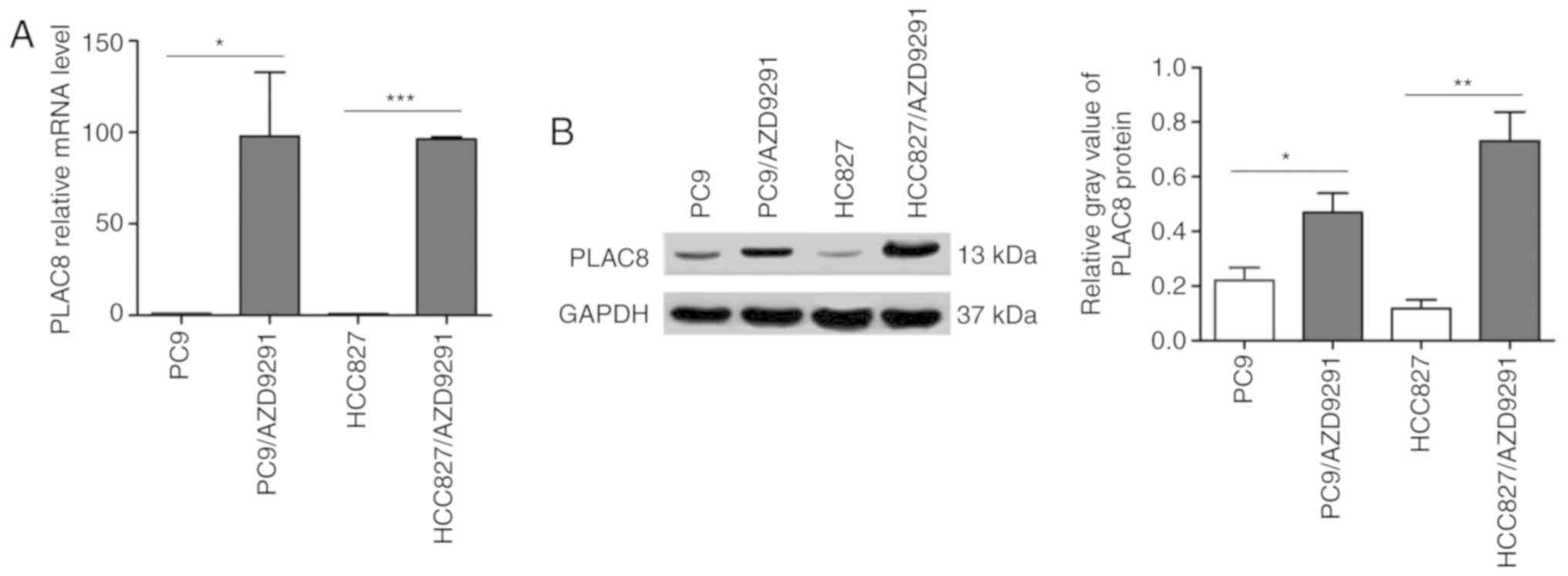
initially assessed by RT-qPCR and western blot analyses. As shown
in Fig. 1A, the PLAC8 mRNA level in
the resistant cells was significantly upregulated, and was almost
100 times higher than that in the parent PC9 and HCC827 cells. To
confirm these results, the protein levels of PLAC8 were then
evaluated. PLAC8 protein expression was notably upregulated in the
resistant cells (Fig. 1B),
consistent with the RT-qPCR results. These findings indicated the
potential correlation between upregulated PLAC8 levels and AZD9291
resistance in NSCLC.
Overexpressed PLAC8 promotes AZD9291
resistance in NSCLC cells
To investigate whether PLAC8 is involved in AZD9291
resistance in NSCLC, PLAC8 overexpression was induced in PC9 and
HCC827 cells (PC9-PLAC8 and HCC827-PLAC8, respectively), and then
the IC50 value for AZD9291 was assessed by an MTT assay.
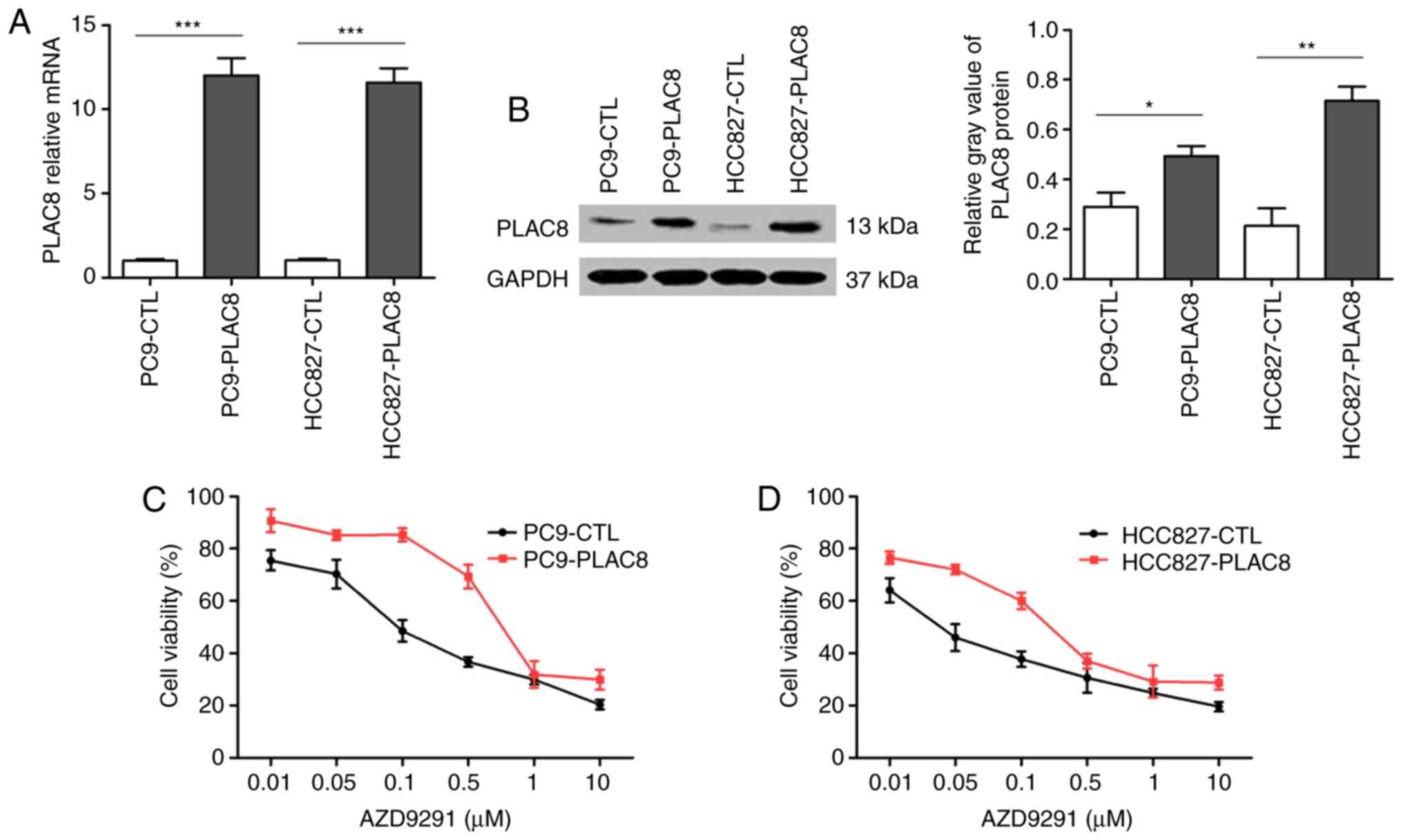
As shown in Fig. 2A, the mRNA levels
of PLAC8 were significantly upregulated in PC9-PLAC8 and
HCC827-PLAC8 cells as compared with those in the control cells
(PC9-CTL and HCC827-CTL). The results for PLAC8 protein expression
shown in Fig. 2B further confirmed
that PLAC8-overexpressing cells were constructed successfully.
The sensitivities of the four cells to AZD9291 were
then detected. As shown in Fig. 2C and
D, resistance trends were observed in PC9-PLAC8 and
HCC827-PLAC8 cells as compared with their corresponding control
cells. The IC50 values of the control and
PLAC8-overexpressing PC9 cells were 0.163 and 1.019 µM,
respectively, while these values in HCC827 cells were 0.035 and
0.245 µM, respectively. These data suggested that the resistance
index reached 6–7, thus PLAC8 may serve an important role in
AZD9291 resistance in NSCLC (21).
Effects of overexpressed PLAC8 on
NSCLC cell proliferation, colony formation and migration
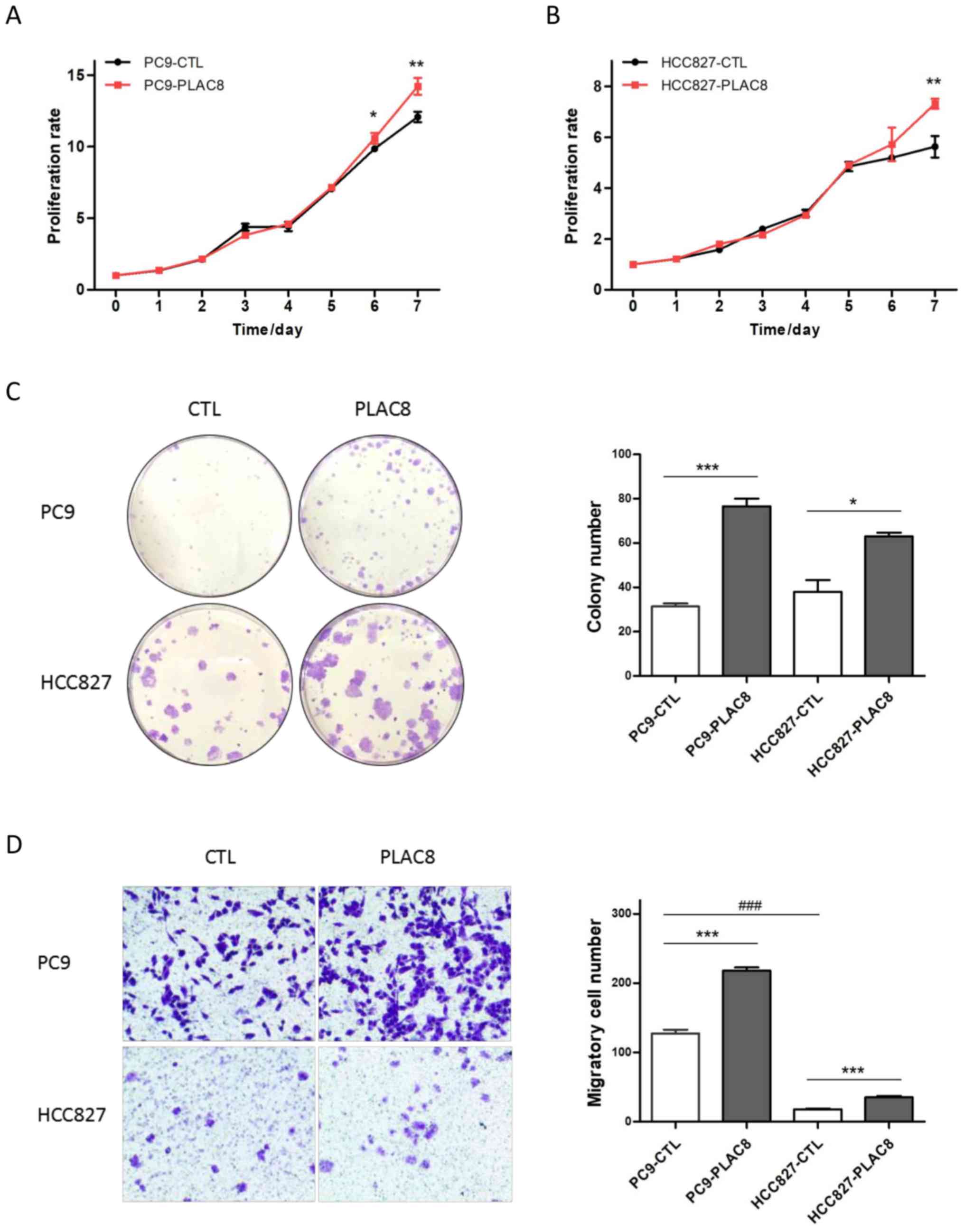
Subsequently, the current study assessed the
proliferation, colony formation and migration abilities of
PLAC8-overexpressing cell lines. The MTT results indicated that the
cell growth patterns of PC9-CTL and PC9-PLAC8 were similar in the
first 5 days of culture, whereas PC9-PLAC8 cells exhibited
significantly increased growth at 6 and 7 days (Fig. 3A). A similar proliferation trend was
observed in Fig. 3B for HCC827
cells. Furthermore, overexpression of PLAC8 significantly elevated
the colony formation ability, with a markedly greater number of
colonies present in PLAC8-overexpressing cells (Fig. 3C). The migratory cell numbers in the
overexpressing cell lines were also significantly increased
compared with the corresponding control cells, despite the
differential migration ability between the two control cell lines
(Fig. 3D).
ALDH1A1 levels are upregulated in
PLAC8-overexpressing cells
ALDH1A1 is a putative marker for CSCs in numerous
types of tumors and is reportedly overexpressed in certain
resistant cells (14). Therefore,
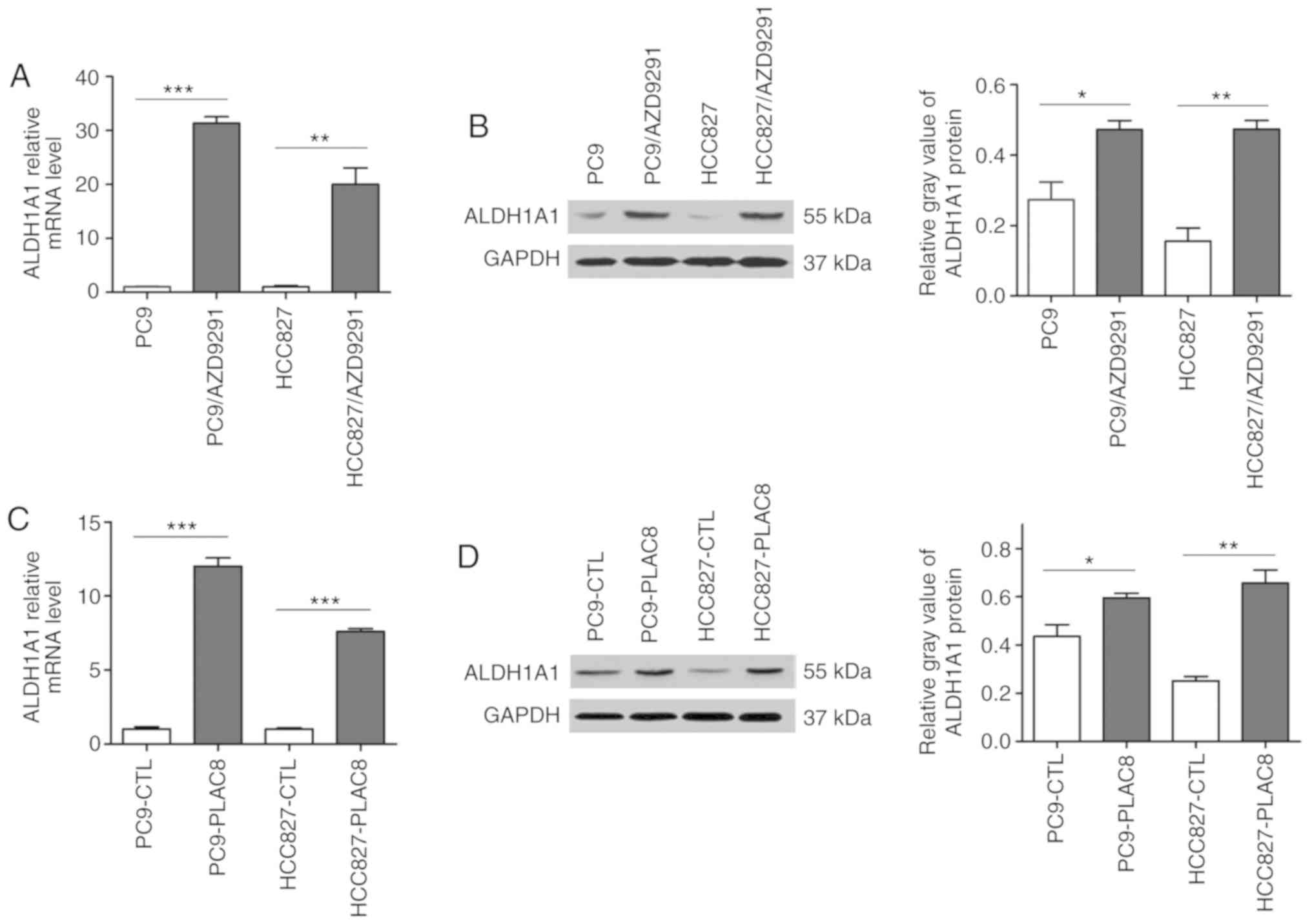
the present study examined whether ALDH1A1 is also upregulated in
AZD9291-resistant cells. As shown in Fig. 4A and B, ALDH1A1 mRNA and protein
levels were significantly increased in resistant cells as compared
with those in parent cells, suggesting an underlying link between
upregulated ALDH1A1 levels and AZD9291 resistance. Based on the
role of PLAC8 in AZD9291 resistance discussed earlier, it was
hypothesized that ALDH1A1 expression may be regulated by PLAC8. To
confirm this, the levels of ALDH1A1 in PLAC8-overexpressing cells
were detected, and it was observed that ALDH1A1 mRNA was
significantly upregulated in the overexpressing cells (Fig. 4C). The results of the western blot
analysis illustrated a similar protein expression pattern (Fig. 4D).
Discussion
EGFR-TKIs have demonstrated significant benefits in
the treatment of NSCLC with EGFR mutations (22). However, the majority of patients
develop resistance within 1 year (23). At present, AZD9291 is the only third
generation TKI approved by the FDA. The inevitable acquired
resistance to AZD9291 is a significant clinical challenge, and
there is an urgent need for the identification of possible
mechanisms involved in AZD9291 resistance.
Accumulating evidence suggests that PLAC8 is an
important player in various cellular processes, thus promoting
cancer progression and resistance. Increased expression of PLAC8
facilitates the pro-survival function of autophagy to allow for
proliferation in cadmium-transformed prostate epithelial cells, and
induces resistance to cadmium toxicity (24). It has been reported that knockdown of
PLAC8 expression in clear cell renal cell carcinoma cells
significantly reduced the proliferation rates and colony formation
ability (13). Additionally, PLAC8
is a critical regulator of autophagy during pancreatic cancer
progression (25). In the present
study, it was observed that the PLAC8 level was substantially
upregulated in AZD9291-resistant cell lines. Overexpression of
PLAC8 in sensitive cells contributed to resistance to AZD9291 and
significantly increased the cell proliferation, colony formation
and migration, which is in agreement with the findings of the
aforementioned studies. Notably, increased migration ability
indicated potentially higher metastatic ability and tumor
malignancy (26). Therefore, PLAC8
may promote increased malignancy in addition to resistance to
AZD9291.
CSCs are a unique population of cancer cells
identified to have stem-like features, including self-renewal,
differentiation and tumorigenesis, which serve an important role in
tumor progression and resistance (27,28). A
recent study suggested that targeting the surface markers of CSCs
may facilitate targeted therapy innovation (29). In the present study, it was
demonstrated that ALDH1A1 was upregulated in AZD9291-resistant
NSCLC cell lines, consistent with a previous study reporting that
overexpression of ALDH1A1 was detected in lung cancer cells that
are resistant to afatinib, a second generation TKI (30). Furthermore, cisplatin-resistant A549
cells exhibited elevated levels of ALDH1A1, while knockdown of
ALDH1A1 significantly decreased A549/DDP proliferation, increased
apoptosis and reduced cisplatin resistance (19). Subsequently, the level of ALDH1A1 in
PLAC8-overexpressing cells was assessed in the current study, and
the results suggested that the ALDH1A1 level was markedly increased
in PC9-PLAC8 and HCC827-PLAC8. These results indicated that the
upregulation of ALDH1A1 may be an important mediator between the
overexpression of PLAC8 and AZD9291 resistance in NSCLC with
increased proliferation, colony formation and migration.
PLAC8 is a relatively small protein, containing a
cysteine-rich domain with several CXXC motifs. Jimenez-Preitner
et al (7) reported that PLAC8
is required for brown fat differentiation via interaction with
C/EBPβ, and binds to the C/EBPβ promoter to induce its
transcription. Another study by Jimenez-Preitner et al
(31) revealed that the
transactivating effect of PLAC8 on the C/EBPβ promoter is critical
for white adipocyte differentiation. Therefore, it can be inferred
that the elevated ALDH1A1 observed in the present study may be
attributed to the transcriptional regulation of the ALDH1A1
promoter by PLAC8, thus inducing resistance. Nevertheless, further
studies need to be conducted to investigate the regulatory
mechanism of PLAC8 on ALDH1A1 expression.
In conclusion, the current study reported that
upregulated PLAC8 prompted NSCLC cell resistance to AZD9291, and
significantly elevated the proliferation, colony formation and
migration potential. Furthermore, ALDH1A1 may provide a link
between the overexpression of PLAC8 and AZD9291 resistance in
NSCLC, while PLAC8 may be a potential target for addressing this
resistance.
Acknowledgements
Not applicable.
Funding
This study was financially supported by the National
Natural Science Foundation of China (grant no. 81673014).
Availability of data and materials
The materials described in this manuscript,
including all relevant raw data, will be freely available to any
researchers wishing to use them for non-commercial purposes.
Authors' contributions
GW and XG put forward the conception of the study.
XG and XF analyzed and interpreted the data. XF and HS collected
and assembled data. All authors contributed to preparation of the
manuscript and approved the final version.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Heigener DF and Reck M: Lung cancer in
2017: Giant steps and stumbling blocks. Nat Rev Clin Oncol.
15:71–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nagasaka M and Gadgeel SM: Role of
chemotherapy and targeted therapy in early-stage non-small cell
lung cancer. Expert Rev Anticancer Ther. 18:63–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mayekar MK and Bivona TG: Current
landscape of targeted therapy in lung cancer. Clin Pharmacol Ther.
102:757–764. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saad N, Poudel A, Basnet A and Gajra A:
Epidermal growth factor receptor T790M mutation-positive metastatic
non-small-cell lung cancer: Focus on osimertinib (AZD9291). Onco
Targets Ther. 10:1757–1766. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jänne PA, Yang JC, Kim DW, Planchard D,
Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al: AZD9291
in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J
Med. 372:1689–1699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thress KS, Paweletz CP, Felip E, Cho BC,
Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A, Kuang Y, et
al: Acquired EGFR C797S mutation mediates resistance to AZD9291 in
non-small cell lung cancer harboring EGFR T790M. Nat Med.
21:560–562. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jimenez-Preitner M, Berney X, Uldry M,
Vitali A, Cinti S, Ledford JG and Thorens B: Plac8 is an inducer of
C/EBPβ required for brown fat differentiation, thermoregulation,
and control of body weight. Cell Metab. 14:658–670. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Q, Lu TF, Liu D, Hu PF, Sun B, Ma JZ,
Wang WJ, Wang KF, Zhang WX, Chen J, et al: Screening and analyzing
genes associated with Amur tiger placental development. Genet Mol
Res. 13:7869–7878. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaistha BP, Lorenz H, Schmidt H, Sipos B,
Pawlak M, Gierke B, Kreider R, Lankat-Buttgereit B, Sauer M,
Fiedler L, et al: PLAC8 localizes to the inner plasma membrane of
pancreatic cancer cells and regulates cell growth and disease
progression through critical cell-cycle regulatory pathways. Cancer
Res. 76:96–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Uehara H, Takahashi T and Izumi K:
Induction of retinol-binding protein 4 and placenta-specific 8
expression in human prostate cancer cells remaining in bone
following osteolytic tumor growth inhibition by osteoprotegerin.
Int J Oncol. 43:365–374. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zou L, Chai J, Gao Y, Guan J, Liu Q and Du
JJ: Down-regulated PLAC8 promotes hepatocellular carcinoma cell
proliferation by enhancing PI3K/Akt/GSK3β/Wnt/β-catenin signaling.
Biomed Pharmacother. 84:139–146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li C, Ma H, Wang Y, Cao Z, Graves-Deal R,
Powell AE, Starchenko A, Ayers GD, Washington MK, Kamath V, et al:
Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon
cancer. J Clin Invest. 124:2172–2187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi L, Xiao L, Heng B, Mo S, Chen W and Su
Z: Overexpression of placenta specific 8 is associated with
malignant progression and poor prognosis of clear cell renal cell
carcinoma. Int Urol Nephrol. 49:1165–1176. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu X, Chai S, Wang P, Zhang C, Yang Y,
Yang Y and Wang K: Aldehyde dehydrogenases and cancer stem cells.
Cancer Lett. 369:50–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koppaka V, Thompson DC, Chen Y, Ellermann
M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD and
Vasiliou V: Aldehyde dehydrogenase inhibitors: a comprehensive
review of the pharmacology, mechanism of action, substrate
specificity, and clinical application. Pharmacol Rev. 64:520–539.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu J, Alharbi A, Shan H, Hao Y, Snetsinger
B, Rauh MJ and Yang X: TAZ induces lung cancer stem cell properties
and tumorigenesis by up-regulating ALDH1A1. Oncotarget.
8:38426–38443. 2017.PubMed/NCBI
|
|
17
|
Duong HQ, You KS, Oh S, Kwak SJ and Seong
YS: Silencing of NRF2 reduces the expression of ALDH1A1 and ALDH3A1
and sensitizes to 5-FU in pancreatic cancer cells. Antioxidants
(Basel). 6(pii): E522017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alamgeer M, Ganju V, Szczepny A, Russell
PA, Prodanovic Z, Kumar B, Wainer Z, Brown T, Schneider-Kolsky M,
Conron M, et al: The prognostic significance of aldehyde
dehydrogenase 1A1 (ALDH1A1) and CD133 expression in early stage
non-small cell lung cancer. Thorax. 68:1095–1104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei Y, Wu S, Xu W, Liang Y, Li Y, Zhao W
and Wu J: Depleted aldehyde dehydrogenase 1A1 (ALDH1A1) reverses
cisplatin resistance of human lung adenocarcinoma cell A549/DDP.
Thorac Cancer. 8:26–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lai YH, Lin SY, Wu YS, Chen HW and Chen
JJW: AC-93253 iodide, a novel Src inhibitor, suppresses NSCLC
progression by modulating multiple Src-related signaling pathways.
J Hematol Oncol. 10:1722017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan CS, Gilligan D and Pacey S: Treatment
approaches for EGFR-inhibitor-resistant patients with
non-small-cell lung cancer. Lancet Oncol. 16:e447–e459. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kolluru V, Pal D, Papu John AMS, Ankem MK,
Freedman JH and Damodaran C: Induction of Plac8 promotes
pro-survival function of autophagy in cadmium-induced prostate
carcinogenesis. Cancer Lett. 408:121–129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kinsey C, Balakrishnan V, O'Dell MR, Huang
JL, Newman L, Whitney-Miller CL, Hezel AF and Land H: Plac8 links
oncogenic mutations to regulation of autophagy and is critical to
pancreatic cancer progression. Cell Rep. 7:1143–1155. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leon G, MacDonagh L, Finn SP, Cuffe S and
Barr MP: Cancer stem cells in drug resistant lung cancer: Targeting
cell surface markers and signaling pathways. Pharmacol Ther.
158:71–90. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Prieto-Vila M, Takahashi RU, Usuba W,
Kohama I and Ochiya T: Drug resistance driven by cancer stem cells
and their niche. Int J Mol Sci. 18(pii): E25742017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
MacDonagh L, Gray SG, Breen E, Cuffe S,
Finn SP, O'Byrne KJ and Barr MP: Lung cancer stem cells: The root
of resistance. Cancer Lett. 372:147–156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hashida S, Yamamoto H, Shien K, Miyoshi Y,
Ohtsuka T, Suzawa K, Watanabe M, Maki Y, Soh J, Asano H, et al:
Acquisition of cancer stem cell-like properties in non-small cell
lung cancer with acquired resistance to afatinib. Cancer Sci.
106:1377–1384. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jimenez-Preitner M, Berney X and Thorens
B: Plac8 is required for white adipocyte differentiation in vitro
and cell number control in vivo. PLoS One. 7:e487672012. View Article : Google Scholar : PubMed/NCBI
|