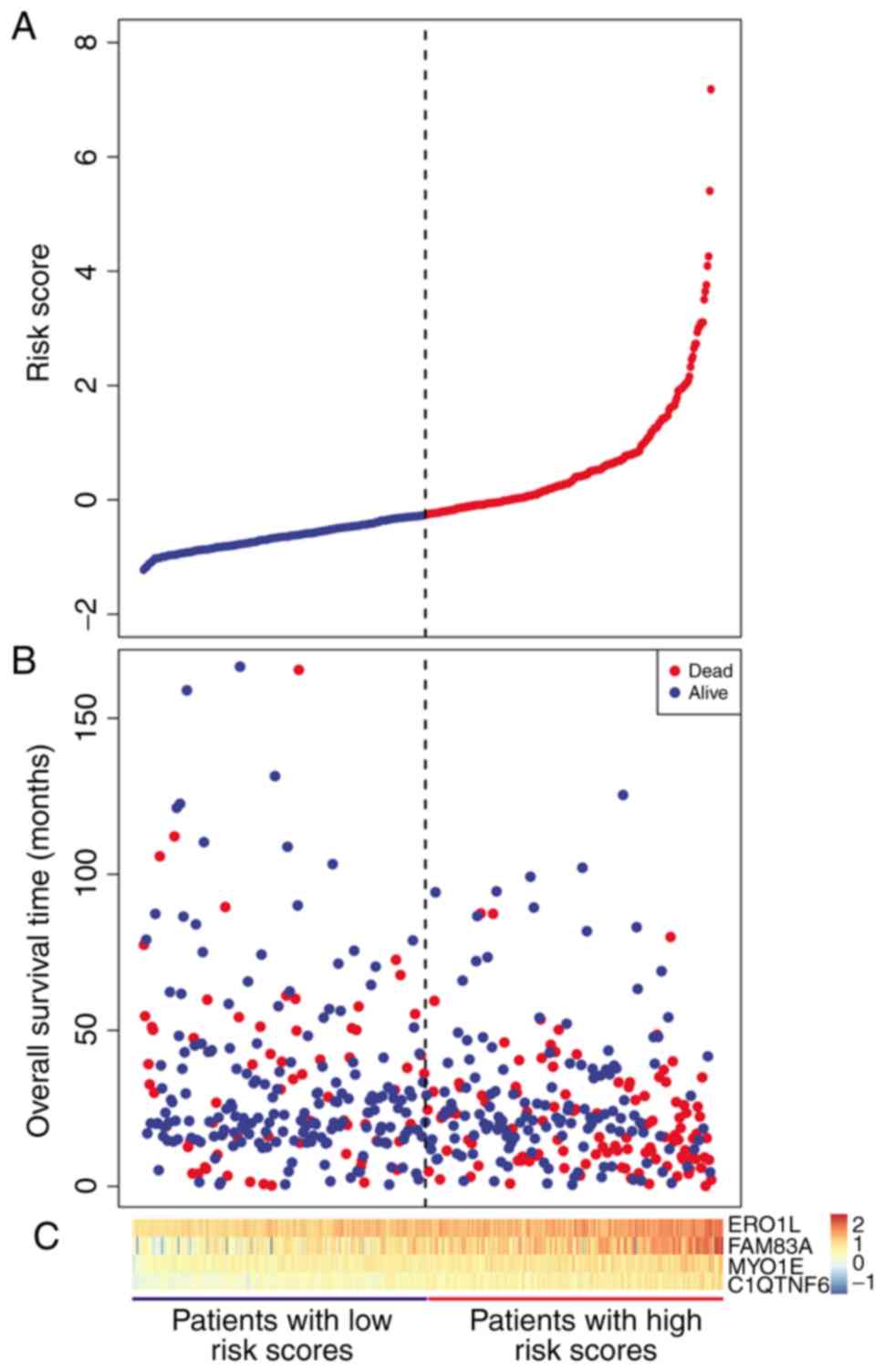
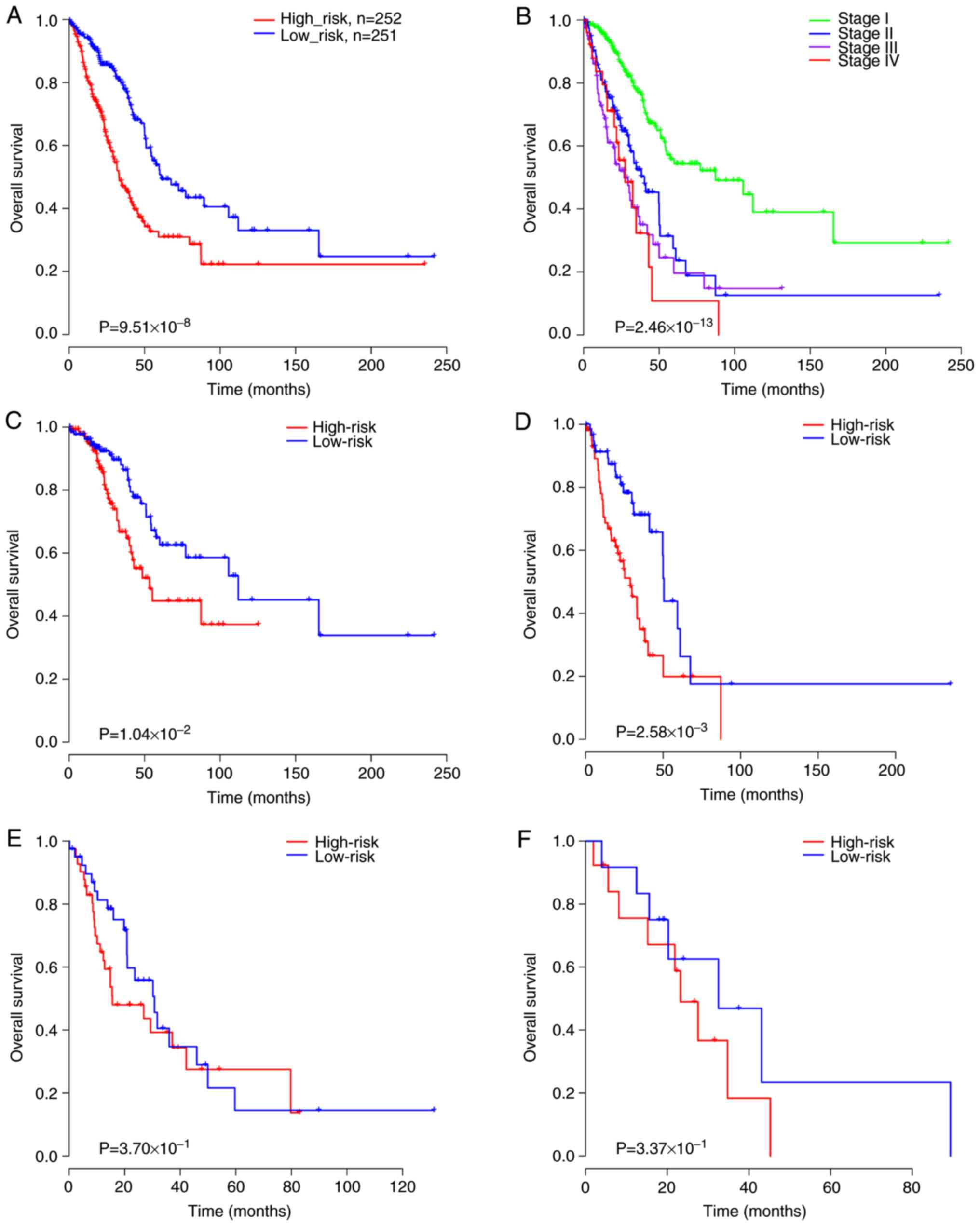
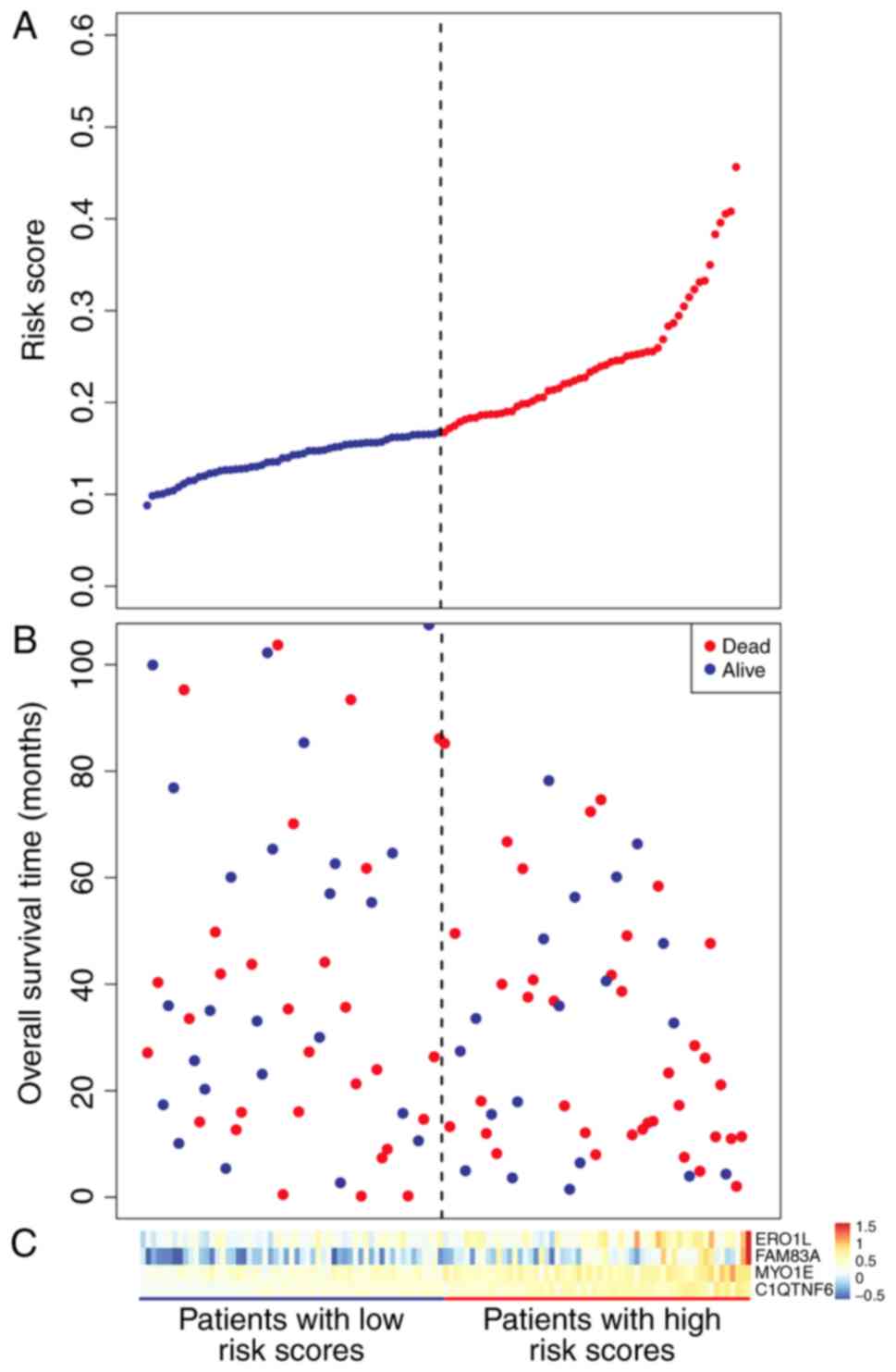
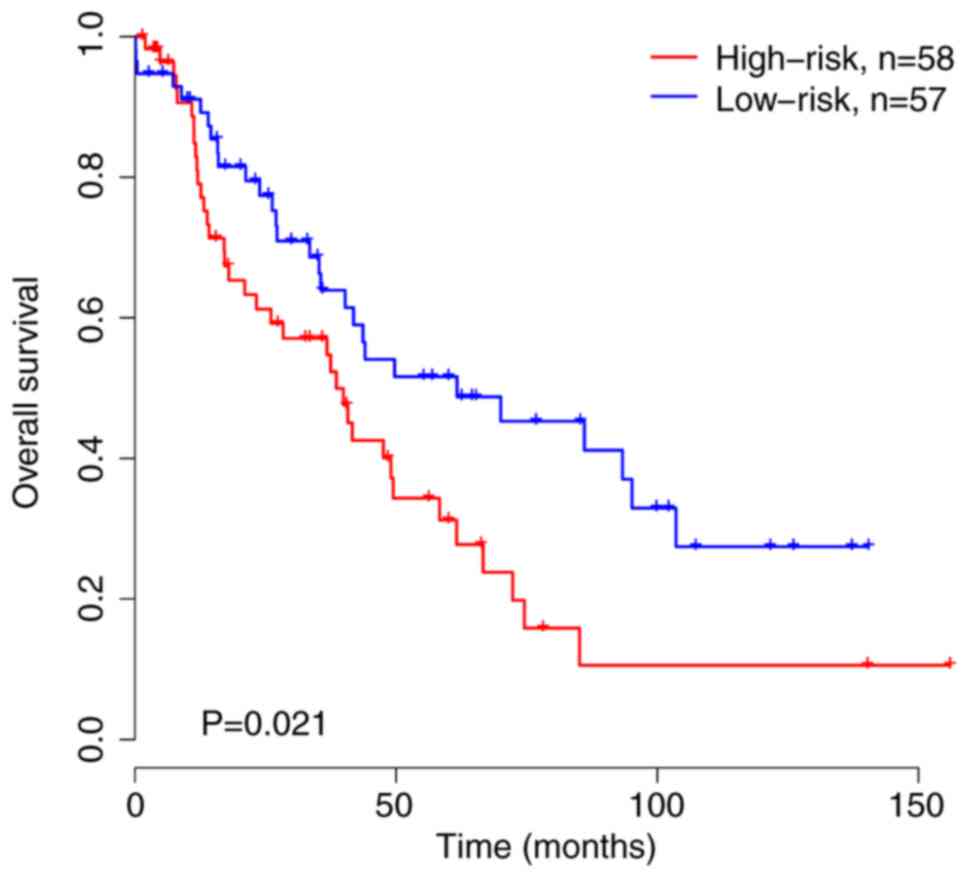
|
1
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuire S: World cancer report 2014.
Geneva, Switzerland: World health organization, international
agency for research on cancer, WHO press, 2015. Adv Nut. 7:418–419.
2016. View Article : Google Scholar
|
|
3
|
Longo DL, Fauci AS, Kasper DL, Hauser SL,
Jameson JL and Loscalzo J: Harrison's principles of internal
medicine. (18th). McGraw Hill Professional. (New York, NYC, USA).
2872011.
|
|
4
|
Subramanian J and Govindan R: Lung cancer
in never smokers: A review. J Clin Oncol. 25:561–570. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marugame T, Sobue T, Nakayama T, Suzuki T,
Kuniyoshi H, Sunagawa K, Genka K, Nishizawa N, Natsukawa S,
Kuwahara O and Tsubura E: Filter cigarette smoking and lung cancer
risk; a hospital-based case-Control study in Japan. Br J Cancer.
90:646–651. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Spiro SG, Tanner NT, Silvestri GA, Janes
SM, Lim E, Vansteenkiste JF and Pirker R: Lung cancer: Progress in
diagnosis, staging and therapy. Respirology. 15:44–50. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fukui T and Mitsudomi T: Small peripheral
lung adenocarcinoma: Clinicopathological features and surgical
treatment. Surgery Today. 40:191–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sholl LM, Barletta JA, Yeap BY, Chirieac
LR and Hornick JL: Sox2 protein expression is an independent poor
prognostic indicator in stage I lung adenocarcinoma. Am J Surg
Pathol. 34:1193–1198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakatsugawa M, Takahashi A, Hirohashi Y,
Torigoe T, Inoda S, Murase M, Asanuma H, Tamura Y, Morita R,
Michifuri Y, et al: SOX2 is overexpressed in stem-like cells of
human lung adenocarcinoma and augments the tumorigenicity. Lab
Invest. 91:1796–1804. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Wang L, Li Y, Pan Y, Wang R, Hu
H, Li H, Luo X, Ye T, Sun Y and Chen H: Protein expression of
programmed death 1 ligand 1 and ligand 2 independently predict poor
prognosis in surgically resected lung adenocarcinoma. Onco Targets
Ther. 7:567–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shimokawa H, Uramoto H, Onitsuka T,
Chundong G, Hanagiri T, Oyama T and Yasumoto K: Overexpression of
MACC1 mRNA in lung adenocarcinoma is associated with postoperative
recurrence. J Thorac Cardiovasc Surg. 141:895–898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Z, Li Z, Wu C, Wang Y, Xia Y, Chen L,
Zhu Q and Chen Y: MACC1 overexpression predicts a poor prognosis
for non-small cell lung cancer. Med Oncol. 31:7902014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen G, Kim SH, King AN, Zhao L, Simpson
RU, Christensen PJ, Wang Z, Thomas DG, Giordano TJ, Lin L, et al:
CYP24A1 is an independent prognostic marker of survival in patients
with lung adenocarcinoma. Clin Cancer Res. 17:817–826. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Minamiya Y, Ono T, Saito H, Takahashi N,
Ito M, Mitsui M, Motoyama S and Ogawa J: Expression of histone
deacetylase 1 correlates with a poor prognosis in patients with
adenocarcinoma of the lung. Lung Cancer. 74:300–304. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Therneau TM and Grambsch PM: Modeling
survival data: Extending the cox model. Springer. (New York). 2000.
View Article : Google Scholar
|
|
17
|
Bland JM and Altman DG: The logrank test.
BMJ. 328:10732004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bera AK and Bilias Y: Rao's score,
Neyman's C(α) and Silvey's LM tests: An essay on historical
developments and some new results. J Stat Plan Inference. 97:9–44.
2001. View Article : Google Scholar
|
|
19
|
Harrell FE: Regression modeling
strategies: With applications to linear models, logistic
regression, and survival analysis. Springer. (New York). 2001.
|
|
20
|
Ishwaran H, Kogalur UB, Blackstone EH and
Lauer MS: Random survival forests. Ann Appl Stat. 2:841–860. 2008.
View Article : Google Scholar
|
|
21
|
Stel VS, Dekker FW, Tripepi G, Zoccali C
and Jager KJ: Survival analysis I: The Kaplan-Meier method. Nephron
Clin Pract. 119:c83–c88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT anaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41:W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li T, Wernersson R, Hansen RB, Horn H,
Mercer J, Slodkowicz G, orkman CT, Rigina O, Rapacki K, Staerfeldt
HH, et al: A scored human protein-protein interaction network to
catalyze genomic interpretation. Nat Methods. 14:61–64. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kutomi G, Tamura Y, Tanaka T, Kajiwara T,
Kukita K, Ohmura T, Shima H, Takamaru T, Satomi F, Suzuki Y, et al:
Human endoplasmic reticulum oxidoreductin 1-α is a novel predictor
for poor prognosis of breast cancer. Cancer Sci. 104:1091–1096.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tanaka T, Kutomi G, Kajiwara T, Kukita K,
Kochin V, Kanaseki T, Tsukahara T, Hirohashi Y, Torigoe T, Okamoto
Y, et al: Cancer-associated oxidoreductase ERO1-α promotes immune
escape through up-regulation of PD-L1 in human breast cancer.
Oncotarget. 8:24706–24718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tanaka T, Kajiwara T, Torigoe T, Okamoto
Y, Sato N and Tamura Y: Cancer-associated oxidoreductase ERO1-α
drives the production of tumor-promoting myeloid-derived suppressor
cells via oxidative protein folding. J Immunol. 194:2004–2010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kajiwara T, Tanaka T, Kukita K, Kutomi G,
Saito K, Okuya K, Takaya A, Kochin V, Kanaseki T, Tsukahara T, et
al: Hypoxia augments MHC class I antigen presentation via
facilitation of ERO1-α-mediated oxidative folding in murine tumor
cells. Eur J Immunol. 46:2842–2851. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tanaka T, Kutomi G, Kajiwara T, Kukita K,
Kochin V, Kanaseki T, Tsukahara T, Hirohashi Y, Torigoe T, Okamoto
Y, et al: Cancer-associated oxidoreductase ERO1-α drives the
production of VEGF via oxidative protein folding and regulating the
mRNA level. Br J Cancer. 114:1227–1234. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Snijders AM, Lee SY, Hang B, Hao W,
Bissell MJ and Mao JH: FAM83 family oncogenes are broadly involved
in human cancers: An integrative multi-omics approach. Mol Oncol.
11:167–179. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bartel CA, Parameswaran N, Cipriano R and
Jackson MW: FAM83 proteins: Fostering new interactions to drive
oncogenic signaling and therapeutic resistance. Oncotarget.
7:52597–52612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cipriano R, Miskimen KL, Bryson BL, Foy
CR, Bartel CA and Jackson MW: Conserved oncogenic behavior of the
FAM83 family regulates MAPK signaling in human cancer. Mol Cancer
Res. 12:1156–1165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin B, Chen T, Zhang Q, Lu X, Zheng Z,
Ding J, Liu J, Yang Z, Geng L, Wu L, et al: FAM83D associates with
high tumor recurrence after liver transplantation involving
expansion of CD44+ carcinoma stem cells. Oncotarget. 7:77495–77507.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Okabe N, Ezaki J, Yamaura T, Muto S, Osugi
J, Tamura H, Imai J, Ito E, Yanagisawa Y, Honma R, et al: FAM83B is
a novel biomarker for diagnosis and prognosis of lung squamous cell
carcinoma. Int J Oncol. 46:999–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Scholz T, Rump A, Uta P, Hinrichs M, Thiel
C and Tsiavaliaris G: Myosin-1 function extends to
microtubule-dependent processes. Biophys J. 98 (Suppl):561a2010.
View Article : Google Scholar
|
|
37
|
Malumbres M and Barbacid M: Cell cycle
kinases in cancer. Curr Opin Genet Dev. 17:60–65. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Asghar U, Witkiewicz AK, Turner NC and
Knudsen ES: The history and future of targeting cyclin-dependent
kinases in cancer therapy. Nat Rev Drug Discov. 14:130–146. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peinado H, Lavotshkin S and Lyden D: The
secreted factors responsible for pre-metastatic niche formation:
Old sayings and new thoughts. Semin Cancer Biol. 21:139–146. 2011.
View Article : Google Scholar : PubMed/NCBI
|