Introduction
Endometrial carcinoma (EC) arises from the inner
lining of the uterus (also termed the endometrium) (1). It is the third most prevalent
gynecological malignancy, second to breast cancer and cervical
cancer (2). In 2016, >10,470
lethal forms of uterine corpus tumors occurred in the USA (3), with an approximate three-fold increase
in the past 25 years (4). The
majority of patients with EC are diagnosed in late clinical stages,
so a poor prognosis is usually received once metastasis or relapse
occurs (5). Histopathological
investigations are the gold standard for the diagnosis of EC
(6). The current study aimed to
identify an alternative diagnostics approach that may be
additionally be used to explore the pathogenesis of EC.
Human T cell lymphotropic virus type 1 (HTLV-1) is
an oncogenic retrovirus first identified in humans (7). It may result in T-cell malignancy,
termed adult T-cell leukemia/lymphoma, as well as chronic
inflammatory disorders including tropical spastic paraparesis,
HTLV-1-associated myelopathy and uveitis (8,9). HTLV-1
affects 15–41 million individuals worldwide (10,11). The
regions with the highest incidence are areas in the Caribbean
islands, Central and South America, Africa and Japan (12,13).
Infectious agents, including parasites, may have oncogenic
capacity. HTLV-1 infection symptoms may occur early in an
individual's lifetime and generally have a long-term latent time
period prior to tumorigenesis (14).
Thus, efforts have been made to prevent HTLV-1-associated
carcinogenesis, and recently, a therapy targeting C-C motif
chemokine receptor 4, which has been identified as an adult T-cell
leukemia-specific marker associated with HTLV-1, has been
clinically tested in Japan with promising results (15). Previous studies explored the
mechanisms underlying HTLV-1-mediated tumorigenesis, including the
promotion of cell proliferation and modulation of multiple host
factors in liver cancer and lymphoma (16–18).
HTLV-1 differs from other acute transforming retroviruses, and may
not rapidly initiate carcinogenesis or alter the expression pattern
of cellular proto-oncogenes (16).
Retroviral infections may result in carcinogenesis by the following
potential mechanisms: Chronic inflammation in the host,
genome-mutagenesis in the host and virally-carried oncogenes that
activate cellular transformation (19–21). As
such, all responses initiate tumorigenesis and promote neoplasia
(19–21). Further studies are required to
elucidate the mechanisms by which HTLV-1 mediates oncogenesis.
The current study hypothesized that HTLV-1 may be
associated with endometrial cancer. To this end, HTLV-1
infection-associated genes that may be linked with endometrial
cancer were identified from The Cancer Genome Atlas database (TCGA;
portal.gdc.cancer.gov/projects).
Publicly available data were analyzed by two-way hierarchical
clustering analysis (HCA) and a support vector machine (SVM)
classifier was constructed to investigate the association between
HTLV-1 infection and EC. Differentially expressed genes (DEGs)
between normal and tumor samples were identified. A total of 41
candidate genes were identified as part of the overlap of HTLV-1
infection-associated pathways and DEGs, and were used to build an
SVM classifier. A log-rank test was used to analyze the association
between the genes and the prognosis of patients with EC. The
predictive power of the genes was verified with an independent
dataset.
Materials and methods
Data acquisition and quality
assessment
Gene expression profile data were downloaded from
TCGA (https://portal.gdc.cancer.gov/projects/TCGA-UCEC/;
Project ID: TCGA-UCEC). A total of 23 normal tissues and 23 matched
cancer tissues were used for DEG identification and the initial
training set. The remaining non-matched TCGA EC samples were used
as a test set, which consisted of a total of 541 samples, including
529 tumour samples and 12 normal samples. The TCGA data were
downloaded in the form of RNA sequencing data on an Illumina HiSeq
4100 RNA Sequencing platform (Illumina, Inc.). In addition, a gene
expression dataset for uterus normal tissues was downloaded from
the Genotype-Tissue Expression project (GTEx; version 7; www.gtexportal.org) and was used as an additional
validation set in the current study. The GTEx dataset contained 111
normal samples. The background correction and normalization were
conducted using the DEseq2 software package (version 1.20.0)
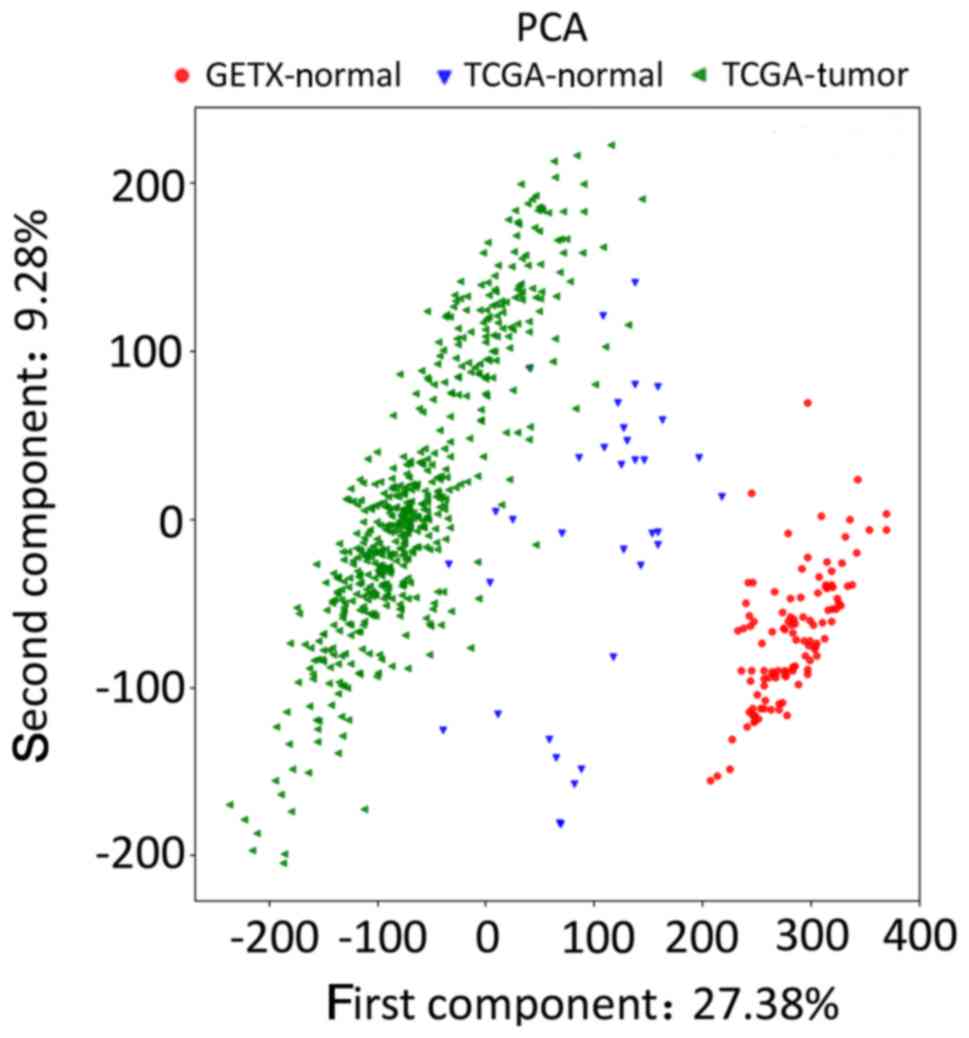
(22). In addition, principal
component analysis (PCA) between TCGA and GETx datasets (of
endometrial samples based on common genes), used to evaluate batch
effects, was performed using the Sklearn.svm package (version 3) of
Python.
Data preprocessing and DEG
screening
Ensembl79 IDs were converted to symbol IDs. Average
expression values were used if different probes were mapped to the
same gene. DEGs between endometrial cancer and healthy matched
controls were analyzed with the DESeq2 package in Bioconductor
(version 3.8) (23), with a cut-off
threshold of P<0.05 and fold change ≥2.0.
Predictive capacity of the proposed
HCA and SVM classifier model
The overlapping genes among the HTLV-1 infection
pathway-associated genes and DEGs were subjected to further
analysis. Two-way HCA was performed based on the expression values
of the candidate genes using the heatmap2 package in R (version
3.5.3 for CentOS Linux; release 7.5.1804) (24,25).
The SVM classifier was constructed using a support
vector classification function in Sklearn.svm package, with the Rbf
Kernel function and a three-fold cross-validation strategy. In
addition, a random seed was set as 100 to shuffle the training set.
The effects of the classification were evaluated based on six
parameters, including accuracy, sensitivity, specificity, positive
predictive value (PPV), negative predictive value (NPV) and area
under the curve (AUC).
Verification of the proposed HCA and
SVM classification model in the test set
The robustness and portability of the two-way HCA
and SVM classifier were based on the overlapping genes feature and
were conducted sequentially to further verify classification
reliability through computing the remaining data resources as a
test set.
Prognostic value of candidate
signature genes
In the TCGA tumor set, associations between the
expression of candidate genes and prognosis were tested using the
survival package (version 2.44) in R. Patient survival time
differences between high and low expression groups (defined by
median expression) of 41 candidate genes were analyzed.
Kaplan-Meier survival curves were constructed with the log-rank
test and P-values were calculated. The cut-off threshold was set to
P<0.05.
Results
Quality assessment of candidate sets
and identification of selected feature genes
PCA analysis of the normalized gene expression
profile data from the two databases was performed to detect
homogeneity between candidate chips, using Sklearn.svm package.
Different groups of data exhibited large differences, showing their
heterogeneity, in PCA analysis (Fig.
1), implying that the quality of the candidate dataset was
suitable for subsequent analysis. A total of 4,381 DEGs were
identified between normal samples and control samples using the
DESeq2 package. Of these, 2,136 were upregulated and 2,245 were
downregulated (data not shown). The 41 overlapping genes on the
candidate HTLV-1 infection pathway and the DEGs were selected for
further analysis. The 41 genes are presented in Table I.
 | Table I.Detailed information of the 41
candidate genes downregulated in patients with endometrial
carcinoma. |
Table I.
Detailed information of the 41
candidate genes downregulated in patients with endometrial
carcinoma.
| Downregulated gene
name | Log2 fold change | P-value | Adjusted P-value |
|---|
| ZFP36 ring finger
protein | −2.89 |
4.22×10−6 |
1.15×10−4 |
| Cyclin D2 | −3.29 |
8.66×10−16 |
2.20×10−13 |
| Early growth response
2 | −1.79 |
7.06×10−4 |
8.43×10−3 |
| Wnt family
member9B | −4.29 |
4.93×10−6 |
1.09×10−4 |
| Major
histocompatibility complex, class I, E | −1.09 |
5.74×10−4 |
7.18×10−3 |
| Ras related | −1.79 |
2.64×10−7 |
1.05×10−5 |
| Serum response
factor | −1.06 |
8.38×10−4 |
9.70×10−3 |
| ETS proto-oncogene 1,
transcription factor | −1.49 |
1.79×10−5 |
3.97×10−4 |
| Mitogen-activated
protein kinase kinase kinase 3 | −1.53 |
4.00×10−5 |
7.93×10−4 |
| Adenylate cyclase
8 | −7.81 |
2.42×10−6 |
7.19×10−5 |
| Early growth response
1 | −3.48 |
1.07×10−8 |
6.08×10−7 |
| Adenylate cyclase
2 | −3.72 |
3.90×10−10 |
3.07×10−8 |
| Talin 1 | −1.74 |
5.95×10−6 |
1.54×10−4 |
| Nuclear factor of
activated T cells 2 | −1.27 |
2.98×10−3 |
2.66×10−2 |
| Mitogen-activated
protein 14 | −1.07 |
5.38×10−3 |
4.14×10−2 |
| Adenylate cyclase
3 | −1.12 |
2.60×10−3 |
2.40×10−2 |
| AKT serine/threonine
kinase 3 | −3.82 |
4.09×10−21 |
3.53×10−18 |
| Fos proto-oncogene,
AP-1 transcription factor subunit | −3.54 |
1.14×10−5 |
2.70×10−4 |
| Frizzled class
receptor 7 | −1.83 |
5.10×10−6 |
1.35×10−4 |
| Transforming growth
factor β 3 | −1.20 |
5.90×10−4 |
7.33×10−3 |
| Adenylate cyclase
4 | −2.16 |
1.62×10−7 |
6.70×10−6 |
| Vascular cell
adhesion molecule 1 | −2.20 |
5.14×10−3 |
4.00×10−2 |
| Adenylate cyclase
9 | −1.51 |
2.54×10−5 |
5.34×10−4 |
| Activating
transcription factor 3 | −2.34 |
2.68×10−4 |
3.92×10−3 |
| Platelet derived
growth factor receptor α | −3.08 |
1.22×10−4 |
2.03×10−3 |
| Frizzled class
receptor 4 | −1.98 |
8.85×10−9 |
5.09×10−7 |
| CD40 molecule | −1.21 |
3.60×10−3 |
3.07×10−2 |
| Neuropilin 1 | −1.39 |
9.01×10−6 |
2.20×10−4 |
| Signal transducer
and activator of transcription 5B | −1.86 |
7.82×10−9 |
4.56×10−7 |
| Signal transducer
and activator of transcription 5A | −1.50 |
3.51×10−5 |
7.08×10−4 |
| SMAD family member
3 | −1.30 |
5.96×10−5 |
1.11×10−3 |
| Nuclear factor of
activated T cells 1 | −1.07 |
6.87×10−3 |
4.97×10−2 |
| WNT family member
2B | −2.59 |
1.07×10−4 |
1.80×10−3 |
| Platelet derived
growth factor receptor β | −1.71 |
2.61×10−6 |
7.65×10−5 |
| Transforming growth
factor β receptor 2 | −2.26 |
8.00×10−6 |
1.98×10−4 |
| Jun proto-oncogene,
AP-1 transcription factor subunit | −1.59 |
5.98×10−5 |
1.11×10−3 |
| Muscle RAS oncogene
homolog | −1.30 |
2.31×10−3 |
2.18×10−2 |
|
Phosphatidylinositol-4,5-bisphosphate
3-kinase catalytic subunit Δ | −1.31 |
3.28×10−3 |
2.86×10−2 |
| WNT family member
4 | −1.98 |
6.85×10−5 |
1.24×10−3 |
| Adenylate cyclase
5 | −2.42 |
4.12×10−8 |
2.01×10−6 |
| MYC proto-oncogene,
bHLH transcription factor | −1.25 |
2.98×10−3 |
2.66×10−2 |
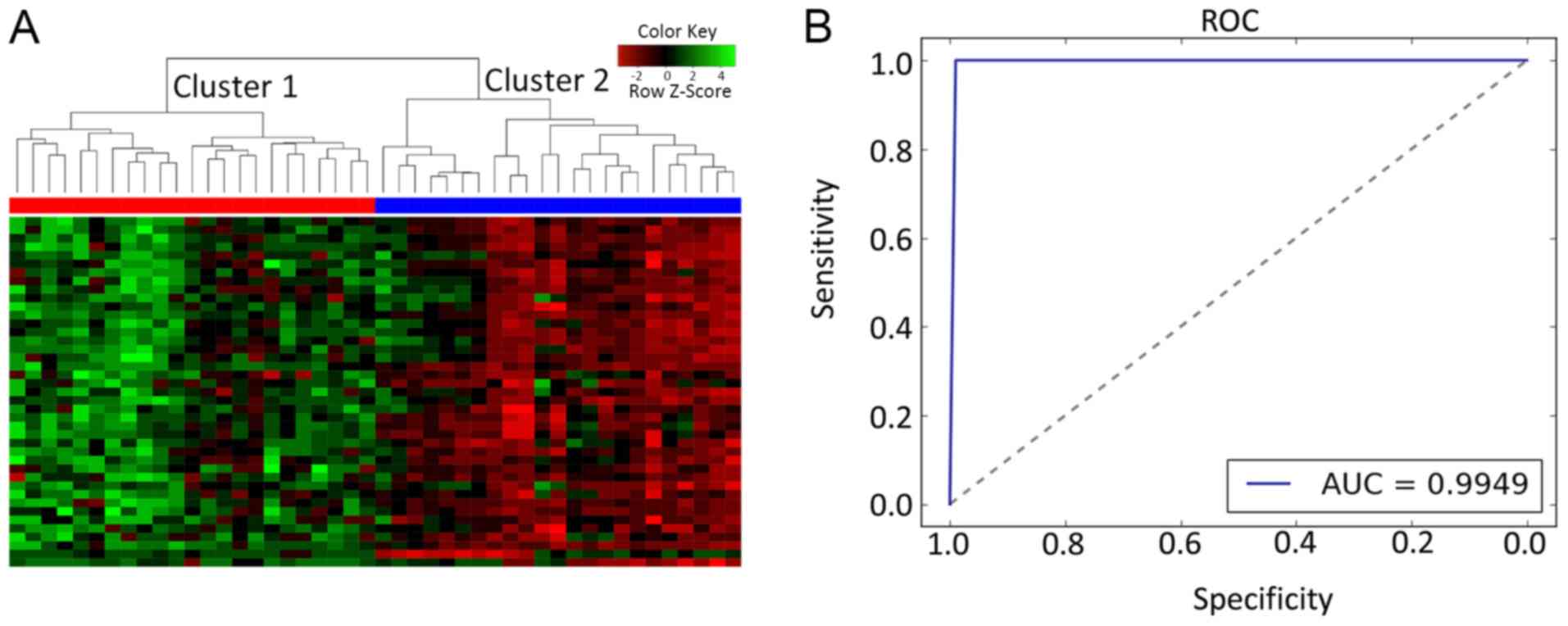
HCA of differentially expressed
mRNAs
A total of 41 candidate HTLV-1 infection
pathway-associated logarithmic expression values were subjected to
HCA on the training set. The results revealed that all samples were
distinctly subdivided into two clusters. The accuracy was 100%
(46/46), and all the tumor samples (n=23) and all the normal
control samples (n=23) were incorporated into two individual
clusters (Fig. 2A).
Assessment on the training dataset
using an SVM-based method
To further confirm whether the candidate genes may
be used to discriminate between tumor and control samples, the SVM
model with equal sensitivity and specificity (sensitivity, 100.00%;
specificity, 100.00%) was proposed. The results indicated that the
41 genes provided an accuracy of 99.49%, with PPV of 100.00%, NPV
of 100.00% and AUC of 99.49% (Fig.
2B).
The 41 candidate genes performed with high
specificity and sensitivity in the training set. The tissue pairs
were classified into a tumor group and a normal match group via the
SVM classifier: Tumor samples were set as a positive group, while
normal samples were set as a negative group.
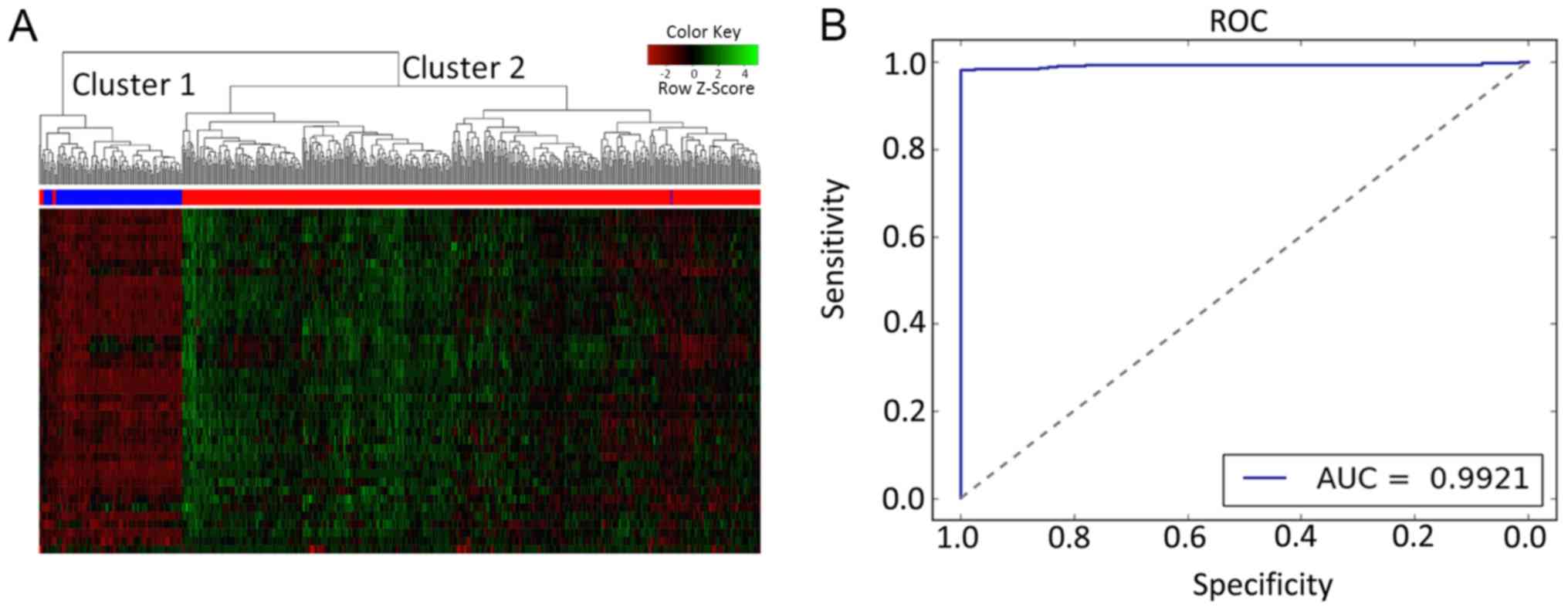
Validation of the proposed HCA and SVM
classification model in the test set
The performance of the two-way HCA and SVM model was
measured on the basis of 41 candidate genes, which were tested on
the merged datasets [remaining TCGA-endometrial cancer samples
(n=541) and GTEx (n=111)]. The results of two-way HCA indicated
that all the samples in the validation dataset were stratified into
two groups. The accuracy was 98.77% (Fig. 3A). A total of seven tumor samples
were incorrectly clustered into the normal group, and one normal
sample was incorrectly clustered into the tumor group. Similarly,
the SVM model correctly distinguished between the tumor and normal
samples. The SVM model attained high accuracy (98.16%), with the
AUC, sensitivity, specificity, PPV and NPV reaching 99.21, 97.73,
100.00, 100.00 and 91.11%, respectively. Taken together, the
results obtained suggested that the 41 candidate genes may reliably
distinguish between normal and tumor samples in EC.
Survival time analysis of the
candidate genes
To further demonstrate the classification
reliability and prognostic potential of the candidate genes, the
survival time of patients with EC was assessed. Of the 529 tumor
samples in the test datasets, 13 out of 41 candidate genes were
associated with poor overall survival time and included Wnt family
member 9B (WNT9B), serum response factor (SRF), mitogen-activated
protein kinase 3 (MAP3K3), adenylate cyclase (ADCY) 3, frizzled
class receptor (FZD) 7, ADCY9, FZD4, SMAD family member 3 (SMAD3),
transforming growth factor beta receptor 2 (TGFBR2), Jun
proto-oncogene, AP-1 transcription factor subunit (JUN), muscle RAS
oncogene homolog (MRAS), ADCY5 and MYC proto-oncogene, bHLH
transcription factor (MYC). The survival time of the low-expression
group was increased compared with the high expression group for all
aforementioned 13 genes (Fig. 4).
The results obtained suggest that the candidate genes identified in
the current study have potential prognostic value in EC.
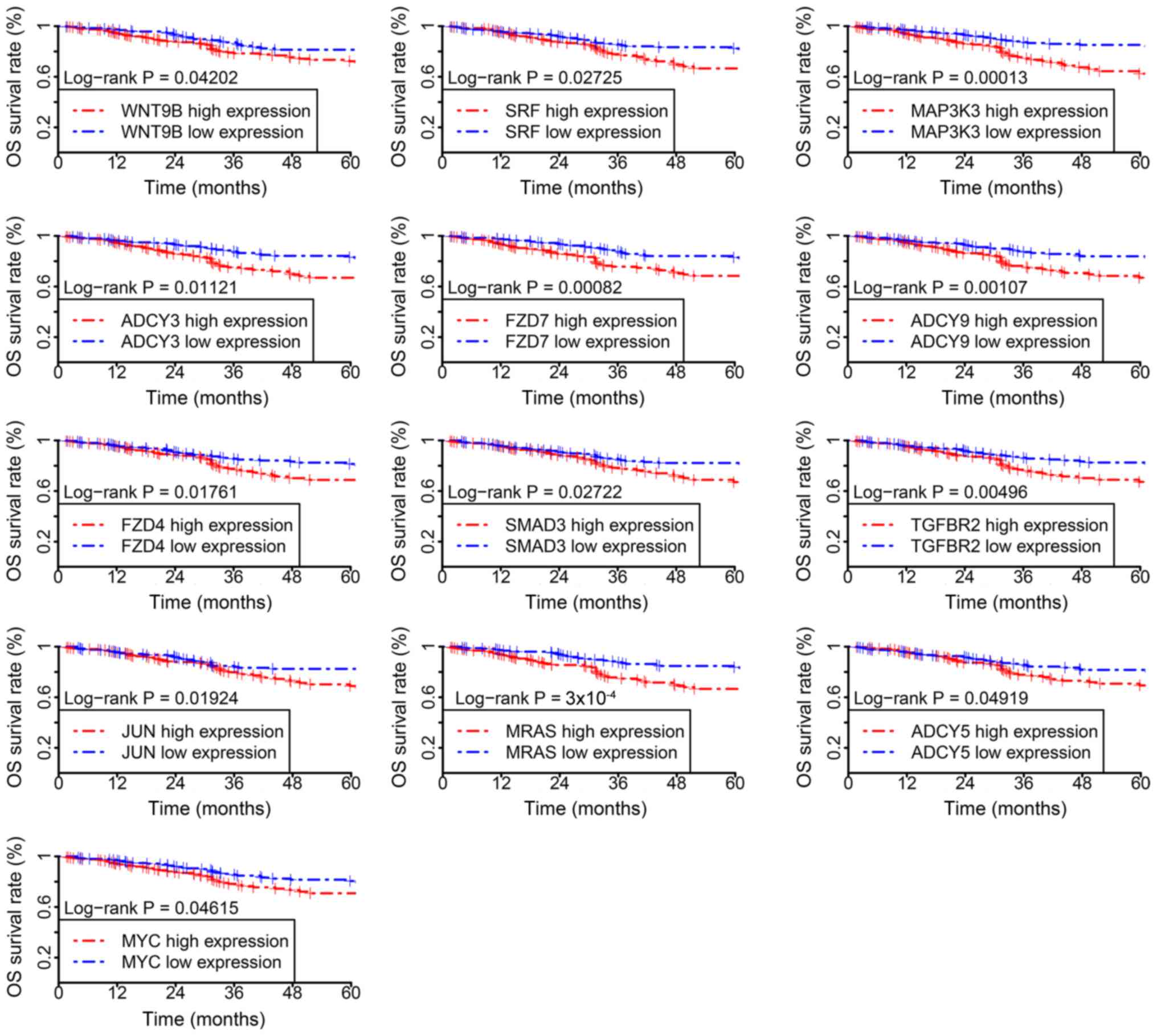 | Figure 4.Kaplan-Meier survival curves for the
high-expression (red) and low-expression (blue) groups. WNT9B, Wnt
family member 9B; SRF, serum response factor; MAP3K3,
mitogen-activated protein kinase 3; ADCY, adenylate cyclase; FZD,
frizzled class receptor; SMAD3, SMAD family member 3; TGFBR2,
transforming growth factor beta receptor 2; JUN, Jun
proto-oncogene, AP-1 transcription factor subunit; MRAS, muscle RAS
oncogene homolog; MYC, MYC proto-oncogene, bHLH transcription
factor. |
Discussion
HTLV-1 is a human retrovirus associated with the
development of various types of cancer, including liver, gastric
and blood cancer (25–28). A 24-year cohort inpatient study in
Japan revealed that the prevalence of HTLV-1 infection in males and
females was 12.3 and 15.5% respectively, and that HTLV-1 infection
was more prevalent in females than in males (16). However, to the best of our knowledge,
the association of HTLV-1 infection with the development of EC has
not been investigated.
In the current study, PCA analysis was used to
assess the data structure of the chosen datasets, and results
indicated that the selected datasets were suitable for further
analysis. Subsequently, comprehensive bioinformatics analyses were
used to identify DEGs between normal samples and endometrial cancer
samples. A total of 4,381 DEGs associated with endometrial cancer
were identified, including 2,136 upregulated and 2,245
downregulated DEGs. A total of 41 candidate genes were found
overlapping between the DEGs and the HTLV-1 infection pathway. The
41 candidate downregulated genes in patients with EC included the
following tumor-associated genes: AKT3, ZFP36, CCND2, EGR2, TGFB3,
JUN, MRAS, PIK3CD, WNT4, ADCY5 and MYC. To further classify the
association between HTLV-1 infection and EC risk, the 41 candidate
genes were selected for two-way HCA and to train the SVM
classifier. The 23 normal samples and the matched tumor samples
were used in the trial. The results obtained demonstrated that the
candidate genes performed well, and the accuracy was 100%. The
classification capability of the feature genes was verified with
the merged-dataset that included 123 normal samples and 529 tumor
samples. Two-way HCA and SVM classifier analysis produced
consistent results, suggesting that the 41 candidate genes
exhibited a potential association between HTLV-1 infection and
EC.
Survival analysis was performed and 13 genes were
associated with the overall survival. These included WNT9B, SRF,
MAP3K3, ADCY3, FZD7, ADCY9, FZD4, SMAD3, TGFBR2, JUN, MRAS, ADCY5
and MYC. These genes may be implicated in the tumorigenesis of
endometrial cancer by serving a role in the regulation of the
HTLV-1 infection-signaling pathway. While the current study did not
demonstrate that HTLV-1 infection is associated with endometrial
cancer directly; it revealed that HTLV-1 infection-associated genes
may be associated with endometrial cancer.
In conclusion, the present study identified 41
HTLV-1 infection-associated DEGs which may be involved in the
development of EC. Future experiments are required to substantiate
the results obtained in the current study in laboratory
experiments. The results obtained in the current study may pave the
way for future experimental research to elucidate the mechanisms
underlying the development of EC. Additionally, it may promote the
advancement of the diagnostic and prognostic tools in EC and
facilitate the development of novel therapeutic targets.
Acknowledgements
Not applicable.
Funding
The current study was funded by the Guangzhou
Science and Technology Projects (grant no. 201707010265) and the
Guangdong Provincial Science and Technology Projects (grant no.
2016ZC0049).
Availability of data and materials
All data used in this study were downloaded from The
Cancer Genome Atlas database (https://portal.gdc.cancer.gov/; Project ID: TCGA-UCEC)
or the Genotype-Tissue Expression project (version 7; www.gtexportal.org).
Authors' contributions
YW conceived and designed the study. GD and ZZ
analyzed and interpreted the data. WZ and MZ were responsible for
data acquisition, processing and analysis. In addition WZ and MZ
were involved in the drafting and critical revision of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Koh WJ, Abu-Rustum NR, Bean S, Bradley K,
Campos SM, Cho KR, Chon HS, Chu C, Cohn D, Crispens MA, et al:
Uterine neoplasms, version 1.2018, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 16:170–199. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahecha AM and Wang H: The influence of
vascular endothelial growth factor-A and matrix metalloproteinase-2
and −9 in angiogenesis, metastasis, and prognosis of endometrial
cancer. Onco Targets Ther. 10:4617–4624. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
sstatistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Malik TY, Chishti U, Aziz AB and Sheikh I:
Comparison of risk factors and survival of type 1 and type II
endometrial cancers. Pak J Med Sci. 32:886–890. 2016.PubMed/NCBI
|
|
5
|
Mittica G, Ghisoni E, Giannone G, Aglietta
M, Genta S and Valabrega G: Checkpoint inhibitors in endometrial
cancer: Preclinical rationale and clinical activity. Oncotarget.
8:90532–90544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fakhar S, Saeed G, Khan AH and Alam AY:
Validity of pipelle endometrial sampling in patients with abnormal
uterine bleeding. Ann Saudi Med. 28:188–191. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yoshida M: Discovery of HTLV-1, the first
human retrovirus, its unique regulatory mechanisms, and insights
into pathogenesis. Oncogene. 24:5931–5937. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
IARC working group on the evaluation of
carcinogenic risks to humans, . Human immunodeficiency viruses and
human T-cell lymphotropic viruses. Lyon, France, 1–18 June 1996.
IARC Monogr Eval Carcinog Risks Hum. 67:1–424. 1996.PubMed/NCBI
|
|
9
|
Human T-cell lymphotropic viruses, . IARC
Monogr Eval Carcinog Risks Hum. 67:261–390. 1996.PubMed/NCBI
|
|
10
|
de Thé G and Kazanji M: An HTLV–I/II
vaccine: From animal models to clinical trials? J Acquir Immune
Defic Syndr Hum Retrovirol. 13 (Suppl 1):S191–S198. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martín-Dávila P, Fortún J, López-Vélez R,
Norman F, Montes de Oca M, Zamarrón P, González MI, Moreno A,
Pumarola T, Garrido G, et al: Transmission of tropical and
geographically restricted infections during solid-organ
transplantation. Clin Microbiol Rev. 21:60–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hollsberg P and Hafler DA: Seminars in
medicine of the Beth Israel Hospital, Boston. Pathogenesis of
diseases induced by human lymphotropic virus type I infection. New
Eng J Med. 328:1173–1182. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Proietti FA, Carneiro-Proietti AB,
Catalan-Soares BC and Murphy EL: Global epidemiology of HTLV–I
infection and associated diseases. Oncogene. 24:6058–6068. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kannian P and Green PL: Human T
lymphotropic virus type 1 (HTLV-1): Molecular biology and
oncogenesis. Viruses. 2:2037–2077. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamanaka S, Nakayama K, Tamai H, Sakamaki
M and Inokuchi K: Adult T-cell leukemia-lymphoma complicated by
Takotsubo cardiomyopathy and HTLV-1-associated myelopathy after
treatment with the anti-CCR4 antibody mogamulizumab. Rinsho
Ketsueki. 58:309–314. 2017.(In Japanese). PubMed/NCBI
|
|
16
|
Tanaka T, Hirata T, Parrott G,
Higashiarakawa M, Kinjo T, Kinjo T, Hokama A and Fujita J:
Relationship among strongyloides stercoralis infection, human
T-cell lymphotropic virus type 1 infection, and cancer: A 24-year
cohort inpatient study in Okinawa, Japan. Am J Trop Med Hyg.
94:365–370. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Howard C: The Interplay between HTLV-1
Viral Factors, Tax and HBZ, During T-cell Transformation. The Ohio
State University. 2016.
|
|
18
|
Vicario M, Mattiolo A, Montini B, Piano
MA, Cavallari I, Amadori A, Chieco-Bianchi L and Calabrò ML: A
preclinical model for the atll lymphoma subtype with insights into
the role of microenvironment in HTLV-1-mediated lymphomagenesis.
Front Microbiol. 9:12152018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen JL, Limnander A and Rothman PB: Pim-1
and Pim-2 kinases are required for efficient pre-B-cell
transformation by v-Abl oncogene. Blood. 111:1677–1685. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo G, Qiu X, Wang S, Chen Y, Rothman PB,
Wang Z, Chen Y, Wang G and Chen JL: Oncogenic E17K mutation in the
pleckstrin homology domain of AKT1 promotes v-Abl-mediated
pre-B-cell transformation and survival of Pim-deficient cells.
Oncogene. 29:3845–3853. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang J, Wang J, Chen K, Guo G, Xi R,
Rothman PB, Whitten D, Zhang L, Huang S and Chen JL: eIF4B
phosphorylation by pim kinases plays a critical role in cellular
transformation by Abl oncogenes. Cancer Res. 73:4898–4908. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Love M, Anders S and Huber W: Differential
analysis of count data - the DESeq2 package. Genome Biol.
15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Therneau TM: Survival analysis [R package
survival version 2.41–3]. Technometrics. 46:111–112. 2015.
|
|
25
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA-seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arisawa K, Soda M, Akahoshi M, Fujiwara S,
Uemura H, Hiyoshi M, Takeda H, Kashino W and Suyama A: Human T-cell
lymphotropic virus type-1 infection and risk of cancer: 15.4 year
longitudinal study among atomic bomb survivors in Nagasaki, Japan.
Cancer Sci. 97:535–539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kozuru M, Uike N, Muta K, Goto T, Suehiro
Y and Nagano M: High occurrence of primary malignant neoplasms in
patients with adult T-cell leukemia/lymphoma, their siblings, and
their mothers. Cancer. 78:1119–1124. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Beltran BE, Quiñones P, Morales D, Revilla
JC, Alva JC and Castillo JJ: Diffuse large B-cell lymphoma in human
T-lymphotropic virus type 1 carriers. Leuk Res Treatment.
2012:2623632012.PubMed/NCBI
|