Introduction
Hepatocellular carcinoma (HCC) is the most common
type of liver cancer, its survival rate ranks only second to lung
cancer and it is a severe threat to human health (1–4).
However, the pathophysiological mechanisms involved remain unclear.
The occurrence of HCC is a complicated process involving multiple
genes and steps. Imbalances in cellular signal transduction
pathways, deficiencies in DNA repair-regulating genes, activation
of protooncogenes, inactivation of tumor suppressor genes and
epigenetic modifications all promote the occurrence of liver cancer
(1–4).
Epigenetics refers to the regulation of gene
expression by affecting a gene's transcription and translation
without changing DNA sequences, including DNA methylation, histone
modification and abnormal miRNA expression (5,6). DNA
methylation has been widely studied in a number of types of tumor
(7). Methylation of DNA leads to the
inactivation of tumor suppressor genes and promotes the occurrence
and development of tumors (8).
Reversion of DNA methylation events has been reported to inhibit
the growth of tumor cells and promote tumor cell apoptosis
(9).
Deuterosome assembly protein 1 (DEUP1) is a new
candidate tumor suppressor gene and is associated with cellular
signal transduction during tumor formation. Bioinformatics methods
revealed that DEUP1expression was closely associated with the
survival time of patients with HCC. Through database analysis, it
was also demonstrated that the inactivation of DEUP1 was correlated
with the methylation of its promoter. DEUP1 expression is absent or
reduced in malignant tumors, such as gastric and thyroid cancer
(10,11). However, its expression in HCC and the
association with clinical information have not been reported on, to
the best of our knowledge.
The present study was undertaken to explore the
effects of DEUP1 in HCC, reverse transcription-polymerase chain
reaction (RT-PCR), bisulfite PCR sequencing (BSP),
immunohistochemistry (IHC) and western blotting were conducted to
detect methylation of the DEUP1 promoter and DEUP1 expression in 60
cases HCC and adjacent non-tumor tissues, and explore the
correlations between DEUP1 and pathological features.
Materials and methods
Clinical information
HCC and adjacent non-tumor tissues (at least 3 cm
from the surgical incision) were collected from 60 patients who
underwent surgical resection between January 2016 and December 2016
at the First Affiliated Hospital of Zhengzhou University. All
specimens were confirmed by pathological diagnosis. No patients
underwent radiotherapy or chemotherapy prior to surgery. A total of
45 males and 15 females, aged 31–75 years old (median, 58 years),
were recruited to the study. According to tumor node and metastasis
(TNM) staging of the AJCC 2018 (12), 32 patients were stage I+II, and 28
were stage III+IV. Informed consent was obtained from each patient
and the study protocol was approved by the Medical Ethics Committee
of the First Affiliated Hospital of Zhengzhou University.
Relationship between DEUP1 mRNA
expression and overall survival
Based on the KM Plotter Online Tool (http://kmplot.com/analysis/), 364 patients with HCC
were divided into two groups according to the median expression of
DEUP1 and Kaplan-Meier survival curve was then plotted. The
best cutoff was auto-selected.
DEUP1 mRNA expression detected by
RT-PCR
HCC and adjacent non-tumor tissues (100 mg each)
were used. Total RNA was extracted using TRIzol (Invitrogen;
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol and the RNA concentration and A260/A280 ratio were
measured using a Nanodrop 2000 (Thermo Fisher Scientific, Inc.).
RNA (1 µg) was transcribed to cDNA using a reverse transcription
kit (cat. no. RR047A; Takara Biotechnology Co., Ltd.) according to
the manufacturer's protocol. The primers were designed based on the
gene coding sequence. DEUP1: 5′-CCTTCGACATTTCAAGCCAAAGA-3′
(forward primer) and 5′-GAAATGCTGTGCAGCCAAAGA-3′ (reverse primer).
GAPDH: 5′-CGCTGAGTACGTCGTGGAGT-3′ (forward primer) and
CATCACGCCACAGTTTCCCG-3′ (reverse primer). TB Green Master Mix kit
(Takara Biotechnology Co., Ltd.) was used with a total reaction
volume of 20 µl containing 10 µl of 2X TB Green Master Mix, 2 µl of
cDNA, 0.8 µl of upstream and downstream primers each, and 6.4 µl of
ddH2O. Reaction conditions were as follows:
Pre-denaturation at 95°C for 30 sec, denaturation at 95°C for 5 sec
and annealing at 59.5°C for 30 sec, for a total of 40 cycles. Human
GAPDH was used as an internal reference (5 µl) and loaded
with PCR product (5 µl) and 6X DNA loading buffer (1 µl). After 2%
agarose gel electrophoresis, the ratio of DEUP1 to GAPDH was
compared using the average value of normal tissue as the standard.
A ratio higher than the value of the standard or within the range
was considered to indicate positive gene expression and no band
present or a band lower than the normal range indicated no gene
expression. Image J version 1.8.0 (National Institutes of Health)
was used to semi-quantitatively analyze the gray scale ratio of
target gene and GAPDH.
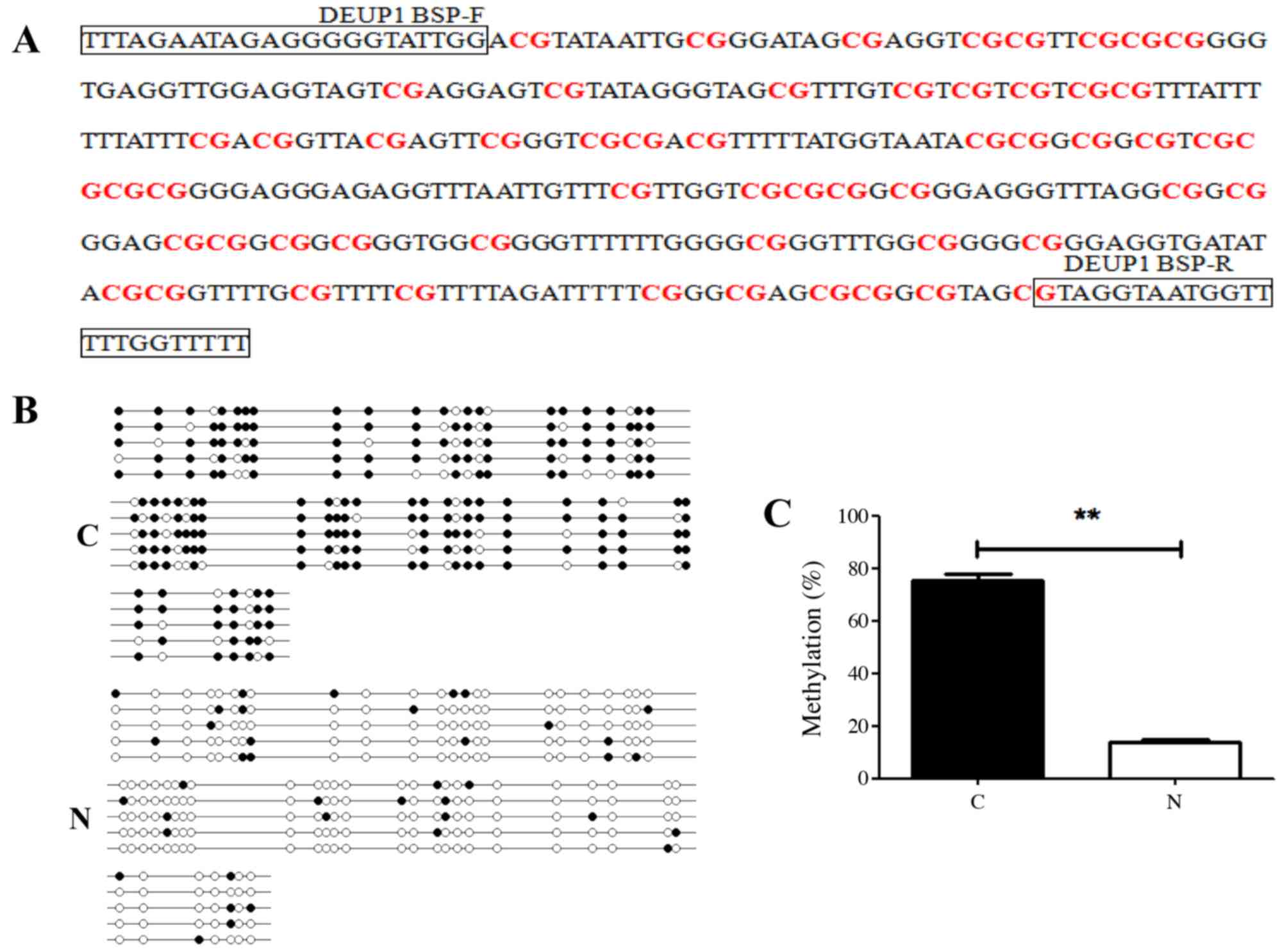
DEUP1 promoter methylation detected by
BSP
DNA in the tissues was extracted using a TINamp
Genomic DNA kit (Sangon Biotech, Co., Ltd.) and resolved via 1%
agarose gel electrophoresis. The absorbance (260/280) was measured
with a UV spectrophotometer to calculate DNA content. The DNA was
modified with sulfite using an EZ DNA Methylation-Gold™ kit D5005
(Zymo Research Corp., Irvine, CA, USA). The primers were designed
using Primer Premier 5 (Premier Biosoft International). BSP primers
were as follows: Upstream: 5′-TTTAGAATAGAGGGGGTATTGG-3′;
downstream: 5′-AAAAACCAAAAACCATTACCTAC-3′. The BSP reaction volume
was 20 µl. Cycle parameters were as follows: 95°C pre-denaturation
for 5 min, 95°C denaturation for 30 sec, 61°C annealing for 30 sec
and 72°C extension for 50 sec, for a total of 35 cycles, followed
by extension at 72°C for 8 min. The integrity of the PCR product (5
µl) was established by 1% agarose gel electrophoresis. The PCR
product was ligated with a T-vector to generate 10 µl linking
product that was transferred to 100 µl SK9307 competent cells using
the Rapid Competent Cell Preps kit (cat. no. B529307; Sangon
Biotech, Co., Ltd.). After screening using LB culture medium
containing ampicillin (cat. no. A600894; Sangon Biotech, Co.,
Ltd.), five independent colonies were picked. The target fragment
was identified by PCR and the products were sequenced.
DEUP1 protein changes detected by
IHC
Paraffin sections of HCC and adjacent non-tumor
tissues at 4 µm thickness were dewaxed and rehydrated with graded
alcohol. Citric acid buffer was used for antigen retrieval under
high temperature (heated to boiling and rested for 15 min at room
temperature) Then, the sections were washed with PBS, blocked with
normal goat serum (Beijing Solarbio Science & Technology Co.,
Ltd.) for 30 min at room temperature and then incubated with a
DEUP1 primary antibody (1:200; cat. no. FLJ25393; Absin) at 4°C
overnight. Afterwards, the sections were washed with PBS, incubated
with a secondary antibody (1:100; cat. no. SP0021; Beijing Solarbio
Science & Technology Co., Ltd.) labeled with biotin for 1 h at
room temperature, washed again with PBS, incubated with horseradish
peroxidase (HRP)-labeled streptavidin and then a DAB chromogenic
reagent, washed with running water, re-stained with hematoxylin at
room temperature until nuclei turned blue, dehydrated with gradient
ethanol, sealed with gum, and then observed under a light
microscope. The IHC results indicated faint yellow or even dark
brown granules. Positive cell counting was scored as follows:
<5% was scored as 0, 5–25% as 1, 25–50% as 2, 50–75% as 3 and
>75% as 4. Color intensity was scored as follows: No color was
scored as 0, faint yellow as 1, pale brown as 2 and dark brown as
3. If the product of the two scores was ≥4, it was deemed
positive.
DEUP1 protein expression detected by
western blotting
A total of 100 mg of liver tissue, 1 ml RIPA
(Beijing Solarbio Science & Technology Co., Ltd.) and 10 µl
PMSF were placed into an EP tube and fully broken with a tissue
breaker. The protein was extracted using RIPA buffer and the
concentration was measured using a bicinchoninic acid Protein Assay
kit (Beijing Solarbio Science & Technology Co., Ltd.). Then, 40
µg of total protein was resolved by SDS-PAGE (10%) and transferred
onto 0.45 µm nitrocellulose membranes. The membranes were blocked
with 5% non-fat milk at room temperature for 1 h, following which
primary antibodies against DEUP1 (1:1,000; cat. no. ab102688;
Abcam) and GAPDH (1:1,000; Cell Signaling Technology, Inc.; cat.
no. 5174) were added and incubated at 4°C overnight. The membranes
were washed three times with 1X TBS containing 1% Tween-20 (10 min
each time) before and after incubation with the secondary antibody
(goat anti rabbit IgG-HRP; Cell Signaling Technology, Inc.; cat.
no. 7074) diluted with 1X TBST at 1:2,000. The blots were
visualized using ECL (cat. no. 32106; Thermo Fisher Scientific,
Inc.) in a dark room. The protein expression levels for each
specimen were calculated using Quantity-One 4.6.6 (Bio-Rad
Laboratories, Inc.).
Statistical analysis
All data are presented as the mean ± standard
deviation. SPSS 22.0 software (IBM, Corps.) was used to analyze the
data. The statistical significance between two groups of
quantitative data were calculated by Student's t-test. A comparison
of constituent ratios was conducted using the χ2 test
and χ2 test of paired quadrilaterals. Correlations in
the data were identified and evaluated using correlation analysis
of paired quadrilaterals. P<0.05 was considered to indicate a
statistically significant difference.
Results
Promoter DEUP1 hypermethylation in HCC
tissues
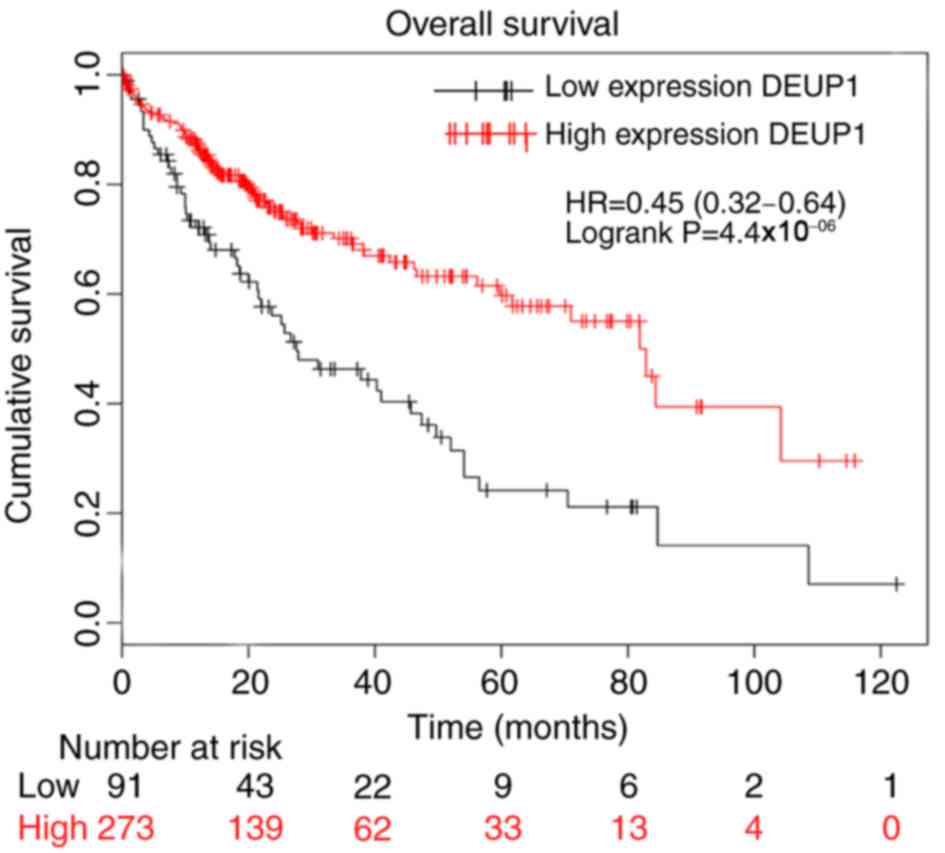
The bioinformatic analysis indicated that increased
expression of DEUP1 was associated with a higher rate of patient
overall survival (Fig. 1) and the
degree of promoter methylation in HCC tissues was significantly
increased compared with the adjacent non-cancerous tissues
(P<0.01; Fig. 2). This suggests
that DEUP1 may be a tumor suppressor gene and promoter methylation
may play an important role in the development of HCC occurrence.
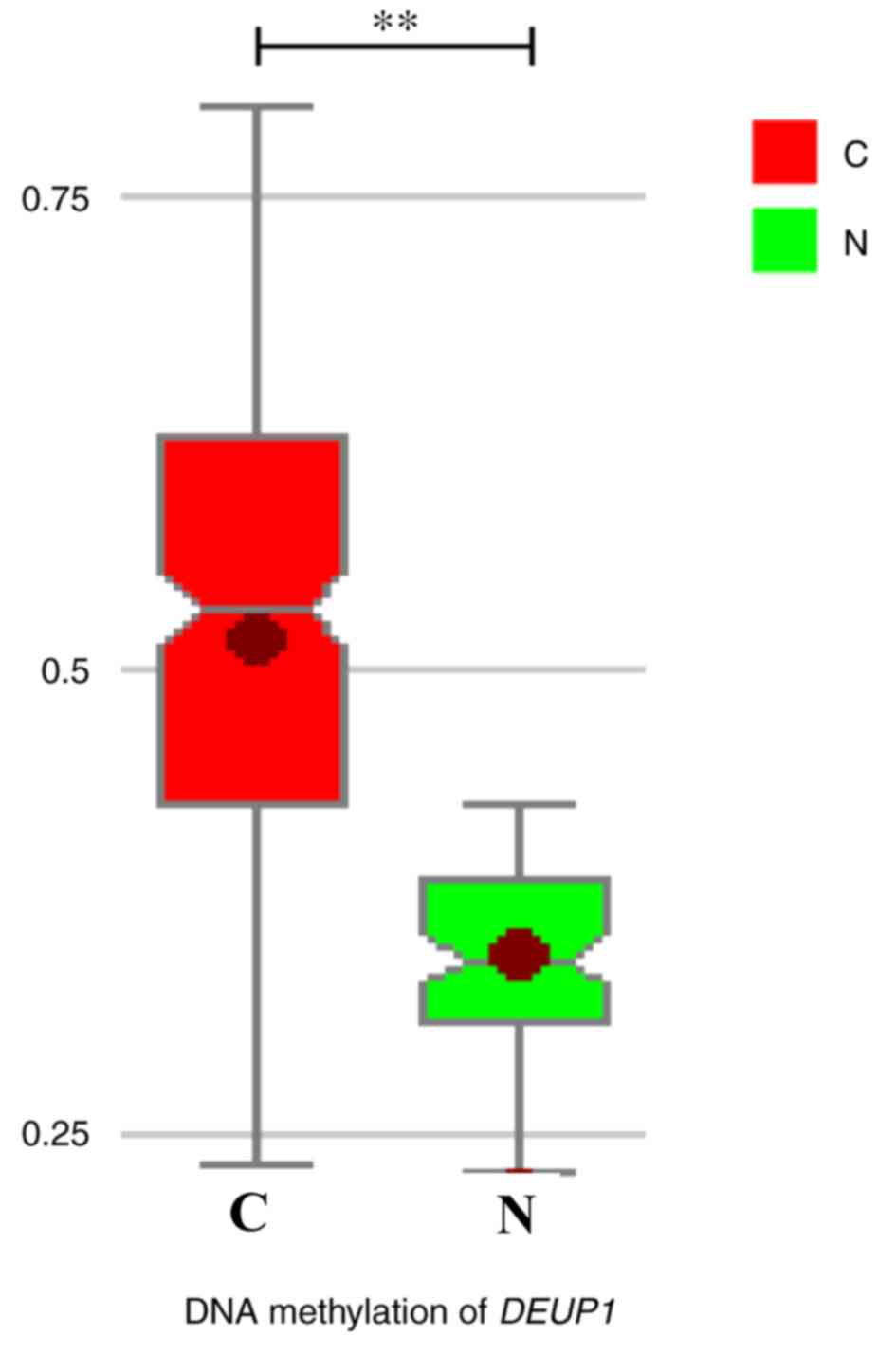
The results of BSP demonstrated that DEUP1 promoter
hypermethylation was detected in 46 of 60 (76.7%) tumors tissues,
while only 5 of 60 in adjacent non-tumor tissues. DEUP1 promoter
methylation levels in HCC were tissues significantly increased
compared with the adjacent non-tumor tissues (P<0.01; Fig. 3).
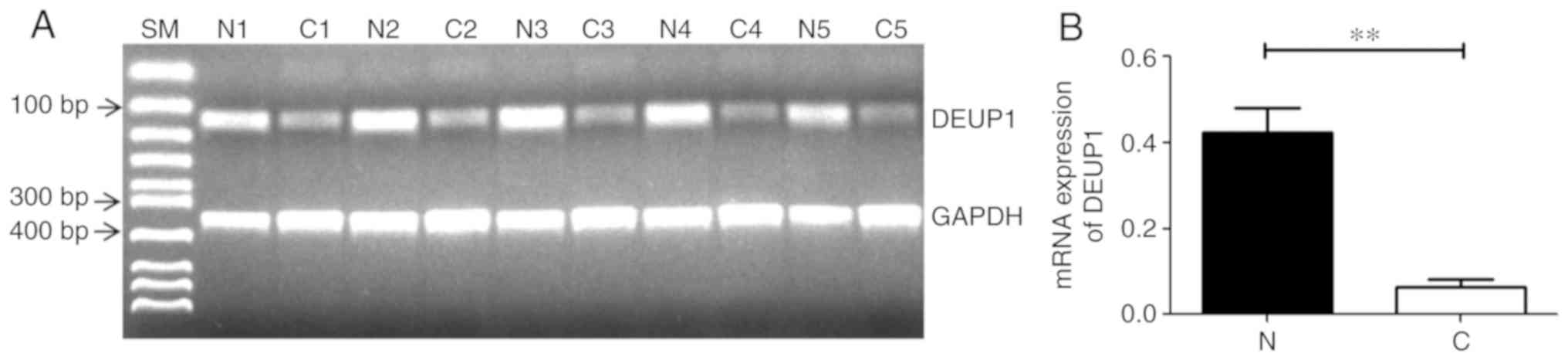
DEUP1 mRNA expression in HCC tumor and
adjacent non-tumor tissues
RT-PCR results demonstrated that 45 of the 60 HCC
tissues had reduced or absent DEUP1 mRNA expression compared with
adjacent non-cancerous tissues. All adjacent non-tumor tissues
showed DEUP1 expression. Representative results are shown in
Fig. 4; the expression of DEUP1 mRNA
in the adjacent non-cancerous tissues was significantly increased
compared with the HCC tissues (P<0.01).
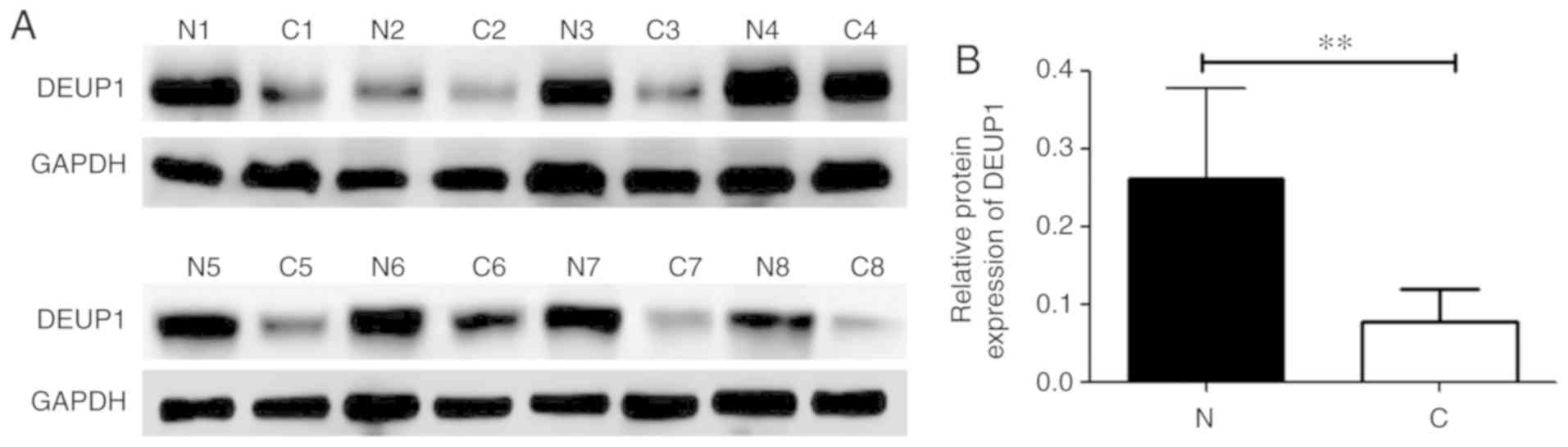
DEUP1 protein expression in HCC tumor
and adjacent non-tumor tissues
The expression of DEUP1 protein was further analyzed
in the 60 HCC tumor and adjacent non-tumor tissues. IHC results
revealed that the positive expression of the DEUP1 protein was
mainly located in the cytoplasm, represented by yellow or
pale-brown granules (Fig. 5). A
total of 48 out of the 60 HCC tumor tissues showed low or no DEUP1
protein expression. Both IHC and western blotting indicated that
the expression of DEUP1 in adjacent non-tumor tissues was
significantly increased compared with in HCC tissues (P<0.01;
Fig. 6).
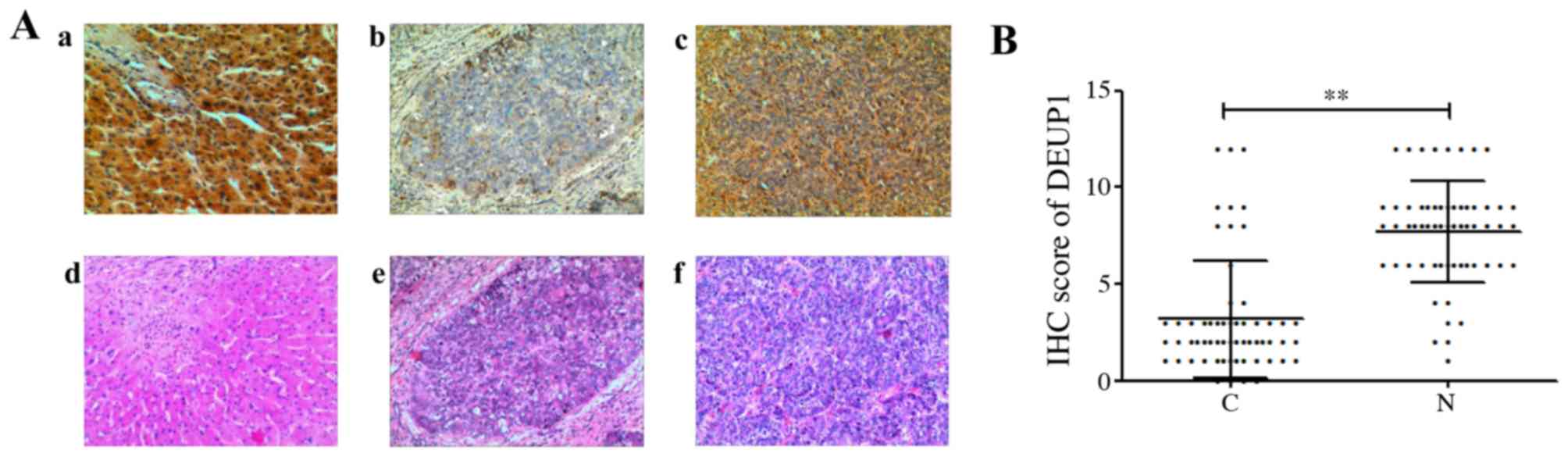 | Figure 5.Results of IHC and HE stain
(magnification, ×100). (Aa) High expression in adjacent non-tumor
tissues, (Ab) negative expression of DEUP1 in HCC tissue, (Ac)
positive expression of DEUP1 in HCC tissue. (Ad-f) are the
corresponding HE stain of adjacent non-tumor and HCC tissues. (B)
IHC scores of DEUP1 in tumor tissues and adjacent non-tumor
tissues. C vs. N, **P<0.01. DEUP1, deuterosome assembly protein
1; HCC, hepatocellular carcinoma; HE, hematoxylin and eosin; IHC,
immunohistochemistry; N, adjacent non-tumor tissues; C, tumor
tissues. |
Association between DEUP1 promoter
methylation and expression, and clinicopathologic parameters in
HCC
Downregulated expression of DEUP1 mRNA and protein
were significantly associated with TNM stage and tumor
differentiation (P<0.05; Tables I
and II). The DEUP1 promoter
hypermethylation were associated with TNM stage and tumor
differentiation. (Table III).
DEUP1 mRNA, protein and the promoter methylation status had no
association with other clinicopathological parameters.
 | Table I.Correlation of deuterosome assembly
protein 1 mRNA expression with clinicopathological features in
hepatocellular carcinoma. |
Table I.
Correlation of deuterosome assembly
protein 1 mRNA expression with clinicopathological features in
hepatocellular carcinoma.
|
|
| Expression of
mRNA |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical data | Number | Positive | Negative | Positive rate
(%) | P-value |
|---|
| Sex |
| Man | 45 | 10 | 35 | 22.3 | 0.606 |
|
Woman | 15 | 5 | 10 | 33.3 |
|
| Age |
| ≤50 | 40 | 8 | 32 | 20.0 | 0.206 |
|
>50 | 20 | 7 | 13 | 35.0 |
|
| Tumor size |
| ≤5
cm | 27 | 6 | 21 | 22.2 | 0.653 |
| >5
cm | 33 | 9 | 24 | 27.3 |
|
| HBsAg |
| + | 44 | 9 | 35 | 20.5 | 0.312 |
| − | 16 | 6 | 10 | 37.5 |
|
| TNM stage |
| I+II | 32 | 12 | 20 | 37.5 | 0.017 |
|
III+IV | 28 | 3 | 25 | 10.7 |
|
| Portal tumor
thrombosis |
| No | 52 | 11 | 41 | 21.2 | 0.188 |
|
Yes | 8 | 4 | 4 | 50.0 |
|
| AFP |
| ≤400
µg/l | 41 | 12 | 29 | 29.3 | 0.423 |
| >400
µg/l | 19 | 3 | 16 | 15.8 |
|
| Tumor
differentiation |
|
Poor | 35 | 5 | 30 | 14.3 | 0.023 |
|
moderate-well | 25 | 10 | 15 | 40.0 |
|
 | Table II.Correlation of deuterosome assembly
protein 1 protein expression with clinicopathological features in
hepatocellular carcinoma. |
Table II.
Correlation of deuterosome assembly
protein 1 protein expression with clinicopathological features in
hepatocellular carcinoma.
|
|
| Expression of
protein |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical data | Number | Positive | Negative | Positive rate
(%) | P-value |
|---|
| Sex |
|
Man | 45 | 8 | 37 | 17.8 | 0.709 |
|
Woman | 15 | 4 | 11 | 26.7 |
|
| Age |
|
≤50 | 40 | 7 | 33 | 17.5 | 0.732 |
|
>50 | 20 | 5 | 15 | 25.0 |
|
| Tumor size |
| ≤5
cm | 27 | 4 | 23 | 14.8 | 0.364 |
| >5
cm | 33 | 8 | 25 | 24.2 |
|
| HBsAg |
| + | 44 | 7 | 37 | 15.9 | 0.343 |
| − | 16 | 5 | 11 | 31.3 |
|
| TNM stage |
|
I+II | 32 | 10 | 22 | 31.3 | 0.020 |
|
III+IV | 28 | 2 | 26 | 7.1 |
|
| Portal tumor
thrombosis |
| No | 52 | 9 | 43 | 17.3 | 0.393 |
|
Yes | 8 | 3 | 5 | 37.5 |
|
| AFP |
| ≤400
µg/l | 41 | 10 | 31 | 24.4 | 0.367 |
| >400
µg/l | 19 | 2 | 17 | 10.5 |
|
| Tumor
differentiation |
|
Poor | 35 | 3 | 32 | 8.6 | 0.009 |
|
moderate-well | 25 | 9 | 16 | 36.0 |
|
 | Table III.Correlation between Methylation of
DEUP1 and clinicopathological features in HCC. |
Table III.
Correlation between Methylation of
DEUP1 and clinicopathological features in HCC.
|
|
| Methylation of
DEUP1 |
|
|---|
|
|
|
|
|
|---|
| Clinical data | Number | Methylated
Unmethylated | Positive rate
(%) | P-value |
|---|
| Sex |
|
Man | 45 | 37 | 8 | 82.2 | 0.159 |
|
Woman | 15 | 9 | 6 | 60.0 |
|
| Age |
|
≤50 | 40 | 33 | 7 | 82.5 | 0.235 |
|
>50 | 20 | 13 | 7 | 65.0 |
|
| Tumor size |
| ≤5
cm | 27 | 22 | 5 | 81.5 | 0.425 |
| >5
cm | 33 | 24 | 9 | 72.7 |
|
| HBsAg |
| + | 44 | 36 | 8 | 81.8 | 0.223 |
| − | 16 | 10 | 6 | 62.5 |
| TNM stage |
|
I+II | 32 | 21 | 11 | 65.6 | 0.031 |
|
III+IV | 28 | 25 | 3 | 89.3 |
|
| Portal tumor
thrombosis |
| No | 52 | 41 | 11 | 78.8 | 0.570 |
|
Yes | 8 | 5 | 3 | 62.5 |
|
| AFP |
| ≤400
µg/l | 41 | 32 | 9 | 78.0 | 0.965 |
| >400
µg/l | 19 | 14 | 5 | 73.7 |
|
| Tumor
differentiation |
|
Poor | 35 | 31 | 4 | 88.6 | 0.010 |
|
Moderate-well | 25 | 15 | 10 | 60.0 |
|
Correlation between DEUP1 promoter
methylation and protein expression
Out of the 60 patients with HCC, 46 had positive
DEUP1 promoter methylation and six had positive protein expression.
Among the 14 patients with a negative methylation status, six
showed protein expression (Table
IV). The protein expression of DEUP1 was negatively correlated
with promoter methylation. Correlations were statistically
significant (P<0.05).
 | Table IV.Correlation of deuterosome assembly
protein 1 promoter methylation with protein expression in
hepatocellular carcinoma. |
Table IV.
Correlation of deuterosome assembly
protein 1 promoter methylation with protein expression in
hepatocellular carcinoma.
|
| Methylation |
|
|---|
|
|
|
|
|---|
| Protein | Positive | Negative | Total |
|---|
| Positive | 6 | 6 | 12 |
| Negative | 40 | 8 | 48 |
| Total | 46 | 14 | 60 |
Discussion
DNA methylation is an epigenetic phenomenon. It is
considered the second strike for inactivation of tumor suppressor
genes after mutation and allele loss (13). DEUP1, also known as coiled-up coil
coiled-coil domain-containing 67 (CCDC67), is located on human
chromosome 11q2.1, encoding 604 amino acids (11). It is a member of CCDC protein family.
CCDC protein is composed of 180–220 amino acids and the quaternary
structure in the coiled coil may be associated with angiogenin and
other protein features and exhibit diverse functions related to
their highly versatile folding motif (10,11), but
little is known about DEUP1 function (14–16). The
results of a bioinformatic predictive analysis indicated that
inactivation of DEUP1 in HCC could be caused by methylation of a
DNA CpG island. The expression of DEUP1 is associated with a high
rate of patient survival. Furthermore, the expression of DEUP1 has
been reported to be absent or significantly reduced in a number of
tumors (10,11). Epigenetic changes, especially the
methylation of DNA CpG islands, are one of the most important
mechanisms behind low or non-expression of mRNA (17,18).
Whether DEUP1 functions as a tumor suppressor gene in HCC as well
as an inactivation mechanism in HCC has not been reported.
DEUP1 mRNA expression was increased in HCC tissues
compared with in adjacent non-tumor tissues in the present study.
Statistical analysis of DEUP1 mRNA expression and
clinicopathological parameters indicated that DEUP1 mRNA expression
in TNM stage I+II and III+IV was 37.5% (12/32) and 10.7% (3/28),
respectively. In the poor and moderate-well differentiation groups,
the expression of DEUP1 mRNA was 14.3% (5/35) and 40.0% (10/25),
respectively, suggesting that mRNA expression is associated with
the degree of malignancy of HCC.
The methylation of the DEUP1 promoter in HCC and
adjacent non-tumor tissues was detected by BSP. The results
indicated that methylation levels in HCC were increased compared
with in the corresponding para-carcinoma tissues, indicating that
methylation might be involved in the occurrence and development of
HCC. Methylation levels in TNM stage I+II and III+IV were 65.6%
(21/32) and 89.3% (25/28), respectively, and 88.6% (31/35) and
60.0% (15/32) in the low and moderate-well groups, respectively.
These data indicated that the methylation status of DEUP1 has the
potential to guide prognostic evaluation for HCC. Yin et al
(10) reported that, as a tumor
suppressor gene, the methylation of the gene promoter led to its
inactivation, playing an important role in the occurrence and
development of papillary thyroid carcinoma. Park et al
(11) found that methylation of the
DEUP1 promoter led to a decrease in DEUP1 expression and had an
important role in gastric cancer. To further confirm the influence
of DEUP1 expression on the development of HCC, the expression of
the DEUP1 protein in HCC was detected by IHC and western blotting.
The DEUP1 protein is located in the cytoplasm and its expression in
HCC tissues was decreased compared with in adjacent non-tumor
tissues. Furthermore, DEUP1 protein expression was associated with
TNM stage and tumor differentiation.
It was also found that in the 46 HCC patients with
promoter methylation, 40 did not have DEUP1 protein expression. The
analysis indicated that methylation of the DEUP1 promoter had a
negative correlation with protein expression, suggesting that gene
promoter methylation may be an important mechanism underlying
non-expression of the protein.
Methylation of one or more tumor suppressor gene CpG
islands occurs in a number of malignant tumors (19–23). The
inactivation of these genes has multiple effects on cellular
processes such as apoptosis and cell cycle regulation, leading to
tumorigenesis. CpG island methylation is a reversible epigenetic
gene modification process (24). In
healthy individuals, genes are in a low-methylation status and
methylation inhibition does not influence gene expression in normal
cells. Methylation of tumor suppressor gene CpG islands can render
normal cells cancerous and demethylation can revert the phenotype
of tumor cells back to normal, therefore providing new avenues for
the therapy of tumors (25).
In conclusion, DEUP1 is a new tumor suppressor gene
in HCC, with important regulatory effects on its occurrence,
development and prognosis. This study lays a foundation for future
studies on DEUP1 functions and the mechanisms of gene silencing in
HCC, and may provide insights into demethylation drugs and new
therapeutic targets. However, if the aim is to better to prove that
DEUP1 is a suppressor gene in HCC and its promoter methylation
results in low expression, cell experiments and animal experiments
can be performed. The lack of further validation makes this study
imperfect and the authors will follow up on cell and animal
experiments to improve this study.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81701946) and the
Science and Technology Department of Henan Province (grants no.
162300410121).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QWY, SLC and HWT performed the experiment, SJZ, WZG
and JL designed the study, QWY and SLC prepared and wrote the
study. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from each patient and
the study protocol was approved by the Medical Ethics Committee of
the First Affiliated Hospital of Zhengzhou University.
Patient consent for publication
Informed consent was obtained from each patient.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
DEUP1
|
deuterosome assembly protein 1
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
BSP
|
bisulfite PCR sequencing
|
|
TNM
|
tumor node metastasis
|
References
|
1
|
Na TY, Ka NL, Rhee H, Kyeong D, Kim MH,
Seong JK, Park YN and Lee MO: Interaction of hepatitis B virus X
protein with PARP1 results in inhibition of DNA repair in
hepatocellular carcinoma. Oncogene. 35:5435–5445. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Branda M and Wands JR: Signal transduction
cascades and hepatitis B and C related hepatocellular carcinoma.
Hepatology. 43:891–902. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wahid B, Ali A, Rafique S and Idrees M:
New insights into the epigenetics of hepatocellular carcinoma.
Biomed Res Int. 2017:16095752017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang X, Cheng Q, Yin H and Yang G:
Regulation of autophagy and EMT by the interplay between p53 and
RAS during cancer progression (Review). Int J Oncol. 51:18–24.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Appleton K, Mackay HJ, Judson I, Plumb JA,
McCormick C, Strathdee G, Lee C, Barrett S, Reade S, Jadayel D, et
al: Phase I and pharmacodynamic trial of the DNA methyltransferase
inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol.
25:4603–4609. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amato RJ: Inhibition of DNA methylation by
antisense oligonucleotide MG98 as cancer therapy. Clin Genitourin
Cancer. 5:422–426. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Zhang J, Xiao X, Liu H, Wang F, Li
S, Wen Y, Wei Y, Su J and Zhang Y: The identification of
age-associated cancer markers by an integrative analysis of dynamic
DNA methylation changes. Sci Rep. 6:227222016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pfeifer GP: Defining driver DNA
methylation changes in human cancer. Int J Mol Sci. 19:E11662018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen Y, Luo D, Tian W, Li Z and Zhang X:
Demethylation of miR-495 inhibits cell proliferation, migration and
promotes apoptosis by targeting STAT-3 in breast cancer. Oncol Rep.
37:3581–3589. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yin DT, Xu J, Lei M, Li H, Wang Y, Liu Z,
Zhou Y and Xing M: Characterization of the novel tumor-suppressor
gene CCDC67 in papillary thyroid carcinoma. Oncotarget.
7:5830–5841. 2016.PubMed/NCBI
|
|
11
|
Park SJ, Jang HR, Kim M, Kim JH, Kwon OH,
Park JL, Noh SM, Song KS, Kim SY, Kim YH and Kim YS: Epigenetic
alteration of CCDC67 and its tumor suppressor function in gastric
cancer. Carcinogenesis. 33:1494–1501. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kamarajah SK, Frankel TL, Sonnenday C, Cho
CS and Nathan H: Critical evaluation of the american joint
commission on cancer (AJCC) 8th edition staging system for patients
with hepatocellular carcinoma (HCC): A Surveillance, Epidemiology,
End Results (SEER) analysis. J Surg Oncol. 117:644–650. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Böck J, Appenzeller S, Haertle L,
Schneider T, Gehrig A, Schröder J, Rost S, Wolf B, Bartram CR,
Sutter C and Haaf T: Single CpG hypermethylation, allele
methylation errors, and decreased expression of multiple tumor
suppressor genes in normal body cells of mutation-negative
early-onset and high-risk breast cancer patients. Int J Cancer.
143:1416–1425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murphy GA, Spedale EJ, Powell ST, Pillus
L, Schultz SC and Chen L: The Sir4 C-terminal coiled coil is
required for telomeric and mating type silencing in saccharomyces
cerevisiae. J Mol Biol. 334:769–780. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tamaki H, Sanda M, Katsumata O, Hara Y,
Fukaya M and Sakagami H: Pilt is a coiled-coil domain-containing
protein that localizes at the trans-Golgi complex and regulates its
structure. FEBS Lett. 586:3064–3070. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Burkhard P, Stetefeld J and Strelkov SV:
Coiled coils: A highly versatile protein folding motif. Trends Cell
Biol. 11:82–88. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng JD, Zhang N, Zhao GJ, Xu LX, Yang Y,
Xu XY, Chen MK, Wang HY, Zheng SX and Li XX: MT1G is silenced by
DNA methylation and contributes to the pathogenesis of
hepatocellular carcinoma. J Cancer. 9:2807–2816. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kishino T, Niwa T, Yamashita S, Takahashi
T, Nakazato H, Nakajima T, Igaki H, Tachimori Y, Suzuki Y and
Ushijima T: Integrated analysis of DNA methylation and mutations in
esophageal squamous cell carcinoma. Mol Carcinog. 55:2077–2088.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kwon H, Song K, Han C, Zhang J, Lu L, Chen
W and Wu T: Epigenetic silencing of miRNA-34a in human
cholangiocarcinoma via EZH2 and DNA methylation Impact on
Regulation of Notch Pathway. Am J Pathol. 187:2288–2299. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alipour M, Zargar SJ, Safarian S,
Fouladdel S, Azizi E and Jafargholizadeh N: The study of DNA
methylation of bax gene promoter in breast and colorectal carcinoma
cell lines. Iran J Cancer Prev. 6:59–64. 2013.PubMed/NCBI
|
|
21
|
Lu Y, Zabihula B, Yibulayin W and Liu X:
Methylation and expression of RECK, P53 and RUNX genes in patients
with esophageal cancer. Oncol Lett. 14:5293–5298. 2017.PubMed/NCBI
|
|
22
|
Zheng J, Mei Y, Xiang P, Zhai G, Zhao N,
Xu C, Liu M, Pan Z, Tang K and Jia D: DNA methylation affects
metastasis of renal cancer and is associated with TGF-β/RUNX3
inhibition. Cancer Cell Int. 18:562018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song L, Yu H and Li Y: Diagnosis of lung
cancer by SHOX2 gene methylation assay. Mol Diagn Ther. 19:159–167.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sajadian SO, Ehnert S, Vakilian H,
Koutsouraki E, Damm G, Seehofer D, Thasler W, Dooley S, Baharvand
H, Sipos B and Nussler AK: Induction of active demethylation and
5hmC formation by 5-azacytidine is TET2 dependent and suggests new
treatment strategies against hepatocellular carcinoma. Clin
Epigenetics. 7:982015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zahnow CA, Topper M, Stone M,
Murray-Stewart T, Li H, Baylin SB and Casero RA Jr: Inhibitors of
DNA methylation, histone deacetylation, and histone demethylation:
A perfect combination for cancer therapy. Adv Cancer Res.
130:55–111. 2016. View Article : Google Scholar : PubMed/NCBI
|