Introduction
Colorectal cancer (CRC) is one of the most common
and serious malignancies in the world, particularly in developed
countries (1). In England and Wales
between 1991 and 1998, the 5-year survival rate of CRC in the
initial stages was 60–95%, markedly decreasing to 35% in stages
where lymph node metastases are detected (2). The lifetime risk of CRC is ~5%, and
almost 45% of patients succumb to CRC, despite treatment (1). In China, although CRC is not the
leading cause of cancer-associated mortality, the incidence and
mortality rates of CRC have been increasing over the past 20 years,
and they have been predicted to continue to increase if no
effective intervention occurs (3).
The initiation and progression of human cancer
depends on both genetic alterations and epigenetic changes
(4). The carcinogenesis of CRC has
been revealed to be involved in epigenetic alterations, including
DNA methylation and histone modifications (5,6). Gene
silencing caused by aberrant promoter methylation acts as one of
the most significant epigenetic mechanisms (7). In CRC, methylation alterations
frequently occur in chromosomes 1, 5, 6, 8, 11, 13, 18, 19, 21 and
22 (8). Differentially methylated
DNA regions have been identified in both primary tumor tissues and
blood samples (9), and more recently
in cell-free DNA (10).
The embryonic ectoderm development (EED) gene,
encodes a member of the Polycomb-group (PcG) family, which has been
demonstrated to maintain the transcriptional repressive states of
genes over successive cell generations (11). PcG genes have been revealed to be
highly mutated in a number of human diseases (12). In addition, PcG proteins have been
identified to be associated with cancer development (13) and proposed as potential targets for
cancer therapeutic strategies (14).
EED has been identified to mediate the repression of gene activity
through histone deacetylation (15).
EED hypermethylation has been detected in cholangiocarcinoma
tumors (16). Furthermore, high
EED expression levels have been demonstrated to be
associated with CRC (17). However,
to the best of our knowledge, no EED methylation in CRC has
been reported. In light of previous findings, the present study
aimed to investigate the association between EED methylation
and CRC.
Materials and methods
Tissue specimens
In the past 5 years, a total of 111 CRC tissue
samples, 111 paired para-tumor tissues and 20 colorectal normal
tissues were collected from patients diagnosed at Shaoxing People's
Hospital (Shaoxing, China), Zhejiang Province Cancer Hospital
(Hangzhou, China) and Nanjing Chinese Medicine Hospital (Nanjing,
China). Clinical diagnosis was determined on the basis of
colonoscopy findings and histological assessment. The types of
cancer were staged according to the seventh edition of the American
Joint Committee on Cancer staging system (18). The mean age of the patients was
60.96±11.66 years and the cohort included 72 males and 39 females.
All individuals were of Han Chinese ethnicity from Eastern China.
The specimens were freshly obtained and stored at −80°C. The study
protocol was approved by the Ethical Committees of Shaoxing
People's Hospital (Shaoxing, China), Zhejiang Province Cancer
Hospital (Hangzhou, China), Nanjing Chinese Medicine Hospital
(Nanjing, China) and Ningbo University (Ningbo, China). The number
of institutional review board approval was IRB-2018-28. Written
informed consent was obtained from all participants.
DNA methylation assay
DNA extraction and bisulfite conversion were
performed as described previously (19). Quantitative methylation-specific
polymerase chain reaction (qMSP) was used to detect the methylation
levels. qMSP was performed as described in our previous studies
(20–22). The percent of methylated reference
(PMR) was used to represent gene methylation (23,24). The
genomic position and function annotations of EED were
obtained from the University of California Santa Cruz genome
browser (GRCh37/hg19; http://genome.ucsc.edu/index.html). The primer
sequences of EED were 5′-GAGGCGGAGGAATATGTT-3′ for the
forward primer and 5′-TCACTACTCAACTTCTACTTCT-3′ for the reverse
primer. The primer sequences of ACTB were
5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ for the forward primer and
5′-AACCAATAAAACCTACTCCTCCCTTAA-3′ for the reverse primer. The
current study used qMSP with internal negative and positive
controls, and a reference gene control for the methylation assay of
EED.
The Cancer Genome Atlas (TCGA) and
Gene Expression Omnibus (GEO) data analysis
EED methylation and EED expression
datasets were retrieved from the online resource [http://www.cbioportal.org/; colorectal Adenocarcinoma
(TCGA, Provisional)] to analyze the association between EED
methylation and EED expression in patients with CRC. The
dataset GSE32323 (25) was
downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds/) to provide data
regarding EED expression with and without
5′-AZA-deoxycytidine treatment in the cell lines COLO320, HT29 and
RKO.
Dual-luciferase reporter gene
assay
293T cells, obtained from the Chinese Academy of
Sciences cell bank (Shanghai, China), were cultured as previously
described (26). The fragment of
EED (−300 bp to +390 bp) was chemically synthesized
according to the manufacturer's protocol (TransLipid HL
Transfection Reagent; TransGen Biotech, Co., Ltd., Beijing, China)
and digested with XhoI and KpnI (New England BioLabs,
Inc., Ipswich, MA, USA). The fragment was then purified by Cycle
Pure kit (Omega Bio-Tek, Inc., Norcross, GA, USA) and the target
DNA fragment was cloned into a pGL3 vector (Promega Cooperation,
Madison, WI, USA) using DNA Ligation kit (Takara Bio, Inc., Otsu,
Japan). The empty pGL3 basic vector was used as the negative
control and the pGL3 promoter vector (both Promega Cooperation,
Madison, WI, USA) was used as the positive control, which contained
an SV40 promoter upstream of the luciferase gene. The plasmid
transfection and the detection of luciferase activity were
performed as previously described (23,27).
Statistical analysis
SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA)
was used for all statistical analysis. Data that were not normally
distributed are presented as the median (interquartile range) and
variables that were normally distributed are presented as the mean
± standard deviation. Mann-Whitney U test, Kruskal-Wallis and
unpaired Student's t-test were applied to analyze the baseline
characteristics among the 111 patients with CRC. Association of PMR
difference with the clinical characteristics in 111 paired samples
was performed by χ2 test. Analysis of variance test was
applied to compare EED methylation levels among tumor,
para-tumor and normal colorectal samples, and the Bonferroni
correction was used for the post hoc test. To analyze the results
of the dual-luciferase reporter assay, an analysis of variance and
Bonferroni's correction were also used. Spearman's correlation test
was applied to evaluate the correlation between EED mRNA
expression level and methylation in 369 patients with CRC from TCGA
data portal. P<0.05 was considered to indicate a statistically
significant difference.
Results
In the present study, a total of 111 CRC tumor
tissues, 111 paired para-tumor tissues and 20 colorectal normal
tissues were obtained to investigate the role of EED
promoter methylation in CRC.
Characteristics of the target sequence
on the EED promoter region
The genomic region and target sequence of EED
are presented in Fig. 1A. One CpG
site was located in the primers of the tested fragment (hg19,
chr11:85956259-85956348). The capillary electrophoresis result
revealed a pMSP product with a length of 90 base pairs, which was
expected, and Sanger sequencing demonstrated that the amplified
fragment matched the target sequence (Fig. 1B).
Baseline characteristics of the
methylation levels among patients
Subsequently, no significant difference was
identified between EED methylation levels and clinical
characteristics, including sex, age, tumor location, tumor size,
differentiation and lymph node metastasis (Table I). However, when the PMR difference
value between methylation levels of tumor and para-tumor samples
was used to perform the χ2 test, the only significant
association of PMR difference with clinical characteristics was
identified for tumor location (P=0.039; Table II).
 | Table I.Baseline characteristics among 111
patients with colorectal cancer. |
Table I.
Baseline characteristics among 111
patients with colorectal cancer.
| Clinical
characteristics | No. (n=111) | Percentage of
methylated reference in tumor | P-value |
|---|
| Sex |
|
| 0.587a |
|
Male | 72 | 4.73
(2.52,6.67) |
|
|
Female | 39 | 4.41
(1.40,6.31) |
|
| Age, years |
|
| 0.165a |
|
≤65 | 74 | 4.79
(2.55,6.63) |
|
|
>65 | 37 | 3.33
(1.42,6.08) |
|
| Tumor location |
|
| 0.929b |
|
Colon | 46 | 4.70
(1.91,6.38) |
|
|
Rectum | 56 | 4.43
(2.50,6.62) |
|
| Colon
and rectum | 9 | 4.78
(1.30,7.69) |
|
| Tumor size, cm |
|
| 0.673a |
| ≤6 | 96 | 4.42
(2.21,6.62) |
|
|
>6 | 15 | 5.76
(2.06,5.94) |
|
|
Differentiation |
|
| 0.222c |
| High
and medium | 15 | 7.58±8.86 |
|
| Low and
none | 96 | 4.63±3.44 |
|
| Lymph node
metastasis |
|
| 0.435a |
|
Positive | 56 | 4.70
(2.60,6.55) |
|
|
Negative | 55 | 4.44
(1.44,6.53) |
|
 | Table II.Associations between the PMR
differences and clinical characteristics. |
Table II.
Associations between the PMR
differences and clinical characteristics.
| Clinical
characteristics | No. (n=111) | P-value |
|---|
| Sex |
| 0.643 |
|
Male | 72 |
|
|
Female | 39 |
|
| Age, years |
| 0.590 |
|
≤65 | 74 |
|
|
>65 | 37 |
|
| Tumor location |
| 0.039 |
|
Colon | 46 |
|
|
Rectum | 56 |
|
| Colon
and rectum | 9 |
|
| Tumor size, cm |
| 0.071 |
| ≤6 | 96 |
|
|
>6 | 15 |
|
|
Differentiation |
| 0.119 |
| High
and medium | 15 |
|
| Low and
none | 96 |
|
| Lymph node
metastasis |
| 0.286 |
|
Positive | 56 |
|
|
Negative | 55 |
|
Notably, a significant difference of EED
methylation levels among tumor, para-tumor and normal colorectal
tissues was revealed (tumor vs. para-tumor vs. normal: 5.03±4.61
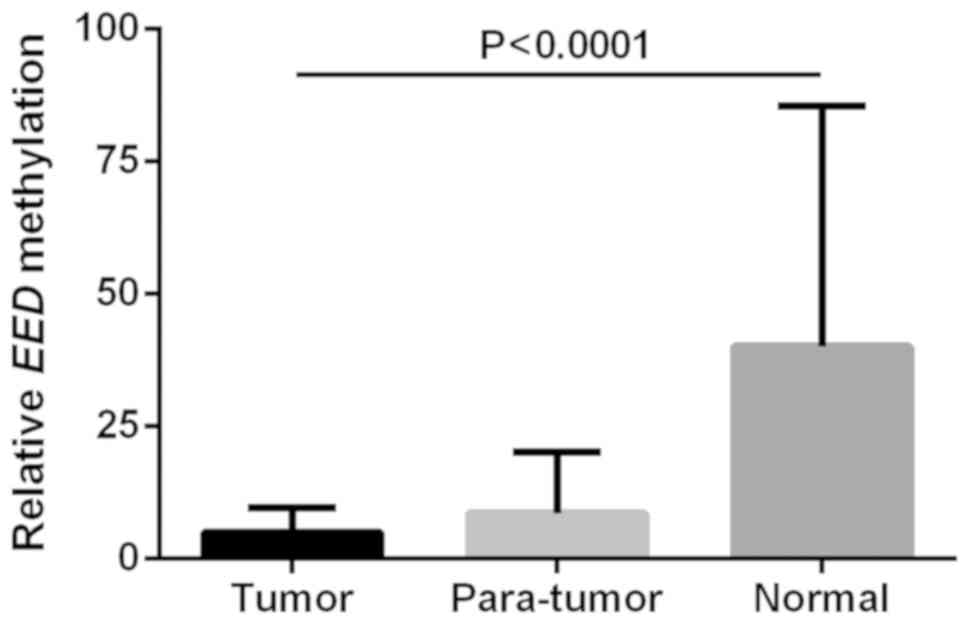
vs. 8.65±11.50 vs. 40.12±45.31; F=45.014; P<0.0001; Fig. 2). Following Bonferroni's correction
the results were as follows: Tumor vs. para-tumor, P=0.237; tumor
vs. normal tissue, P<0.0001; and para-tumor vs. normal tissue,
P<0.0001 (Fig. 2). Previous
studies have demonstrated that bisulfite sequencing PCR and qMSP
can yield similar conclusions (28,29).
Bisulfite sequencing PCR is a reliable and accurate method;
however, it is labor intensive and therefore only applicable for
methylation in a limited number of samples.
Diagnostic value of EED
methylation
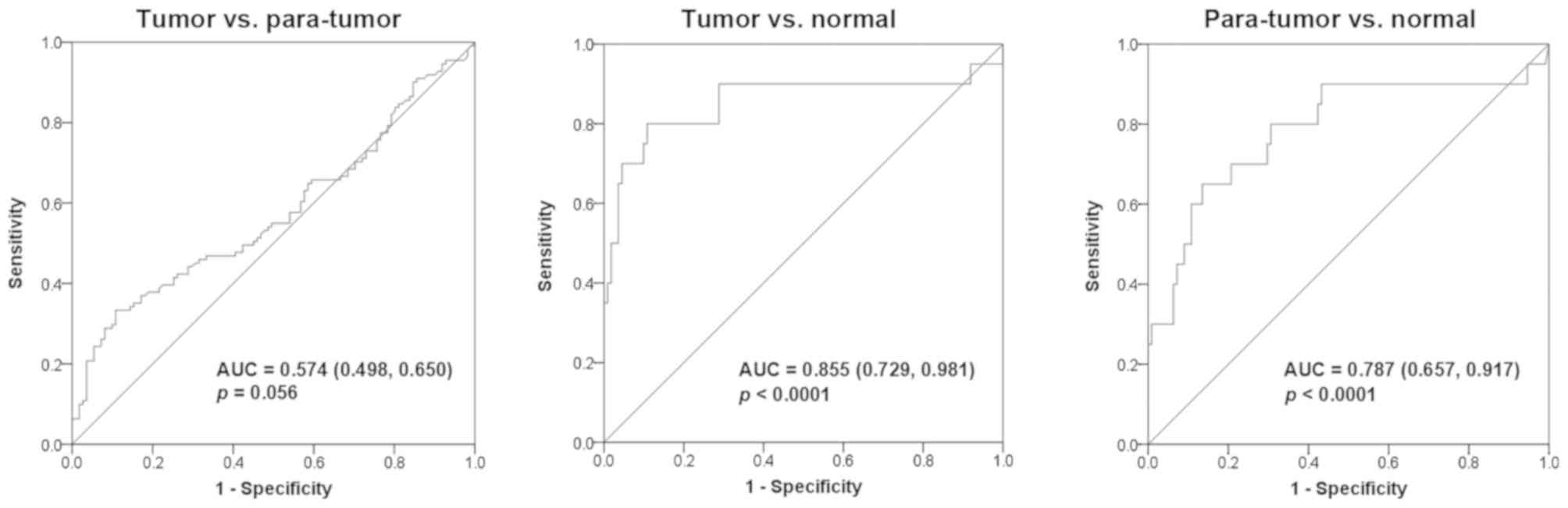
A further estimation of the diagnostic value of
EED methylation for CRC revealed an area under the curve
(AUC) of 0.574 (95% CI, 0.498–0.650) with a sensitivity of 33.3%
and a specificity of 89.2% between tumor and para-tumor tissues
(median PMR, 4.54 vs. 4.90%; P=0.056; Fig. 3). Furthermore, an AUC of 0.855 (95%
CI, 0.729–0.981) with a sensitivity of 80% and a specificity of
89.2% was identified between tumor tissues and normal colorectal
tissues (median PMR, 4.54 vs. 18.97%; P<0.0001; Fig. 3). In addition, an AUC of 0.787 (95%
CI, 0.657–0.917) with a sensitivity of 65% and a specificity of
86.5% was revealed between para-tumor tissues and normal colorectal
tissues (median PMR, 4.90 vs. 18.97%; P<0.0001; Fig. 3).
Characteristics of EED expression and
methylation
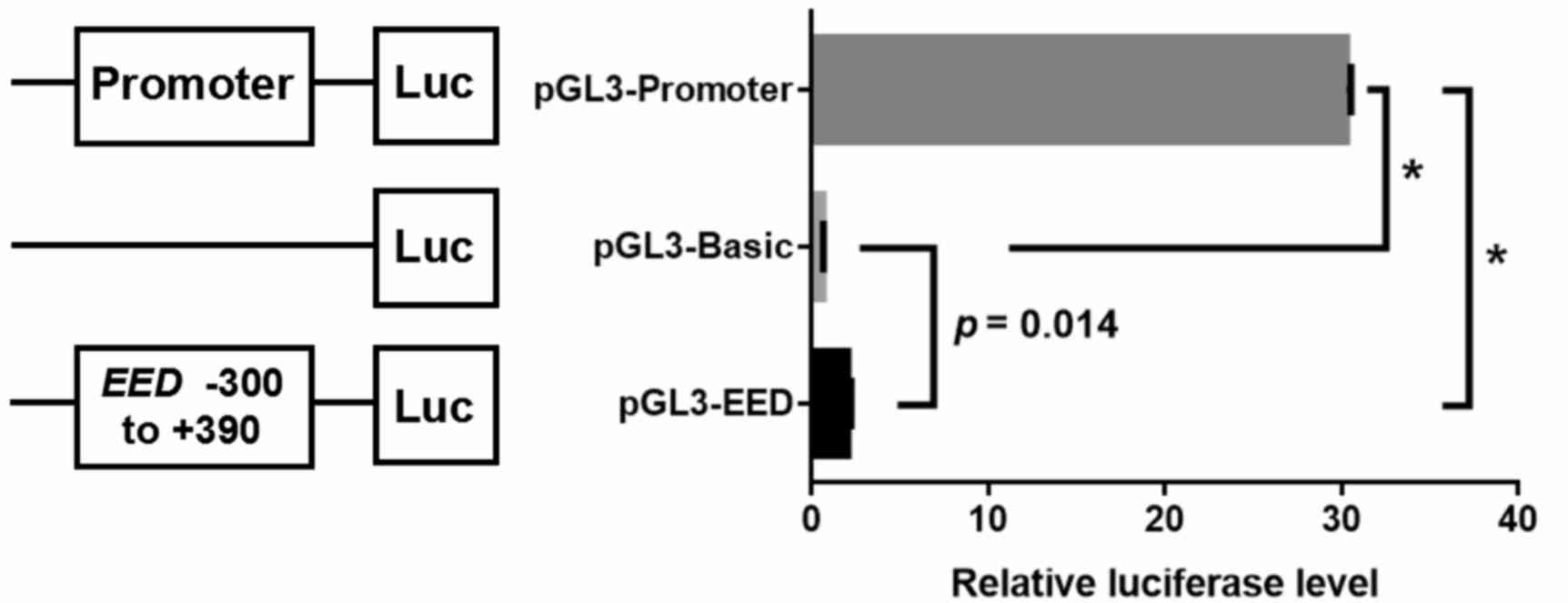
A dual-luciferase reporter gene assay was performed
to evaluate whether the EED fragment (−300 bp to +390 bp)
was able to regulate gene expression. The dual-luciferase assay
demonstrated that the transcriptional activity of recombinant
pGL3-EED plasmid was significantly higher compared with the
pGL3-Basic control vector (pGL3-EED vs. pGL3-Basic vs.
pGL3-Promoter: 2.05±0.19 vs. 0.65±0.02 vs. 30.27±0.26; F=16589.76;
P<0.0001; pGL3-EED vs. pGL3-Basic: fold-change, 3.15;
P=0.014; pGL3-EED vs. pGL3-Promoter: P<0.0001; pGL3-Basic
vs. pGL3-Promoter: P<0.0001; Fig.
4), which indicates that the EED fragment is able to
promote gene expression.
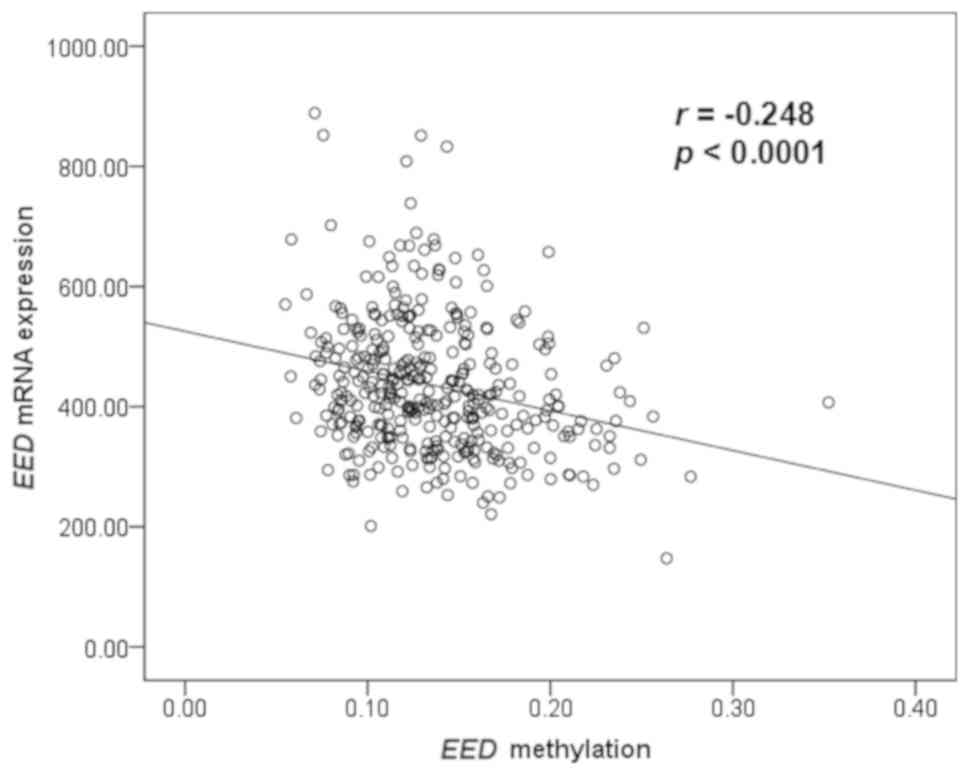
Furthermore, EED mRNA expression and
methylation data of patients with CRC were obtained from TCGA
online database to investigate their correlation. Spearman
correlation test revealed that EED mRNA expression level was
inversely correlated with methylation (r=−0.248; P<0.0001;
Fig. 5). In addition, a further
analysis of GEO data (GSE32323) demonstrated that EED
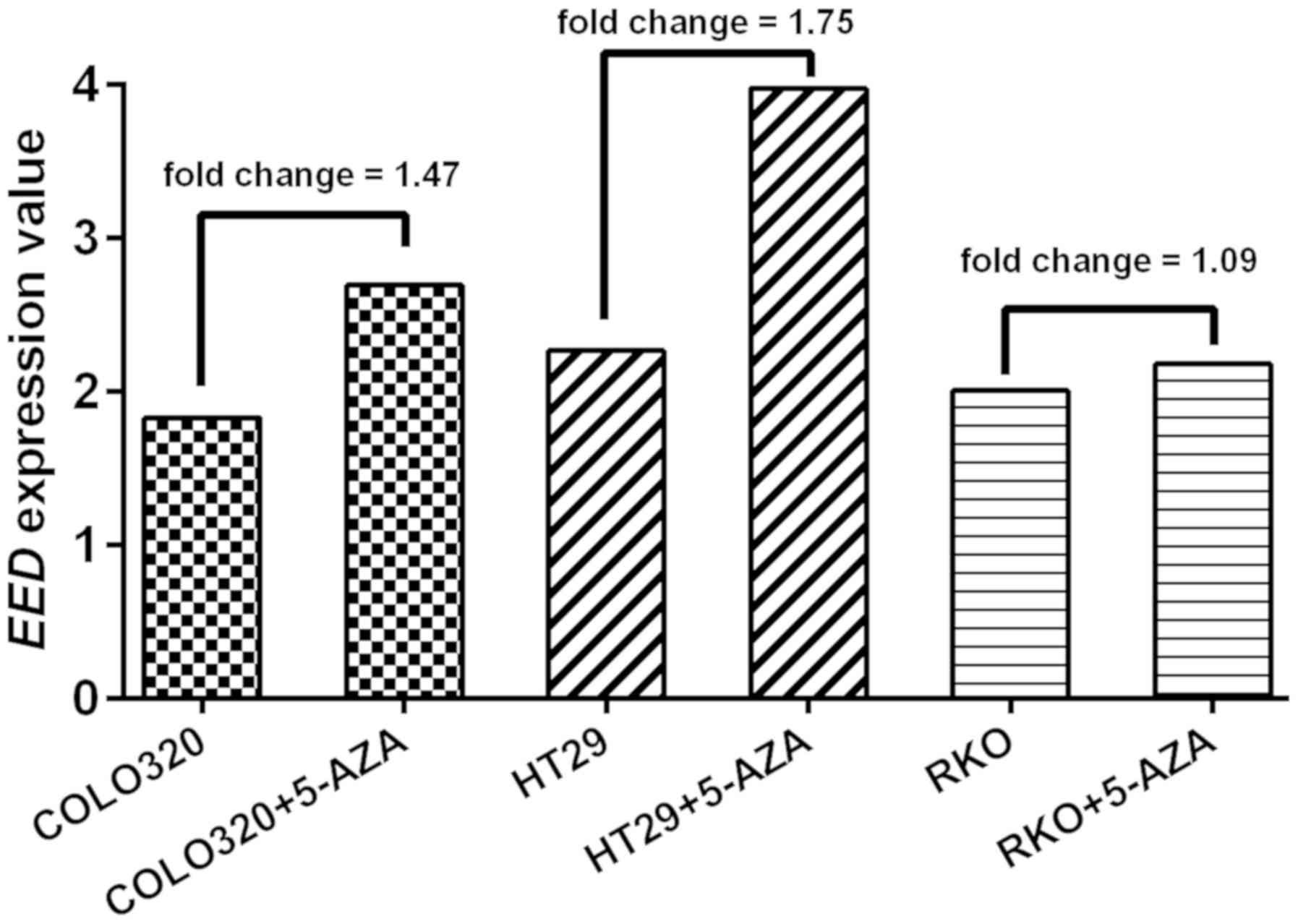
expression level in three CRC cell lines (COLO320, HT29 and RKO)
was increased following 5′-AZA-deoxycytidine treatment (average
fold-change, 1.44; Fig. 6). In
summary, the present results indicate that EED
hypomethylation is likely to upregulate EED expression and
eventually increase the risk of CRC.
Discussion
In the current study, EED methylation levels
were detected in 111 pairs of CRC tumor and para-tumor tissues from
patients with CRC and 20 colorectal tissues from normal controls.
The results revealed that EED hypomethyation was
significantly associated with the risk of CRC. In addition, the
dual-luciferase reporter gene assay demonstrated that the
EED fragment exhibited promoter activity. Further
bioinformatics analyses revealed that EED methylation was
inversely correlated with EED expression and demethylation
treatment was identified to upregulate EED expression.
EED is a key component of the polycomb repressive
complex 2 (PRC2) (30), which can
mediate epigenetic silencing of genes associated with worse
survival of patients with colon cancer (31). Higher expression of EED and two other
PRC2 components has been demonstrated to contribute to the
progression of CRC (17).
Polymorphism of EED gene has been identified
to be associated with the lymph node metastatic process of CRC
(32). In addition, EED
expression has been demonstrated to be regulated by interleukin-22
and signal transducer and activator of transcription 3 (33). EED mRNA levels are
significantly higher in CRC tissues compared with non-cancerous
tissues (17). The dual-luciferase
reporter gene assay performed in the current study demonstrated an
enhanced transcriptional activity of the cloned EED
fragment. Furthermore, data from TCGA and GEO databases suggested
that EED methylation was associated with decreased
expression levels of EED. This supports the hypothesis that
EED hypomethylation may promote CRC via upregulation of
EED expression. The present study identified that EED
expression may be regulated by the methylation of its promoter,
which may promote understanding of the role of EED in
CRC.
Previous studies have revealed that DNA cytosine
modifications serve an important role in cancer biology, and
provide promising biomarkers for cancer diagnosis and prognosis
evaluation (10,34). Several epigenetic biomarkers have
been studied for the diagnosis of CRC, including SEPT9
(35), BMP3, NDRG4 (36) and hMLH1 (37). The ROC curves generated in the
current study demonstrated that EED hypomethylation is a
good biomarker for the diagnosis of CR. Tumor vs. para-tumor
revealed a sensitivity of 33.3% and a specificity of 89.2%. Tumor
vs. normal tissue analysis demonstrated a sensitivity of 80% and a
specificity of 89.2%. Finally, para-tumor vs. normal tissue
revealed a sensitivity of 65% and a specificity of 86.5%.
However, there were a number of limitations of the
present study. Firstly, the GEO analyses to investigate the effect
of demethylation agent on EED expression involved data from
only three cell lines. Furthermore, a comparison of EED mRNA
expression level between 17 paired tumor and non-cancerous tissues
from TCGA database yielded an insignificant result (4.45±0.58 vs.
4.32±0.50; P=0.393), although this discrepancy may be due to
different ethnic samples and a small sample size. Future studies
are required to confirm the current findings and further address
the functional roles of EED methylation in CRC. In
conclusion, the present results demonstrated that EED
hypomethylation might be an important risk factor associated with
CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Institutes of Health (grant no. U01CA217078) and the K. C.
Wong Magna Fund in Ningbo University.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
SD and WZ made substantial contributions to the
conception and design. XY and RP analyzed and interpreted the data,
drated and revised the manuscript, and agreed to be accountable for
all aspects of the work. JZ, BW, JY and YJ contributed to the
interpretation of data and completion of figures and tables. SZ,
YS, CZ and JD contributed to performing the experiments and
analyzing the data. All the authors have read and approved the
final manuscript.
Ethical approval and consent to
participate
Informed consent was provided by all participants
prior to their inclusion within the study. The study was approved
by the Ethical Committees of the Shaoxing People's Hospital,
Zhejiang Province Cancer Hospital, Nanjing Chinese Medicine
Hospital and Ningbo University.
Patient consent for publication
All patients have provided informed consent for the
publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanthan R, Senger JL and Kanthan SC:
Molecular events in primary and metastatic colorectal carcinoma: A
review. Patholog Res Int. 2012:5974972012.PubMed/NCBI
|
|
3
|
Zheng ZX, Zheng RS, Zhang SW and Chen WQ:
Colorectal cancer incidence and mortality in China, 2010. Asian Pac
J Cancer Prev. 15:8455–8460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fukushige S and Horii A: DNA methylation
in cancer: A gene silencing mechanism and the clinical potential of
its biomarkers. Tohoku J Exp Med. 229:173–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Engeland M, Derks S, Smits KM, Meijer
GA and Herman JG: Colorectal cancer epigenetics: Complex
simplicity. J Clin Oncol. 29:1382–1391. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sakai E, Nakajima A and Kaneda A:
Accumulation of aberrant DNA methylation during colorectal cancer
development. World J Gastroenterol. 20:978–987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sipos F, Mũzes G, Patai AV, Fũri I,
Péterfia B, Hollósi P, Molnár B and Tulassay Z: Genome-wide
screening for understanding the role of DNA methylation in
colorectal cancer. Epigenomics. 5:569–581. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song BP, Jain S, Lin SY, Chen Q, Block TM,
Song W, Brenner DE and Su YH: Detection of hypermethylated vimentin
in urine of patients with colorectal cancer. J Mol Diagn.
14:112–119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li W, Zhang X, Lu X, You L, Song Y, Luo Z,
Zhang J, Nie J, Zheng W, Xu D, et al: 5-Hydroxymethylcytosine
signatures in circulating cell-free DNA as diagnostic biomarkers
for human cancers. Cell Res. 27:1243–1257. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schumacher A, Lichtarge O, Schwartz S and
Magnuson T: The murine Polycomb-group gene eed and its human
orthologue: Functional implications of evolutionary conservation.
Genomics. 54:79–88. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Piunti A and Shilatifard A: Epigenetic
balance of gene expression by Polycomb and COMPASS families.
Science. 352:aad97802016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du J, Li L, Ou Z, Kong C, Zhang Y, Dong Z,
Zhu S, Jiang H, Shao Z, Huang B and Lu J: FOXC1, a target of
polycomb, inhibits metastasis of breast cancer cells. Breast Cancer
Res Treat. 131:65–73. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scelfo A, Piunti A and Pasini D: The
controversial role of the Polycomb group proteins in transcription
and cancer: How much do we not understand Polycomb proteins? FEBS
J. 282:1703–1722. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu JI, Kang IH, Seo GS, Choi SC, Yun KJ
and Chae SC: Promoter polymorphism of the EED gene is associated
with the susceptibility to ulcerative colitis. Dig Dis Sci.
57:1537–1543. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sriraksa R, Zeller C, Dai W, Siddiq A,
Walley AJ, Limpaiboon T and Brown R: Aberrant DNA methylation at
genes associated with a stem cell-like phenotype in
cholangiocarcinoma tumors. Cancer Prev Res (Phila). 6:1348–1355.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu YL, Gao X, Jiang Y, Zhang G, Sun ZC,
Cui BB and Yang YM: Expression and clinicopathological significance
of EED, SUZ12 and EZH2 mRNA in colorectal cancer. J Cancer Res Clin
Oncol. 141:661–669. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pike JG, Berardinucci G, Hamburger B and
Kiruluta G: The surgical management of urinary incontinence in
myelodysplastic children. J Pediatr Surg. 26:466–471. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Yang Y, Liu J, Li B, Xu Y, Li C,
Xu Q, Liu G, Chen Y, Ying J and Duan S: NDRG4 hypermethylation is a
potential biomarker for diagnosis and prognosis of gastric cancer
in Chinese population. Oncotarget. 8:8105–8119. 2017.PubMed/NCBI
|
|
20
|
Chen R, Hong Q, Jiang J, Chen X, Jiang Z,
Wang J, Liu S, Duan S and Shi S: AGTR1 promoter hypermethylation in
lung squamous cell carcinoma but not in lung adenocarcinoma. Oncol
Lett. 14:4989–4994. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang Y, Chen X, Hu H, Jiang Y, Yu H, Dai
J, Mao Y and Duan S: Elevated UMOD methylation level in peripheral
blood is associated with gout risk. Sci Rep. 7:111962017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li B, Chen X, Jiang Y, Yang Y, Zhong J,
Zhou C, Hu H and Duan S: CCL2 promoter hypomethylation is
associated with gout risk in Chinese Han male population. Immunol
Lett. 190:15–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu H, Chen X, Wang C, Jiang Y, Li J, Ying
X, Yang Y, Li B, Zhou C, Zhong J, et al: The role of TFPI2
hypermethylation in the detection of gastric and colorectal cancer.
Oncotarget. 8:84054–84065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang B, Du Z, Gao YT, Lou C, Zhang SG, Bai
T, Wang YJ and Song WQ: Methylation of Dickkopf-3 as a prognostic
factor in cirrhosis-related hepatocellular carcinoma. World J
Gastroenterol. 16:755–763. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khamas A, Ishikawa T, Shimokawa K, Mogushi
K, Iida S, Ishiguro M, Mizushima H, Tanaka H, Uetake H and Sugihara
K: Screening for epigenetically masked genes in colorectal cancer
Using 5-Aza-2′-deoxycytidine, microarray and gene expression
profile. Cancer Genomics Proteomics. 9:67–75. 2012.PubMed/NCBI
|
|
26
|
Shen Z, Chen X, Li Q, Zhou C, Xu Y, Yu R,
Ye H, Li J and Duan S: Elevated methylation of CMTM3 promoter in
the male laryngeal squamous cell carcinoma patients. Clin Biochem.
49:1278–1282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji H, Wang Y, Liu G, Xu X, Dai D, Chen Z,
Zhou D, Zhou X, Han L, Li Y, et al: OPRK1 promoter hypermethylation
increases the risk of Alzheimer's disease. Neurosci Lett.
606:24–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Demokan S, Chuang AY, Pattani KM,
Sidransky D, Koch W and Califano JA: Validation of nucleolar
protein 4 as a novel methylated tumor suppressor gene in head and
neck cancer. Oncol Rep. 31:1014–1020. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park JY, Kim D, Yang M, Park HY, Lee SH,
Rincon M, Kreahling J, Plass C, Smiraglia DJ, Tockman MS and Kim
SJ: Gene silencing of SLC5A8 identified by genome-wide methylation
profiling in lung cancer. Lung Cancer. 79:198–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Margueron R and Reinberg D: The Polycomb
complex PRC2 and its mark in life. Nature. 469:343–349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagarsheth N, Peng D, Kryczek I, Wu K, Li
W, Zhao E, Zhao L, Wei S, Frankel T, Vatan L, et al: PRC2
epigenetically silences Th1-Type chemokines to suppress effector
T-cell trafficking in colon cancer. Cancer Res. 76:275–282. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Seo GS, Yu JI, Chae SC, Park WC, Shin SR,
Yoo ST, Choi SC and Lee SH: EED gene polymorphism in patients with
colorectal cancer. Int J Biol Markers. 28:274–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun D, Lin Y, Hong J, Chen H, Nagarsheth
N, Peng D, Wei S, Huang E, Fang J, Kryczek I and Zou W: Th22 cells
control colon tumorigenesis through STAT3 and polycomb repression
complex 2 signaling. Oncoimmunology. 5:e10827042016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Das PM and Singal R: DNA methylation and
cancer. J Clin Oncol. 22:4632–4642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ravegnini G, Zolezzi Moraga JM, Maffei F,
Musti M, Zenesini C, Simeon V, Sammarini G, Festi D, Hrelia P and
Angelini S: Simultaneous analysis of SEPT9 promoter methylation
status, micronuclei frequency, and folate-related gene
polymorphisms: The potential for a novel blood-based colorectal
cancer biomarker. Int J Mol Sci. 16:28486–28497. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kadiyska T and Nossikoff A: Stool DNA
methylation assays in colorectal cancer screening. World J
Gastroenterol. 21:10057–10061. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Li D, Li X, Teng C, Zhu L, Cui B,
Zhao Y and Hu F: Prognostic significance of hMLH1/hMSH2 gene
mutations and hMLH1 promoter methylation in sporadic colorectal
cancer. Med Oncol. 31:392014. View Article : Google Scholar : PubMed/NCBI
|