Introduction
Prostate cancer (PC) is the most commonly diagnosed
form of cancer and the sixth leading cause of cancer-related deaths
among men worldwide (1). The 5-year
survival rate is approximately 100% in patients with localized PC,
but it drops to 31% in those with distant metastasis (2). Similar to most other solid
malignancies, PC may metastasize to distant organs such as the
liver, lungs, and brain; however, it has an abnormally high
propensity to metastasize to bones (3). Thus, it is desirable to gain a better
understanding of mechanisms underlying PC metastasis to the bone
for development and use of therapies to improve patient
survival.
Studies have provided new insights into this
advanced disease based on critical molecular and cellular events
surrounding tumor progression, invasion, and metastasis to the bone
and other sites (4).
Epithelial-to-mesenchymal transition (EMT), in which cellular,
morphological, and functional switches may transform adherent
epithelial cells to migratory mesenchymal cells, is critical for
tumorigenic progression and cancer metastasis. EMT may promote stem
cell-related properties and generate cells with features related to
tumor initiation (5). During EMT,
tumor cells exhibit stem cell characteristics and confer invasive
and migratory properties to primary tumor cells, ultimately
resulting in metastasis (5).
It is hypothesized that tumors rely on a small
portion of cells called cancer stem cells (CSCs) that have the
ability to self-renew and generate multiple ‘mature’ tumor
progenies (6). CSCs are typically
dormant, and their regrowth is responsible for metastases (7). Primary tumors comprise a majority of
differentiated epithelial cells and a small number of cells
expressing stem cell markers (i.e., PC stem cells) (8). A previous study showed that CSCs
derived from primary tumors exhibited increased invasion,
self-renewal, and clonogenic properties (9). These cells may be identified using
several CSC markers such as CD133, c-Met, and prostate stem cell
antigens (10). A recent analysis
involving a cohort of high-risk PC patients showed that the number
of positive cancer cells for the a-6 and a-2 integrin subunits and
the c-Met receptor in primary PC correlated with bone metastatic
progression (11).
CD133 (prominin-1), a 5-transmembrane glycoprotein,
was originally recognized as a hematopoietic stem cell marker
(12). CD133 is regarded as an
important cell surface marker used to identify cancer-initiating
cell subpopulations in brain tumors, colon carcinoma, head and neck
cancer, hepatocellular carcinoma, thyroid carcinoma, and prostate
carcinoma (13,14). Previous experiments have demonstrated
a correlation between upregulated CD133 expression and progression
of EMT in head and neck cancer cells and head and neck squamous
cell carcinoma tissues (15). The
CD133-mediated molecular mechanisms regulating cancer-initiating
cells in bone metastasis of PC remain unclear.
In the present study, we have demonstrated that
CD133 plays a significant role by increasing stemness, enhancing
EMT, and promoting bone metastasis in a PC cell line, LNCaP.
Furthermore, we aimed to identify cytokines secreted by CD133+
LNCaP cells that play a critical role during bone metastasis of PC
cells. Our results suggest that CD133+ PC cells play a central role
in bone metastasis.
Materials and methods
Cell culture
LNCaP and 293T cells were purchased from the Korean
Cell Line Bank (KCLB; no. 21573), Korean Cell Line Research
Foundation, Seoul, Korea, in 2017. The cells were maintained in
RPMI-1640 (Welgene, Daegu, Korea) supplemented with 10%
heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and 0.1% antibiotic/antimycotic
solution (Welgene) at 37°C in a 5% CO2 humidified
chamber.
Cloning of human CD133 and
establishment of stably transfected LNCaPVec control and
LNCapCD133+ cells
HT29 colon cancer cells were used as a source of
CD133. Cloning of CD133 was performed as previously described
(15). Plasmid pcDNA3.1/N-terminal
green fluorescent protein (NT-GFP) lacking the CD133 insert was
used as a transfection control. 293T cells were used as a positive
control to evaluate the translated fusion product.
Stable cell lines overexpressing the CD133 protein
were obtained through co-transfection of confluent LNCaP-luciferase
cells in 100-mm plates with 20 µg pcDNA3.1/NT-GFP:CD133 or
pcDNA3.1/NT-GFP plasmid using the FuGENE HD transfection reagent,
as previously described (15).
Western blot analysis
After reaching approximately 80% confluency, the
medium was removed, and the cells were washed twice with
phosphate-buffered saline (PBS, pH 7.4). Cell lysates were prepared
in 200 µl cold lysis buffer [1% NP-40, 50 mM Tris-HCl, pH 7.5, 150
mM sodium chloride (NaCl), 0.02% sodium azide, 150 mg/ml of
phenylmethylsulfonyl fluoride (PMSF), 2 mg/ml of aprotinin, 20
mg/ml of leupeptin, and 1 mg/ml of pepstatin A]. Approximately 30
mg of tissue lysate was separated via 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a polyvinylidene difluoride (PVDF) membrane
(Amersham, Piscataway, NJ, USA). Each membrane was blocked for 30
min with a blocking solution containing 5% skim milk in
Tris-buffered saline containing Tween-20 (TBST, 2.42 g/l Tris-HCl,
8 g/l NaCl, 0.1% Tween-20, pH 7.6) and rinsed with TBST. The
membrane was incubated overnight at 4°C with appropriate primary
antibodies, including anti-CD133 (1:1,000; cat. no. MBS850595;
MyBioSource, San Diego, CA, USA), anti-octamer-binding
transcription factor 4 (Oct-4, 1:1,000; cat. no. 2750, Cell
Signaling Technology, Beverly, MA, USA), anti-NANOG (1:1,000; cat.
no. 3580; Cell Signaling Technology), anti-(sex determining region
Y)-box 2 (SOX2, 1:1,000; cat. no. 2748; Cell Signaling Technology),
anti-E-cadherin (1:1,000; cat. no. sc-8426; Santa-Cruz
Biotechnology Inc., Dallas, TX, USA), anti-transcription factor 4
(TCF-4, 1:1,000; cat. no. 2953; Cell Signaling Technology),
anti-vimentin (1:1,000; cat. no. sc-6260; Santa-Cruz Biotechnology
Inc.), anti-β-catenin (1:1,000; cat. no. 9562; Cell Signaling
Technology), and anti-macrophage migration inhibitory factor (MIF,
1:1,000; cat. no. 88186; Cell Signaling Technology). A mouse
monoclonal immunoglobulin G (IgG) specific for
glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 1:1,000; cat. no.
sc-47724; Santa-Cruz Biotechnology Inc.) was used as a control. The
membrane was rinsed with TBST, and protein immunoreactivity was
detected using an enhanced chemiluminescence detection kit (ECL,
Amersham).
Confocal microscopic analyses
Immunolabeled cells were counterstained with
4′,6-diamidino-2-phenylindole (DAPI) provided in ProLong Gold
antifade mounting medium (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) to visualize nuclear morphology. Digital
images were acquired at the Korea Basic Science Institute Gwangju
Center using a TCS SP5 AOBS laser-scanning confocal microscope
(Leica Microsystems, Heidelberg, Germany).
Colony-forming assays
Cells were seeded at a density of 1,000 cells/well
in non-adherent 24-well culture plates coated with a 10% poly
(2-hydroxyethyl methacrylate) (polyHEMA) solution (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) in absolute ethanol and then dried
overnight. After seeding, cells were incubated in serum-free
Dulbecco's modified Eagle's medium (DMEM) supplemented with 200
ng/ml epidermal growth factor (Sigma-Aldrich; Merck KGaA), 20 ng/ml
of basic fibroblast growth factor (Sigma-Aldrich; Merck KGaA), and
B-27 supplement (Invitrogen; Thermo Fisher Scientific, Inc.). After
5 days of incubation, the number of spheroids in each well was
counted using a light microscope (Zeiss, Zena, Germany).
Cell migration assay
Cells were seeded at 1×105 cells/well in
6-well plates and cultured to approximately 90% confluency in 1 ml
of DMEM supplemented with 10% FBS. The media were removed from the
wells, and a straight transverse line was drawn through the
adherent cells using a ruler and sterile 200-µl plastic
micropipette tip to produce a uniform gap. Serum-free DMEM was
added, and the distance between the gaps was measured immediately
and 24 h later, following image capture of six random microscopic
fields. The distance measured immediately was defined as 100%.
Cell invasion assay
Cell invasion was assessed using a chemotaxis
chamber (Neuro Probe, Gaithersburg, MD, USA). The cells
(5×104 cells in 0.35-ml serum-free DMEM per well) were
seeded into the top chamber of a 10-well invasion chamber assay
plate. DMEM supplemented with 10% FBS was placed in the lower
chamber, and a matrigel-coated membrane was inserted between the
two chambers. After incubation for 24 h at 37°C, the membranes were
fixed and stained with a Hemacolor rapid staining kit (Merck KGaA).
Cells from five random microscopic fields (each 0.5 mm2)
were counted using a hematocytometer under a light microscope
(Zeiss).
Immunofluorescence
To confirm morphological changes, cells were washed
thrice with PBS, fixed in 4% paraformaldehyde (PFA) for 10 min at
room temperature, and permeabilized with PBS containing 0.25%
Triton X-100 (PBST) for 10–15 min at room temperature. After three
washes with PBS, cells were blocked with 1% bovine serum albumin
for 30 min. Samples were incubated overnight at 4°C with the
relevant antibodies anti-MIF (1:500; Cell Signaling Technology) and
anti-F actin (1:500; Santa-Cruz Biotechnology Inc.) for 2 h at room
temperature. After three washes with PBS, immune-labeled cells were
counterstained with 50 µl of DAPI at 37°C for 10 min. Cells were
analyzed using a laser scanning confocal microscope (Leica
Microsystems).
Immunoprecipitation
Immunoprecipitation analysis was carried out as
previously described (16). Briefly,
cells were lysed using a protein lysis buffer [1% (v/v) Triton
X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM
ethylenediaminetetraacetic acid (EDTA), and protease inhibitors],
and cell lysates were incubated overnight with primary
anti-β-catenin and protein A or G beads (GE Healthcare, Uppsala,
Sweden) at 4°C on a rotator. The beads were washed, boiled in
Laemmli buffer, and processed for western blotting.
Animals
Five-week-old male athymic nude mice (BL-6/Nu, 20–25
g; Orient Bio Co., Ltd., Seoul, Korea) were housed under controlled
light conditions and provided water ad libitum. All
experimental procedures involving animals were performed in
compliance with institutional and governmental requirements and
approved by the Institutional Animal Care and Use Committee
(approval no. CIACUC2015-A0032), Chosun University, Gwangju,
Korea.
Intracardiac xenograft model
Twenty mice were subjected to intracardiac
injections of LNCaPVec or LNCaPCD133+ cells
and 10 mice subjected PBS as mock mice to examine the ability of PC
cells to metastasize to bone, respectively. Before injection, the
cells were transfected with the pGL4.5 vector plasmid encoding
luciferase (Promega Corporation, Madison, WI, USA). The stably
transfected cells were grown in Hank's balanced salt solution
(HBSS; Welgene) supplemented with hygromycin (2 mg/ml;
Sigma-Aldrich; Merck KGaA) and, luciferase activity was measured
using the Luciferase Assay System (Promega Corporation) and
detected by luminometer GENios Plus (Tecan Group Ltd., Salzburg,
Austria). The severe combined immunodeficient mice were
anesthetized using isoflurane gas. Subsequently,
LNCaPVec+ or LNCaPCD133+ cells
(1×105 cells per mouse) were injected into the left
cardiac ventricle, as per the technique first described by Arguello
et al (17) with some
modifications. After 4 weeks, images of tumor-bearing tissues
excised from mice during necropsy were obtained.
Bioluminescence imaging
After intracardiac injection, mice were weekly
imaged via bioluminescence for 4–6 weeks using the IVIS®
imaging system (PerkinElmer, Waltham, MA, USA) at the Korea Basic
Science Institute (Gwangju, Korea). Images were captured and
processed using the Living Image® v.4.2 software.
Anesthesia was induced using inhaled isoflurane and maintained with
2% isoflurane mixed with oxygen/nitrogen via nose cone delivery.
d-Luciferin (3 mg dissolved in water) was administered at 150 mg/kg
in Dulbecco's phosphate-buffered saline (DPBS) via intraperitoneal
injection. Optical imaging was acquired approximately 10–15 min
later.
Histological analysis of mouse
tissues
Tumor-bearing tissues were fixed in cold 4% PFA.
Bone tissue was first decalcified using a sodium citrate solution
before processing onto histological slides. Decalcified bones were
cut at the midpoint and embedded in paraffin blocks. Fluorescence
from serial paraffin sections was monitored via fluorescence
microscopy (Leica Microsystems). Tissues were stained with
hematoxylin and eosin (H&E) stain, and images were acquired
using a microscope slide scanner (3D-HISTECH Ltd., Budapest,
Hungary).
Cytokine profiling
Supernatants from LNCaPVec and
LNCaPCD133+ cells and sera from tumor-bearing mice were
collected and assayed using a cytokine array kit (R&D Systems,
Minneapolis, MN, USA), according to the manufacturer's
instructions. The cytokines examined using this technique are
listed in Table I. Cytokines were
detected using the ECL detection kit (Amersham) and quantified via
densitometric analysis using ImageJ software (Scion Corp, MD,
USA).
 | Table I.List of human inflammatory cytokines
examined using the antibody array (R&D Systems). |
Table I.
List of human inflammatory cytokines
examined using the antibody array (R&D Systems).
| Coordinate |
Analyte/control | Entrez gene ID | Alternate
nomenclature |
|---|
| A1, A2 | Reference
spots | N/A | RS |
| A3, A4 | Adiponectin | 9370 | Acrp30 |
| A5, A6 | Apolipoprotein
A-I | 335 | ApoA1 |
| A7, A8 | Angiogenin | 283 | – |
| A9, A10 | Angiopoietin-1 | 284 | Ang-1, ANGPT1 |
| A11, A12 | Angiopoietin-2 | 285 | Ang-2, ANGPT2 |
| A13, A14 | BAFF | 10673 | BLyS, TNFSF13B |
| A15, A16 | BDNF | 627 | Brain-derived
neurotrophic factor |
| A17, A18 | Complement
component C5/C5a | 727 | C5/C5a |
| A19, A20 | CD14 | 929 | – |
| A21, A22 | CD30 | 943 | TNFRSF8 |
| A23, A24 | Reference
spots | N/A | RS |
| B3, B4 | CD40 ligand | 959 | CD40L, TNFSF5,
CD154, TRAP |
| B5, B6 | Chitinase 3-like
1 | 1116 | CHI3L1, YKL-40 |
| B7, B8 | Complement factor
D | 1675 | Adipsin, CFD |
| B9, B10 | C-Reactive
protein | 1401 | CRP |
| B11, B12 | Cripto-1 | 6997 |
Teratocarcinoma-derived growth factor |
| B13, B14 | Cystatin C | 1471 | CST3, ARMD11 |
| B15, B16 | Dkk-1 | 22943 | Dickkopf-1 |
| B17, B18 | DPPIV | 1803 | CD26, DPP4,
dipeptidyl-peptidase IV |
| B19, B20 | EGF | 1950 | Epidermal growth
factor |
| B21, B22 | Emmprin | 682 | CD147, basigin |
| C3, C4 | ENA-78 | 6374 | CXCL5 |
| C5, C6 | Endoglin | 2022 | CD105, ENG |
| C7, C8 | Fas ligand | 356 | TNFSF6, CD178,
CD95L |
| C9, C10 | FGF basic | 2247 | FGF-2 |
| C11, C12 | FGF-7 | 2252 | KGF |
| C13, C14 | FGF-19 | 9965 | – |
| C15, C16 | Flt-3 ligand | 2323 | FLT3LG |
| C17, C18 | G-CSF | 1440 | CSF3 |
| C19, C20 | GDF-15 | 9518 | MIC-1 |
| C21, C22 | GM-CSF | 1437 | CSF2 |
| D1, D2 | GROα | 2919 | CXCL1, MSGA-α |
| D3, D4 | Growth hormone | 2688 | GH,
somatotropin |
| D5, D6 | HGF | 3082 | Scatter factor,
SF |
| D7, D8 | ICAM-1 | 3383 | CD54 |
| D9, D10 | IFN-γ | 3458 | IFNG |
| D11, D12 | IGFBP-2 | 3485 | – |
Reverse transcription-quantitative PCR
(qPCR) analysis
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). cDNA was synthesized from 2 µg total RNA using the
Super-Script II First-Strand Synthesis System (Invitrogen; Thermo
Fisher Scientific, Inc.). mRNA levels were measured via qPCR, and
the Gapdh gene was used as an endogenous control. Sequences
of the primers used to target various genes are listed in Table II.
 | Table II.Gene primer sequences. |
Table II.
Gene primer sequences.
| Gene name | Upstream primer
(5′-3′) | Downstream primer
(3′-5′) |
|---|
| MIF |
GCAGAACCGCTCCTACAGCA |
GGCTCTTAGGCGAAGGTGGA |
| GAPDH |
TGGAATCCACTGGCGTCTTC |
GGTTCACGCCCATCACAAAC |
Immunohistochemical analysis of bone
specimens
Paraffin sections were deparaffinized in three
solutions of xylene and rehydrated in a graded series of ethanol
solutions. For antigen retrieval, slides were placed in 0.01 M
citrate buffer at pH 6.0 and heated in a steamer for 30 min.
Endogenous peroxidases were quenched by incubating the samples with
3% hydrogen peroxide for 20 min at room temperature. Sections were
incubated overnight at 4°C using a 1:50 dilution of the primary
antibody against MIF (Santa-Cruz Biotechnology Inc.). Sections were
then incubated for 30 min with a biotinylated secondary antibody
(LSAB system HRP kit; DakoCytomation, Glostrup, Denmark), rinsed in
PBS, and incubated for 30 min with a streptavidin-peroxidase
conjugate (LSAB; DakoCytomation). The reaction was developed for 5
min using 3, 30-diaminobenzidine tetrahydrochloride (Sigma-Aldrich;
Merck KGaA). Slides were counterstained in hematoxylin, dehydrated,
and covered with a cover slip. Negative and positive controls were
simultaneously analyzed. Positive controls comprised mammary
tissues. The slides were captured using a microscope slide scanner
(3D-HISTECH Ltd., Budapest, Hungary).
Statistical analysis
One-way analysis of variance (ANOVA) followed by
Sidak's multiple comparison test (unless specifically mentioned
otherwise) were used for statistical analyses. P<0.05 was
considered to indicate a statistically significant difference. Data
are expressed as the mean ± standard deviation (SD) unless
specified otherwise. Data were analyzed using the SPSS v.20.0
software program for Windows (SPSS, Inc., Chicago, IL, USA).
GraphPad Prism v.6.00 software program for Windows (GraphPad, La
Jolla, CA, USA) was used to analyze data from in vitro and
in vivo experiments.
Results
Stable overexpression of CD133 in
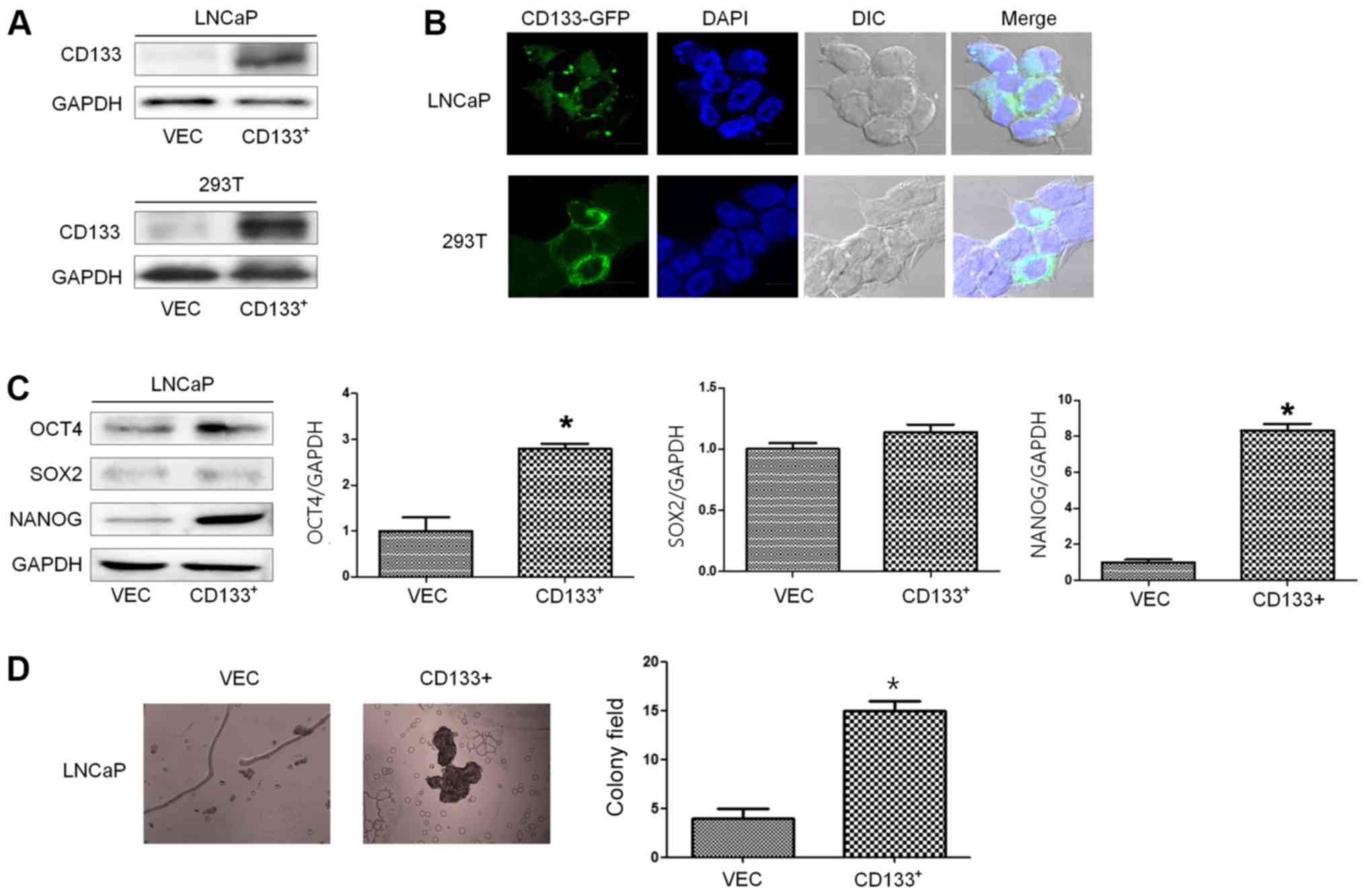
LNCaP cells
To evaluate the effect of CD133 overexpression in
vitro, we established a stable cell line overexpressing CD133
under the control of a constitutive promoter. We transfected LNCaP
cells with either CD133 or an empty vector tagged with GFP. A
representative CD133 clone was selected from LNCaPCD133+
cells and compared against control cells transfected with the empty
vector (LNCaPVec). Transiently transfected
293TVec or 293TCD133+ cells were used as a
positive control. Basal expression of CD133 protein in
LNCaPVec cells was very low; however, the basal
expression of CD133 was high in stable CD133-transfected cells
(LNCaPCD133+) and transiently transfected 293T cells
(Fig. 1A). Green fluorescence was
observed in the cytosol and membrane of LNCaPCD133+
cells (Fig. 1B).
To explore the function of CD133 in cancer stemness,
we evaluated the acquisition of CSC properties such as increased
ability to form tumor spheres and expression of stem cell markers
such as Oct-4, SOX2, and NANOG. The expression levels of Oct-4 and
NANOG were significantly elevated in LNCaPCD133+ cells,
consistent with the increase in stemness properties (Fig. 1C). Stemness was also confirmed by a
colony-formation assay. Colony-forming ability was significantly
elevated in LNCaPCD133+ cells, in line with the elevated
expression of stemness factors (Fig.
1D). These results suggest that the increase in colony-forming
ability and Oct-4 and NANOG expression conferred stem cell-like
properties during ectopic overexpression of CD133.
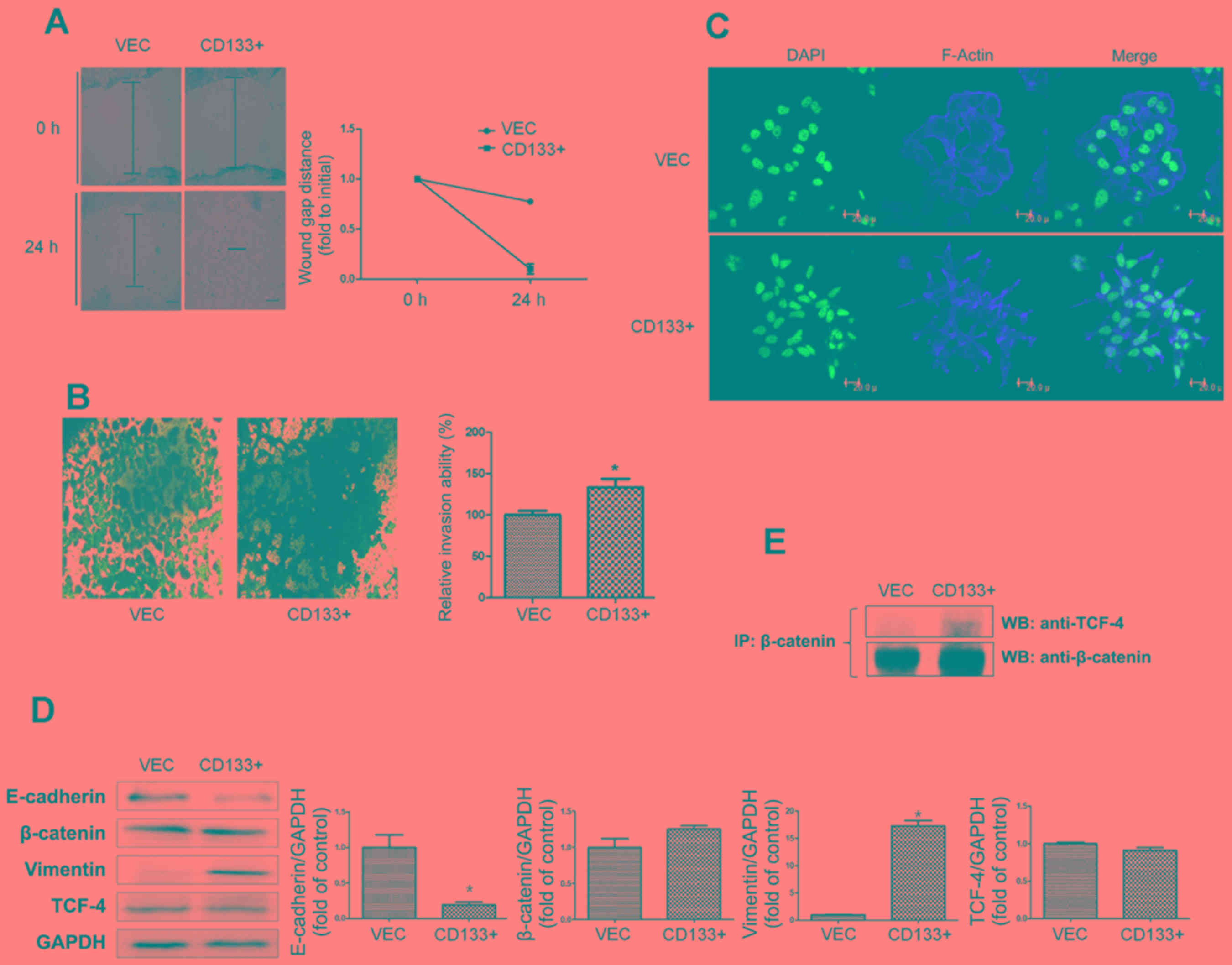
EMT-related properties in
CD133-overexpressing LNCaP cells
CD133 overexpression in LNCaP cells, as determined
by wound healing and cell invasion assays, induced a significant
increase in cell migration and invasion. After 24 h, the artificial
wound gaps became significantly narrower in LNCaPCD133+
cells than in LNCaPVec cells (Fig. 2A). Furthermore, the invasive
abilities of LNCaPCD133+ cells significantly increased
(Fig. 2B). To monitor morphological
changes in LNCaP cells, immunofluorescence analysis for F-actin was
performed using confocal microscopy. The number of spindle and
discontiguous cells observed in the LNCaPCD133+ group
was higher than that observed in the LNCaPVec group
(Fig. 2C).
To identify factors associated with altered
invasiveness and migration caused by ectopic overexpression of
CD133, we monitored levels of epithelial and mesenchymal markers in
PC cells. Protein expression analyses revealed a slightly lower
expression of epithelial marker E-cadherin in
LNCaPCD133+ cells than in LNCaPVec cells
(Fig. 2D). In contrast, expression
of the mesenchymal marker vimentin was significantly upregulated in
LNCaPCD133+ cells. Further, analysis of
β-catenin-associated signal transduction was carried out via
immunoprecipitation in LnCaPCD133+ cells (Fig. 2E). CD133 overexpression led to a
significant increase in binding between β-catenin and TCF-4, an
EMT-inducing transcriptional factor in cancer cells in compared
with LnCaPvec cells as a negative control. These data
strongly support the idea that CD133 is essential for increased and
decreased expression of vimentin and E-cadherin, respectively,
leading to subsequent EMT and metastasis in PC cells.
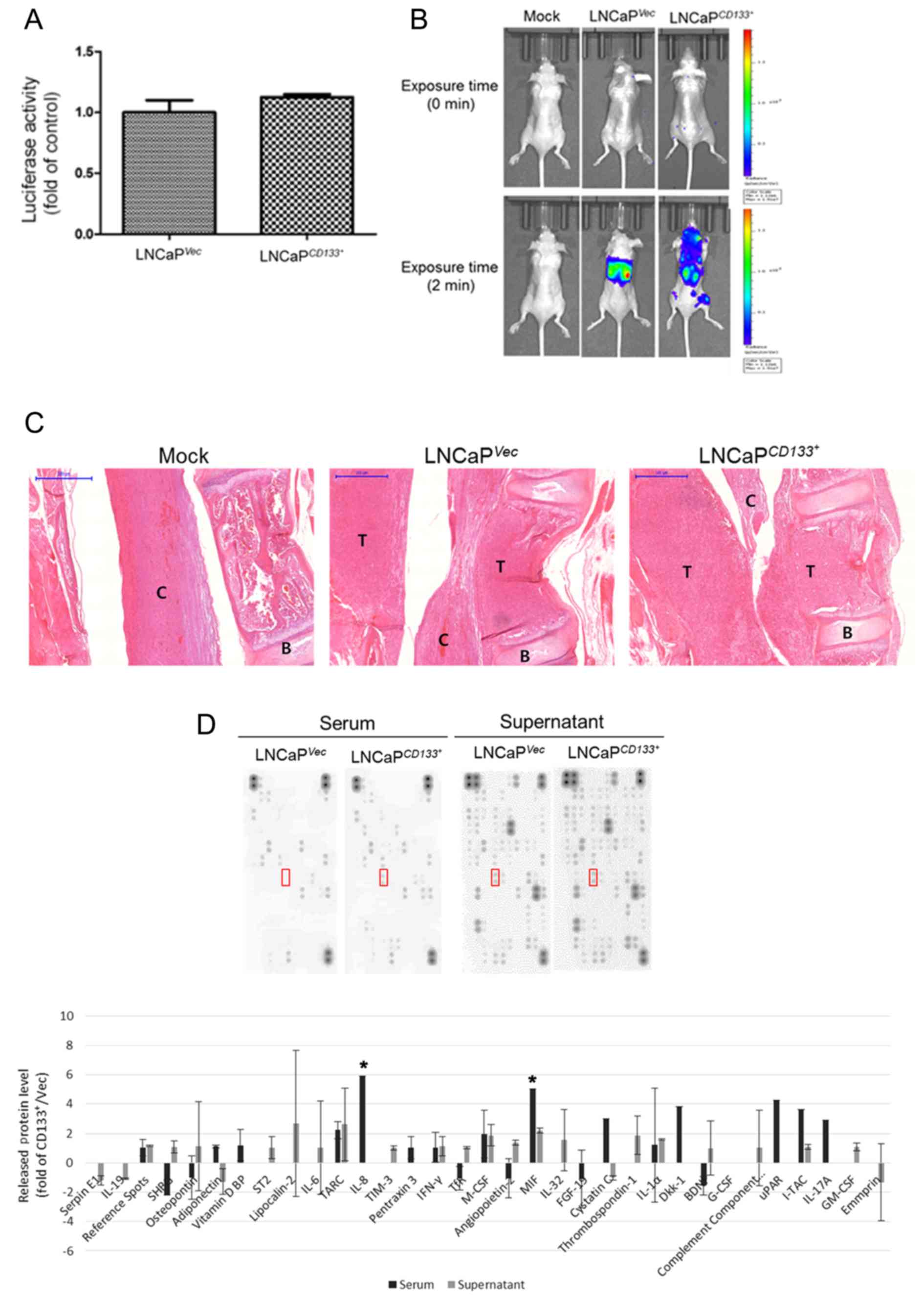
CD133 promotes LNCaP-induced bone
metastasis in vivo
Both of LNCaPVec and
LNCaPCD133+ cells were labeled with luciferase, and the
cells showed a similar luciferase activities (Fig. 3A). An then we injected
LNCaPVec and LNCaPCD133+ cells into the left
cardiac ventricle of male nude mice at 0.5×106
cells/mouse. The mice were imaged weekly, starting from week 4
after intracardiac injection. The results of PBS (mock) injection
and injection of LNCaPVec and LNCaPCD133+
cells are shown in the whole-mouse image in Fig. 3B. A large hot-spot and a small spot
of bioluminescence were observed in the whole body following
LNCaPCD133+ cell injection. Only 20% of animals
inoculated with LNCaPVec cells developed skeletal tumors
(n = 10) as compared with 80% of those inoculated with
LNCaPCD133+ cells (Table
III).
 | Table III.Characteristics of the tumor
formations. |
Table III.
Characteristics of the tumor
formations.
| Cell Line | Injection | Cell no. | Tumor
formation |
|---|
|
LNCaPVec | Intracardiac |
1×105 | 2/10 (20%) |
|
LNCaPCD133+ | Intracardiac |
1×105 | 8/10 (80%) |
We performed H&E staining to examine the
histological features of skeletal tumors formed by
LNCaPVec/LNCaPCD133+ cells inoculated into
mice via intracardiac injection. As shown in Fig. 3C, gross examination of excised spines
in mice injected with LNCaPVec or LNCaPCD133+
cells showed that the tumor mass occupied the primary spongiosum
(trabecular epiphysis) and displaced the bone marrow cells. In
particular, an apparent margin between the spinal cord and
metastatic tumors was observed in LNCaPVec-inoculated
mice. However, metastatic tumors invading the spinal cord and
discursive osteolytic features of vertebrae were observed in
LNCaPCD133+ cell-inoculated mice. We aimed to identify
the secreted molecules that played a critical role in
LNCaPCD133+ cell-induced bone metastasis through a
cytokine screening using a proteome array cytokine kit (Fig. 3D). CD133 overexpression in LNCaP
cells led to a marked increase in the release of interleukin 8 and
MIF from mouse serum. MIF expression increased in the supernatants
of LNCaPCD133+ cells. These data suggest that ectopic
overexpression of CD133 might lead to a significantly increased
risk of bone metastatic cancer. Further, increased MIF expression
may contribute to PC metastasis in the bone.
Expression of MIF in primary PC and
metastatic lesions
To investigate the role of MIF in bone metastasis of
LNCaP cells, its mRNA and protein expression levels were analyzed.
MIF protein (Fig. 4A) and mRNA
(Fig. 4B) levels were higher in
LNCaPCD133+ cells than in LNCaPVec cells. To
monitor the cellular distribution of MIF in LNCaP cells, MIF
immunofluorescence was performed. MIF expression in the membrane
and cytosol was higher in LNCaPCD133+ cells than in
LNCaPVec cells (Fig.
4C).
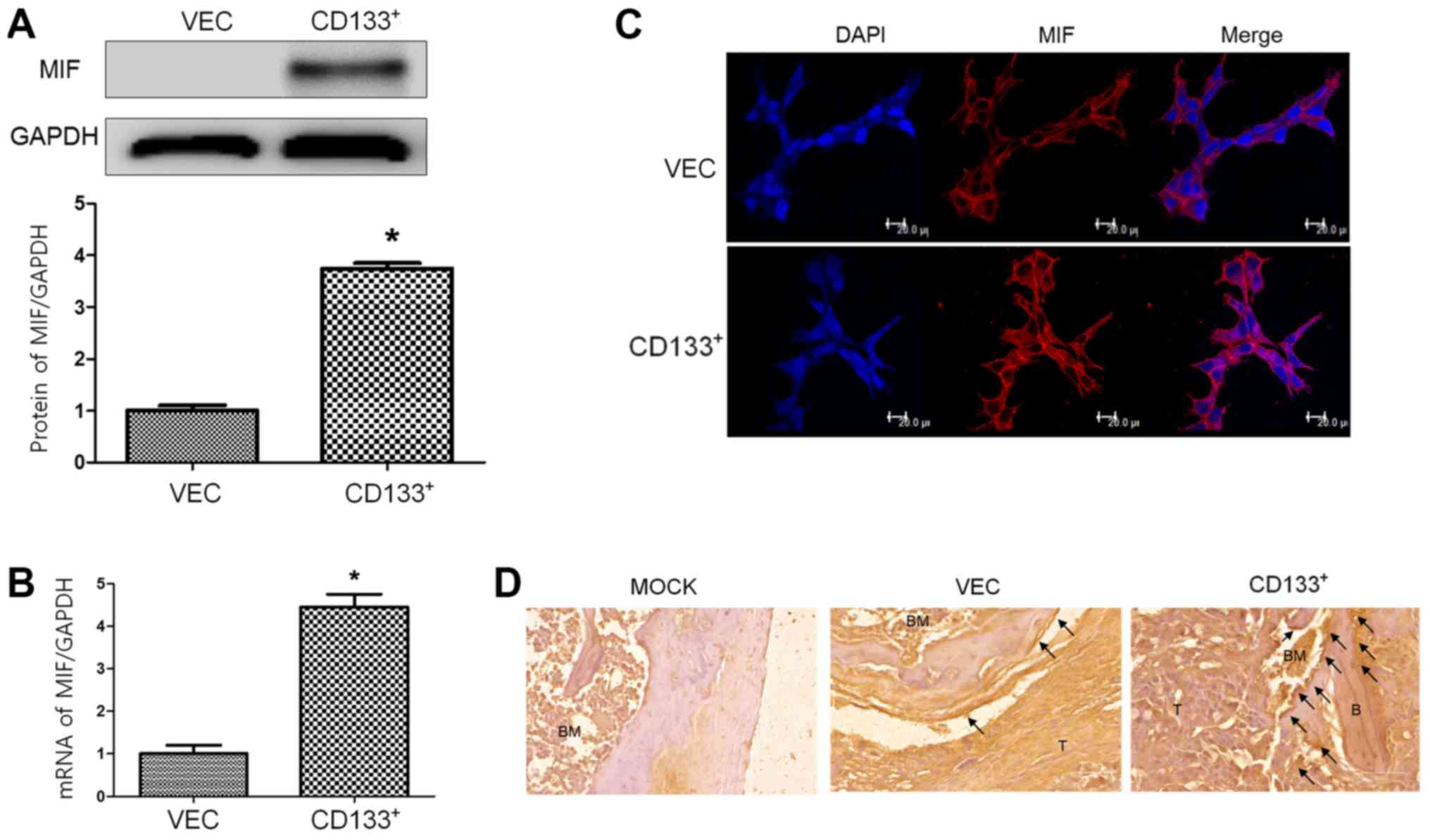 | Figure 4.In vitro and in vivo
expression of MIF in LNCaPVec and LNCaPCD133+
cells. (A) Expression of OPN in LNCaPVec (Vec) and
LNCaPCD133+ (CD133+) cells was measured by western
blotting. GAPDH was used as a loading control. (B) MIF mRNA
expression was characterized in LNCaPVec (Vec) and
LNCaPCD133+ (CD133+) cells via reverse
transcription-quantitative PCR. (C) Confocal microscopy of MIF
expression in LNCaP cells. Nuclei were stained with DAPI (blue).
Magnification, ×630; scale bar, 20 µm. (D) Immunohistochemical
analysis of MIF in mouse tissue sections at the end of the study
period. Metastases in representative histological sections of spine
tissues are shown (magnification, ×200; scale bar, 20 µm) and black
arrows indicate strong MIF positive cells near bone tissue.
*P<0.05 vs. VEC. MIF, macrophage migration inhibitory factor, B,
bone; T, tumor mass; C, spinal cord; OPN, osteopontin; MIF,
macrophage migration inhibitory factor. |
In the immunohistochemical study, MIF expression
increased around the tumor mass and trabecular epiphysis region of
bones inoculated with LNCaPCD133+ cells. These
observations suggest that ectopic overexpression of CD133 leads to
increased MIF expression in CD133+ cells and activation of bones
adjacent to the tumor, particularly in LNCaP cell-injected bones,
and that CD133 expression is associated with PC metastasis to the
bone.
Discussion
Bone is a common site of distant metastasis in PC.
Association of bone and metastatic cancer cells is important in
site-specific manifestation of PC (18). There are no curative treatments for
the disease at this stage, and bone metastasis remains a
devastating complication of advanced PC, despite advances in
understanding of mechanisms underlying the basic molecular biology
of this process. The existence and identification of CSCs reveal
the pivotal roles that these cells play in metastasis (19). In the present study, we identified
CSC-like cells following ectopic overexpression of CD133 in the PC
cell line LNCaP. This phenomenon gave rise to genetic variation in
PC cells and altered expression of several factors related to
stemness, including NANOG and Oct-4, resulting in increased
colony-forming abilities. Furthermore, genetic modulation of CD133
altered expression of EMT-related factors increased vimentin
expression and decreased E-cadherin expression, resulting in the
generation of a complex with TCF-4 and β-catenin. Although the role
of EMT in PC remains unclear, several factors involved in EMT or
related pathways have been identified as key molecules in cancer
cell invasion and metastasis. There is the strong correlation
between CD133 and EMT factors that may explain the invasiveness and
migration of LNCaPCD133+ cells.
Recent studies have shown that EMT may induce
differentiation of cancer cells into a CSC-like state. EMT plays a
critical role in the process of development and wound healing;
however, recent studies have linked EMT with human pathology,
including cancer metastasis (20).
However, no research has yet revealed the mechanisms underlying
this phenomenon or identified the source of cells with both EMT and
stem cell properties. In the present study, we show for the first
time that PC cells via ectopic overexpression of CD133 may present
mesenchymal characteristics and stem cell properties. In addition,
LNCaP cells were originated from not bone metastatic PC but lymph
node, it confirmed that ectopic overexpression of CD133 in LNCaP
led to promote the ability of bone metastasis in the present study.
These results imply that CSCs may induce local and distant
metastases to the bone by acquiring mesenchymal features, which may
greatly facilitate systemic dissemination from the primary mass.
Furthermore, the redundant regulation of β-catenin/TCF-4 signaling
during tumor metastasis through several EMT-inducing signals
suggests that this expression is critical in the initiation and
maintenance of EMT. According to recent studies, the
microenvironment at the invasive front of cancer cells, especially
secretion of factors such as hepatocyte growth factor by stromal
myofibroblasts near dedifferentiated cancer cells, plays a key role
in the nuclear translocation of β-catenin and activation of
β-catenin/TCF-4 signaling (16).
We explored factors that play a critical role in
CD133+ PC cell-induced bone metastasis. LNCaPVec and
LNCaPCD133+ cells were inoculated into athymic nude mice
via intracardiac injection, and showed an osteolytic bone
metastasis with a similar result of previous study (21). And then, secreted cytokines with
paracrine and endocrine actions were evaluated using mouse serum in
array kits. Histological analysis revealed that CD133+ cells
generated larger and more aggressive tumors than their wild-type
counterparts, while LNCaPCD133+ cells showed higher
expression of MIF than LNCaPVec cells. Moreover, MIF is
a pleiotropic inflammatory cytokine with chemokine-like functions
and may bind and signal via several receptors, including CD74,
C-X-C motif chemokine receptor 2 (CXCR2), and CXCR4. MIF may play a
central role in bone metastasis of PC and perform critical
biological functions (22). MIF is
known to induce osteoclast differentiation and contribute to the
progression of various bone and periodontal diseases (23). A previous study has shown that
binding of both MIF and stromal cell-derived factor 1 chemokines to
CXCR4 may trigger chemotaxis of osteoclast precursors and enhance
osteoclastogenesis (24). The
interaction of MIF with chemokine receptors may explain its ability
to induce macrophage migration and osteoclastogenesis. Our current
findings support the conclusion that while a physiological target
is yet unidentified, tautomerase activity is important for MIF
functionality and may contribute to tumor growth and metastasis.
Tropism of LNCaPCD133+ cells to bone suggests that CSCs
may preferentially interact with specific cells in the bone
microenvironment, and the most likely candidates are osteoclasts
that cause an increase in bone lysis at sites of bone metastases,
as demonstrated by histomorphometric evidence.
With respect to clinical applications, CD133 may
serve as a candidate marker to detect bone metastasis risk in
patients with PC. The inability to predict development of
metastatic disease in patients is a major challenge in PC
management and has resulted in excessive therapy in some patients
and delayed or insufficient therapy in others (25). The nature of the association between
circulating tumor cells and bone metastasis remains controversial.
The presence of circulating PC cells in the blood is an indication
of dissemination of tumor cells from the primary site. Only a
subset of circulating tumor cells may possess the necessary
properties to target bone.
In light of the significant progress in the field of
metastasis and stem cell research, we suggest a CSC-based model for
bone metastasis in PC. CD133 regulates CSC and EMT properties in PC
and sustains acquisition of osteolytic features through MIF
secretion. These data demonstrate the role of EMT and CSCs in
cancer metastasis to the bone. Thus, our findings may facilitate
development of a novel classification system and therapeutic
strategies for bone metastasis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea,
funded by the Ministry of Education (grant no.
2017R1D1A1A02018589), and by the Clinical Medicine Research
Institute of the Chosun University Hospital (2015).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
HS, BK, MP, YKo, YM, BJ, JS, YKi and WL take
responsibility for the integrity of the data analysis. HS, BK, MP,
YKo, YM, BJ and WL performed the experiments. HS and BK reviewed,
analyzed, and interpreted the data. JS and YKi performed additional
experiments. HS, BK and WL wrote the paper. All authors discussed
the results and commented on the manuscript.
Ethics approval and consent to
participate
All experimental procedures involving animals were
performed in compliance with institutional and governmental
requirements and were approved by the Institutional Animal Care and
Use Committee (approval no. CIACUC2015-A0032), Chosun University,
Gwangju, Korea.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PC
|
prostate cancer
|
|
MIF
|
macrophage migration inhibitory
factor
|
|
CSC
|
cancer stem cell
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
TCF-4
|
anti-transcription factor 4
|
References
|
1
|
Hassanipour-Azgomi S,
Mohammadian-Hafshejani A, Ghoncheh M, Towhidi F, Jamehshorani S and
Salehiniya H: Incidence and mortality of prostate cancer and their
relationship with the Human Development Index worldwide. Prostate
Int. 4:118–124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jin JK, Dayyani F and Gallick GE: Steps in
prostate cancer progression that lead to bone metastasis. Int J
Cancer. 128:2545–2561. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakazawa M and Kyprianou N:
Epithelial-mesenchymal-transition regulators in prostate cancer:
Androgens and beyond. J Steroid Biochem Mol Biol. 166:84–90. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen YS, Wu MJ, Huang CY, Lin SC, Chuang
TH, Yu CC and Lo JF: CD133/Src axis mediates tumor initiating
property and epithelial-mesenchymal transition of head and neck
cancer. PLoS One. 6:e280532011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shiozawa Y, Berry JE, Eber MR, Jung Y,
Yumoto K, Cackowski FC, Yoon HJ, Parsana P, Mehra R, Wang J, et al:
The marrow niche controls the cancer stem cell phenotype of
disseminated prostate cancer. Oncotarget. 7:41217–41232. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klonisch T, Wiechec E, Hombach-Klonisch S,
Ande SR, Wesselborg S, Schulze-Osthoff K and Los M: Cancer stem
cell markers in common cancers-therapeutic implications. Trends Mol
Med. 14:450–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu C, Yao Z, Jiang Y and Keller ET:
Prostate cancer stem cell biology. Minerva Urol Nefrol. 64:19–33.
2012.PubMed/NCBI
|
|
10
|
Kryczek I, Liu S, Roh M, Vatan L, Szeliga
W, Wei S, Banerjee M, Mao Y, Kotarski J, Wicha MS, et al:
Expression of aldehyde dehydrogenase and CD133 defines ovarian
cancer stem cells. Int J Cancer. 130:29–39. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Colombel M, Eaton CL, Hamdy F, Ricci E,
van der Pluijm G, Cecchini M, Mege-Lechevallier F, Clezardin P and
Thalmann G: Increased expression of putative cancer stem cell
markers in primary prostate cancer is associated with progression
of bone metastases. Prostate. 72:713–720. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shmelkov SV, St Clair R, Lyden D and Rafii
S: AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 37:715–719.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Major AG, Pitty LP and Farah CS: Cancer
stem cell markers in head and neck squamous cell carcinoma. Stem
Cells Int. 2013:3194892013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moon Y, Kim D, Sohn H and Lim W: Effect of
CD133 overexpression on the epithelial-to-mesenchymal transition in
oral cancer cell lines. Clin Exp Metastasis. 33:487–496. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sanchez-Tillo E, de Barrios O, Siles L,
Cuatrecasas M, Castells A and Postigo A: β-catenin/TCF4 complex
induces the epithelial-to-mesenchymal transition (EMT)-activator
ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci USA.
108:19204–19209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arguello F, Baggs RB and Frantz CN: A
murine model of experimental metastasis to bone and bone marrow.
Cancer Res. 48:6876–6881. 1988.PubMed/NCBI
|
|
18
|
Valta MP, Tuomela J, Bjartell A, Valve E,
Väänänen HK and Härkönen P: FGF-8 is involved in bone metastasis of
prostate cancer. Int J Cancer. 123:22–31. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee KH, Ahn EJ, Oh SJ, Kim O, Joo YE, Bae
JA, Yoon S, Ryu HH, Jung S and Kim KK: KITENIN promotes glioma
invasiveness and progression, associated with the induction of EMT
and stemness markers. Oncotarget. 6:3240–3253. 2015.PubMed/NCBI
|
|
20
|
Xu MH, Gao X, Luo D, Zhou XD, Xiong W and
Liu GX: EMT and acquisition of stem cell-like properties are
involved in spontaneous formation of tumorigenic hybrids between
lung cancer and bone marrow-derived mesenchymal stem cells. PLoS
One. 9:e878932014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dai J, Hensel J, Wang N, Kruithof-de Julio
M and Shiozawa Y: Mouse models for studying prostate cancer bone
metastasis. Bonekey Rep. 5:7772016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pasqualon T, Lue H, Groening S,
Pruessmeyer J, Jahr H, Denecke B, Bernhagen J and Ludwig A: Cell
surface syndecan-1 contributes to binding and function of
macrophage migration inhibitory factor (MIF) on epithelial tumor
cells. Biochim Biophys Acta. 1863:717–726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Madeira MF, Queiroz-Junior CM, Costa GM,
Santos PC, Silveira EM, Garlet GP, Cisalpino PS, Teixeira MM, Silva
TA and Souza Dda G: MIF induces osteoclast differentiation and
contributes to progression of periodontal disease in mice. Microbes
Infect. 14:198–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Movila A, Ishii T, Albassam A,
Wisitrasameewong W, Howait M, Yamaguchi T, Ruiz-Torruella M,
Bahammam L, Nishimura K, Van Dyke T and Kawai T: Macrophage
migration inhibitory factor (MIF) supports homing of osteoclast
precursors to peripheral osteolytic lesions. J Bone Miner Res.
31:1688–1700. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chu K, Cheng CJ, Ye X, Lee YC, Zurita AJ,
Chen DT, Yu-Lee LY, Zhang S, Yeh ET, Hu MC, et al: Cadherin-11
promotes the metastasis of prostate cancer cells to bone. Mol
Cancer Res. 6:1259–1267. 2008. View Article : Google Scholar : PubMed/NCBI
|