Introduction
Ovarian cancer has the third highest incidence of
all malignant tumor types in the female reproductive system
globally, but is the leading cause of cancer-associated mortality
worldwide (1). A variety of
mechanisms may underlie the occurrence and development of ovarian
cancer; however, further investigation is required. Novel
diagnostic and prognostic markers and therapeutic targets are
urgently required. Suppressor of cytokine signaling (SOCS) is a
class of negative regulators that block the cytokine signaling
process, and are also negative regulators of Janus kinase (JAK) and
signal transducers and activators of transcription (STAT), which
are involved in the cytokine signaling pathway (2). Among the SOCS family, SOCS1 is the most
important cytokine that induces the JAK/STAT signaling pathway
(3,4), and its main function is to regulate the
activation of STAT3 and STAT5 (5,6). Altered
SOCS1 expression has been reported in a wide range of human cancer
types and may function as a diagnostic or prognostic biomarker. In
a study by Scutti et al (7),
the potential for the mutation and abnormal expression of the SOCS1
gene were observed in liver cancer, non-small cell lung cancer and
malignant melanoma. The mutation usually occurs in the promoter and
the first exon region, and often in the gene regulatory region at
the 5′ end of the gene; the abnormal methylation of the SOCS1 gene
may be involved in the occurrence and development of tumors
(8). However, this phenomenon has
not been clearly reported in ovarian cancer. Therefore, the aim of
the present study was to determine the methylation status and
differential expression of the CpG island of the SOCS1 gene in
ovarian cancer and normal ovarian tissue. We also investigated the
role of the abnormal methylation and expression of SOCS1 in ovarian
cancer, and the potential underlying mechanisms.
Materials and methods
Patients and samples
A total of 26 ovarian cancer samples and 8 normal
ovarian tissues were obtained from patients who underwent
laparoscopic oophorectomy at The First Clinical Hospital of Harbin
Medical University (Harbin, China) between 2014 and 2016. All
patients, with a median age of 40 years (age range, 35–45), were
women and were confirmed to have ovarian cancer by
immunohistochemistry (IHC) testing. According to the classification
of the International Federation of Gynecology and Obstetrics in
2009 (9), there were 6 cases of
stage I, 4 of stage II, 12 of stage III and 4 of stage IV. The
histological grades of the tumor were classified as GI
(well-differentiated) in 9 cases, GII (moderately differentiated)
in 11 cases and GIII (poorly differentiated) in 6 cases. None of
the patients received any radiotherapy or chemotherapy prior to
surgery. Before the specimens were collected, informed consent was
obtained from patients and their families, and approval was granted
from the HMU Medical Science Ethics Committee. All the specimens
were stored at −80°C prior to analysis to avoid repeated
freeze/thawing.
Methylation-specific polymerase chain
reaction (MSP)
Using a DNA Extraction kit (Takara Bio, Inc.)
according to the manufacturer's protocol, DNA was extracted from
the epithelium of the ovarian cancer tissue. Sodium bisulfite
modification was performed using a fast DNA bisulfite kit (Takara
Bio, Inc.), according to the manufacturer's protocols, following
the quantification of the extracted DNA. MSP experiments were
performed using the modified genomic DNA and primers for
non-methylated SOCS1 (forward, 5′-AACCAAAAAAATAAACCATAACATC-3′ and
reverse, 5′-TAGTTGTGTTTATTGAGGTTGAATG-3′) and primers for
methylated SOCS1 (forward, 5′-ACCGAAAAAATAAACCATAACGTC-3′ and
reverse, 5′-TAGTTGTGTTTATTGAGGTTGAACG-3′). The primers were
designed using MethPrimer v2.0 software (http://www.urogene.org/methprimer2) (10). A total volume of 50 µl reaction
solution containing 2 µl DNA, 2 µl primers, 25 µl SYBR Premix Ex
Taq™ (Takara Bio, Inc.) and 21 µl ddH2O was used. The
ABI PRISM 7500 FRT sequence detection system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) was used for the MSP reaction. The
thermocycling conditions were 95°C for 5 sec and 60°C for 30 sec
(40 cycles). The data were collected and analyzed using the
sequence detection system. The sequence detection system was
performed using 7500 Software for 7500 and 7500 Fast Real-Time PCR
systems v2.3 (Thermo Fisher Scientific, Inc.). In order to
standardize the methylated or non-methylated status of the SOCS1
gene in all specimens, all values were expressed as a multiple of
the increase or decrease in methylated SOCS1 gene relative to the
non-methylated SOCS1 gene. The gene replication values of each
specimen were expressed by quantification cycle (Cq). The
methylation levels of the gene (ΔCq) were expressed as the
difference between the methylation value of the SOCS1 gene and the
non-methylation value of the SOCS1 gene; the methylation rate of
genes was determined using the following formula: 2−(Cq
methylation-Cq non-methylation)/2−(Cq methylation-Ct
non-methylation) +1.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The RNA from the tumor tissues and corresponding
normal tissues were extracted using the RNeasy® Mini Kit
(Qiagen, Inc.), according to the manufacturer's protocol. The
purity ratio of the optical density at 260/280 for the extracted
RNA was between 1.9–2.0. RT-qPCR was performed using the
SuperScript III kit 2-Step RT-PCR system (Invitrogen; Thermo Fisher
Scientific, Inc.). The total RNA used was 2 µg. The thermocycling
conditions were as follows: 65°C for 15, 5 min on ice, 50°C for 60
min and 70°C for 15 min. The total reaction volume used was 20 µl.
Applied Biosystems® GeneAmp®PCR System 9700
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for
the RT-qPCR. A primer set (a pair of qPCR primers for each set)
specific for SOCS1 were designed by HaiGene. Primers used were as
follows: SOCS1 forward, 5′-GGAACTGCTTTTTCGCCCCTTA-3′ and reverse,
5′-AGCAGCTCGAAGAGGCAGTC-3′. β-actin forward,
5′-AAACTGGAACGGTGAAGGTG-3′ and reverse, 5′-GTGGACTTGGGAGAGGACTG-3′.
The reaction system was as follows: 2 µl cDNA, 4 µl SOCS1
primers/β-actin primers (10 µM), 25 µl SYBR Premix Ex Taq™ (Takara
Bio, Inc.) and 19 µl ddH2O; the total reaction volume
was 50 µl. The ABI PRISM7500 Sequence detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used for the qPCR.
The thermocycling conditions were as follows: 95°C for 15 sec and
60°C for 30 sec (40 cycles). The data was collected and analyzed
using the sequence detection system. The sequence detection system
was performed using 7500 Software for 7500 and 7500 Fast Real-Time
PCR systems 2.3 software (Thermo Fisher Scientific, Inc.). The
β-actin gene was used as an endogenous reference to standardize the
relative expression of the SOCS1 gene in all specimens. All values
were expressed as a multiple of increase or decrease in SOCS1 mRNA
relative to the β-actin gene. The value of gene replication in each
specimen was expressed by the quantification cycle (Cq limit cycle
number). The differences between the Cq value of the target gene
and the endogenous reference gene β-actin (ΔCq) were determined.
The relative expression level of the gene (ΔΔCq) was expressed as
the difference between the ΔCq value of the test specimen and the
reference specimen, and the relative expression quantity of the
target gene was expressed as 2−ΔΔCq (11).
IHC analysis
SP link IHC detection kits (biotin-streptavidin HRP
detection systems, including goat anti-rabbit secondary antibodies,
endogenous peroxidase blockers and normal goat serum) was purchased
from Zhongshan Golden Bridge Biotechnology, Co., Ltd. Endogenous
peroxidase blockers (100 µl; cat. no., SP-9001) was purchased from
Zhongshan Golden Bridge Biotechnology, Co., Ltd. and used as
blocking agent at room temperature for 10 min. Normal goat serum
(100 µl; cat. no., SP-9001) was purchased from Zhongshan Golden
Bridge Biotechnology, Co., Ltd. and sealed for 15 min at room
temperature. Concentrated rabbit anti-SOCS1 polyclonal antibody
(dilution, 1:100; cat. no., SC-9021) was purchased from Santa Cruz
Biotechnology, Inc. and used as the primary antibody. A total of 4%
paraformaldehyde (Biosharp; REF:BL539A) was used as a fixative at
room temperature for 48 h. The thickness of the sections was 5 µm.
Tissue sections were incubated with primary antibody at 4°C
overnight. Then, tissue sections were incubated with HRP-conjugated
goat anti-rabbit secondary antibody (100 µl; cat. no., SP-9001) at
37°C for 60 min. The protein location and expression intensity were
visualized by 3,3-diaminobenzidine tetrahydrochloride (Boster
Biological Technology) and hematoxylin staining at room temperature
for 2 min. Finally, a binocular light microscope (×400
magnification) was used to analyze staining. IHC was performed as
previously reported (12). The
staining results for the SOCS1 protein were semi-quantitatively
calculated by multiplying the staining intensity and the percentage
of positive ovarian cancer cells. The staining result was
considered to be positive for SOCS1 if brown-yellow granules were
observed and if they were located in the cytoplasm.
The Cancer Genome Atlas (TCGA) gene
expression data
The data of ovarian cancer were downloaded by R
(TCGA-Assembler 2 R package) from TCGA (http://cancergenome.nih.gov). The expression of SOCS1
in RNA-HTSeq-FPKM-UQ data and overall survival time in
clinicopathological parameters were obtained for
survival-associated comparison using R survival package (survival
2.42–6.tar.gz).
Statistical analyses
All quantitative data are expressed as (mean ±
standard deviation). Overall survival time analysis was estimated
by the Kaplan-Meier method using the ‘survival’ package (13) of R (14). The Kaplan-Meier method was performed
for visualization purposes and the differences between survival
curves were calculated by the log-rank test. Pearson's correlation
coefficient test was used for correlation analysis. Statistical
analysis was performed using GraphPad Prism v.6 Software (GraphPad
Software, Inc.). The unpaired t-test was used for statistical
analysis and repeated in triplicate. P<0.05 was considered to
indicate a statistically significant difference.
Results
Methylation rate of SOCS1 mRNA is
decreased in ovarian cancer tissues
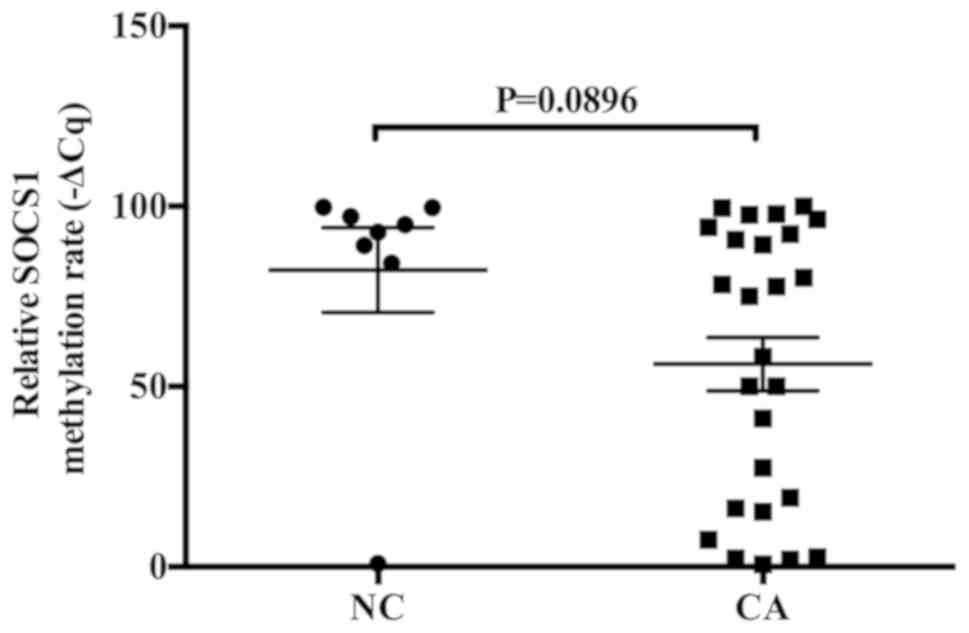
MSP was used to detect the relative methylation rate
of SOCS1 mRNA in 26 cases of ovarian cancer and 8 cases of normal
ovarian tissue. The methylation rate of SOCS1 mRNA was decreased in
ovarian cancer tissues compared with that in the normal ovarian
tissues. The methylation rate of SOCS1 mRNA in 13 cases of ovarian
cancer was <56% (Fig. 1).
Expression of SOCS1 mRNA is
significantly increased in ovarian cancer tissues and its
expression level is associated with the overall survival time in
ovarian cancer
The present study selected two sections of each
sample each from 19 ovarian cancer tissues and 5 normal ovarian
specimens. The 19 ovarian cancer samples were taken from 26 ovarian
cancer samples, and the 5 normal ovarian samples were taken from 8
normal ovarian samples, then applied RT-qPCR to detect the relative
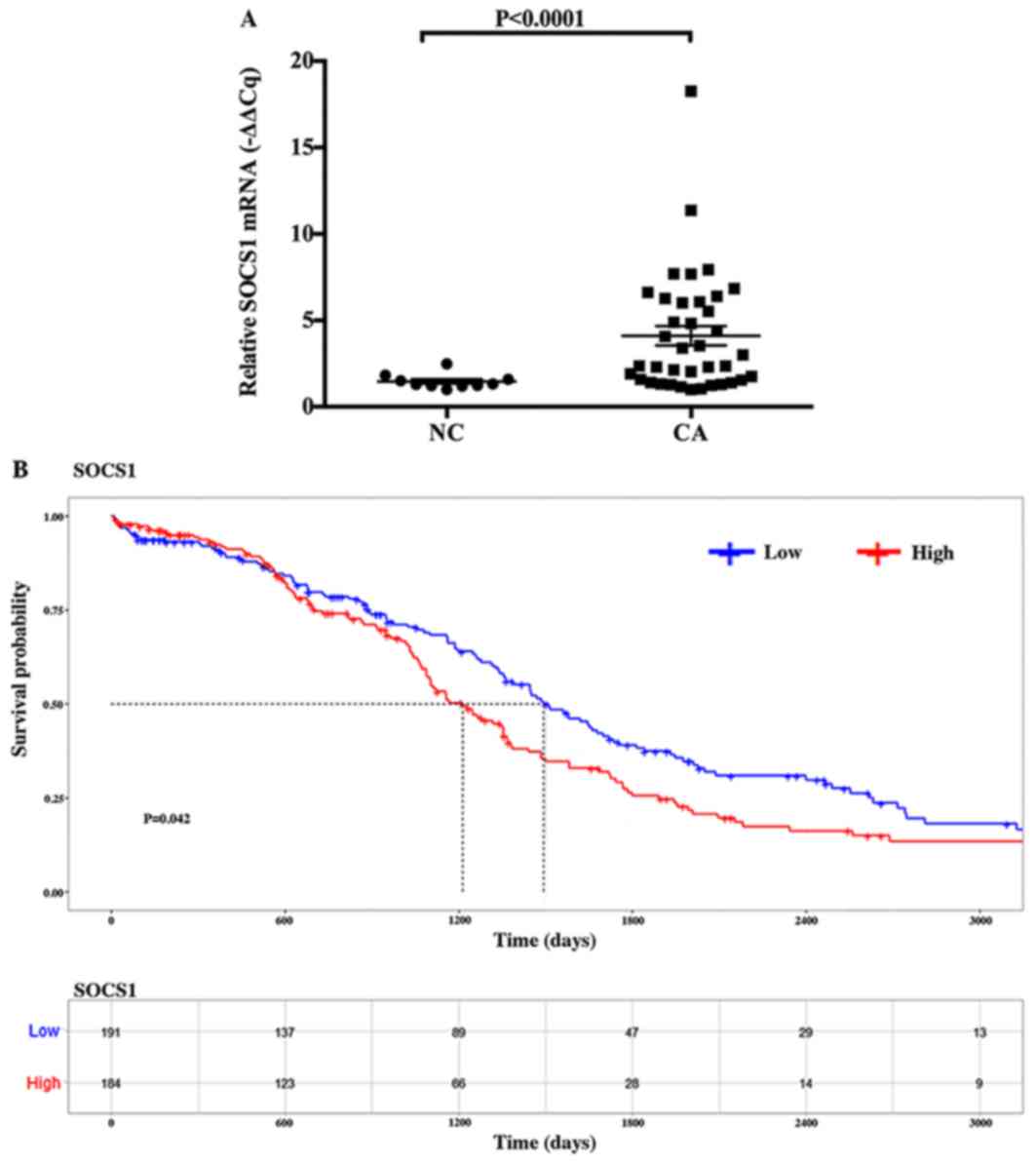
expression of SOCS1 mRNA. The mean value of the relative expression
of SOCS1 mRNA in ovarian cancer specimens was 4.0633, and that in
normal ovarian tissues was 1.4681. The expression of SOCS1 was
significantly increased in ovarian cancer tissues compared with in
the normal tissues (P<0.0001; Fig.
2A). Additionally, in order to further clarify the association
between the expression of SOCS1 mRNA and the occurrence and
development of ovarian cancer, the present study investigated the
expression of SOCS1 mRNA and overall survival time in ovarian
cancer using a dataset downloaded from TCGA database. The results
of the survival prediction table showed that SOCS1 mRNA expression
in ovarian cancer is significantly associated with overall survival
time (P=0.042; Fig. 2B). When the
predicted overall survival time was 0, 600, 1,200, 1,800, 2,400 and
3,000 days, the number of ovarian cancer samples with low SOCS1
expression corresponded to 191, 137, 89, 47, 29 and 13 cases,
respectively; the number of ovarian cancer samples with high SOCS1
expression corresponded to 184, 123, 66, 28, 14 and 9 cases,
respectively (Fig. 2B).
Expression of SOCS1 protein is
positive in ovarian cancer
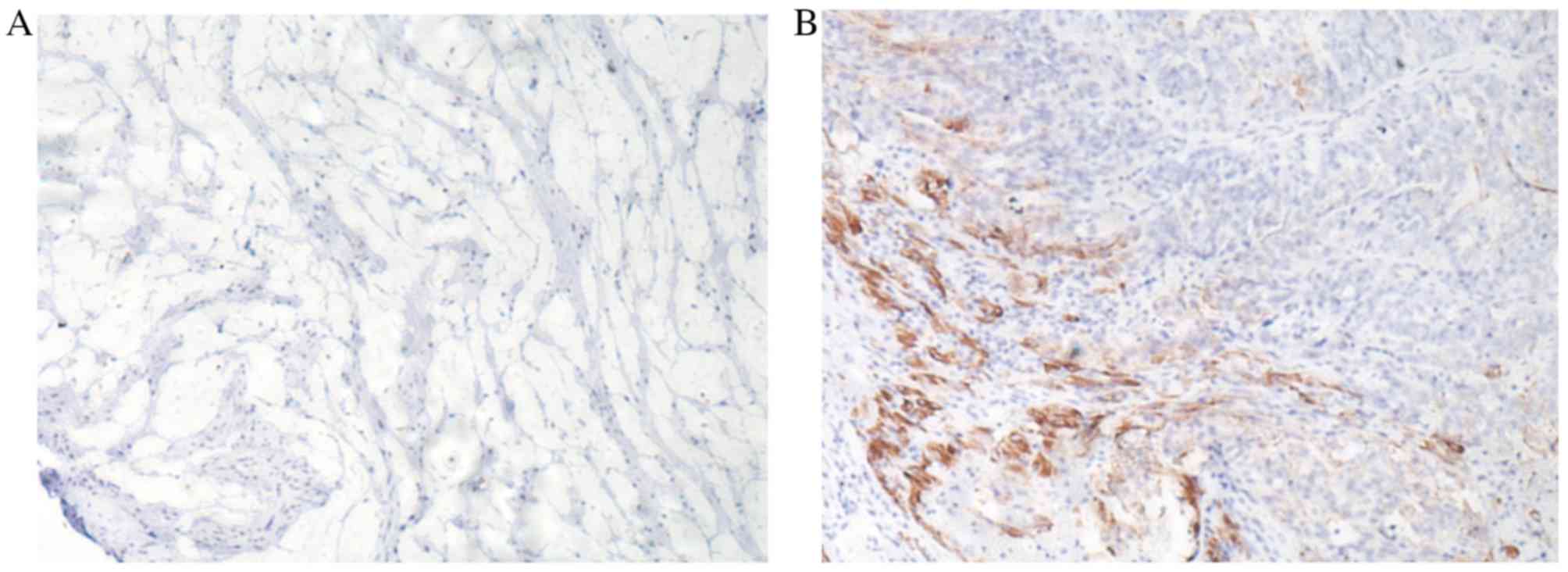
IHC analysis revealed that the expression of SOCS1
protein was positive in ovarian cancer specimens, but negative for
normal ovarian tissue. Positive SOCS1 protein staining presented
brown-yellow granules, while negative SOCS1 protein staining
presented light yellow granules, which were located in the
cytoplasm (Fig. 3).
Methylation of SOCS1 mRNA correlates
with the rising expression of the SOCS1 gene
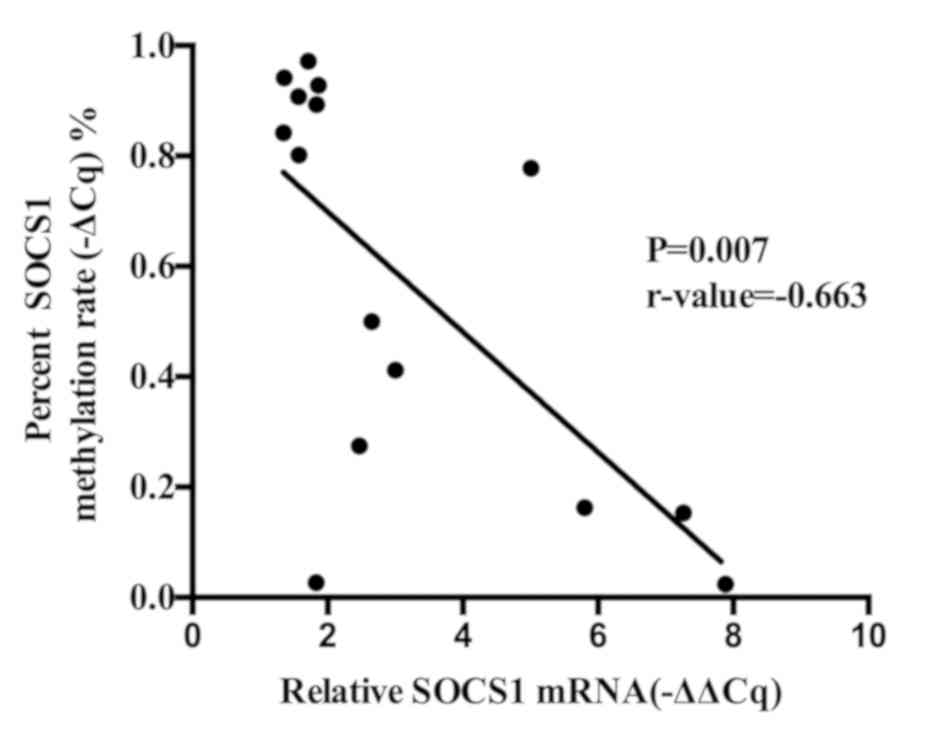
Pearson's correlation coefficient test was performed
to study the association between the methylation rate of SOCS1 and
the relative expression of SOCS1 mRNA, which was revealed to be
negative in ovarian cancer specimens and normal tissues (P=0.007,
r-value=−0.663; Fig. 4).
Discussion
SOCS1 is the earliest discovered member of the SOCS
family and a key molecule in cancer development (5,7,15). As a major signal transduction
pathway, the JAK/STAT signaling pathway serves a notable role in
numerous biological reactions (3).
As one of the important negative regulatory proteins in the
JAK/STAT signaling pathway, SOCS1 has a strong inhibitory effect on
this signaling pathway via three mechanisms: i) It is able to
inactivate the N-terminal of JAK kinase by binding its SH2 domain
to the phosphorylated tyrosine residues of the target protein, so
that signal transduction is inhibited (16). Additionally, the N-terminal of SOCS1
contains a kinase inhibitory region that directly inhibits the
activity of JAK tyrosine kinase (17–20); ii)
the SOCS1 promoter contains a STAT binding site that can inhibit
STAT from binding to the receptor site; and iii) the SOCS cassette
is able to interact with elongin B, elongin C, cullin-2 and
RINGbox-2, and recruits E3 ubiquitin transferase, to promote the
degradation of the bound protein via the proteasome-dependent
pathway (21). The silencing of the
SOCS1 gene may result from its methylation and the inactivation of
the JAK/STAT signaling pathway in the negative feedback control of
tumor cells (22–25). In contrast, the hypomethylation of
the SOCS1 gene may restore the transcription and protein expression
of the SOCS1 gene, which is able to induce the apoptosis and growth
inhibition of cells (26,27).
It has been reported that the expression of the
SOCS1 gene is increased in breast cancer (28), prostate cancer (29), acute myeloid leukemia and other
malignancies (30). However, a study
by Li et al (31), the
expression of SOCS1 was revealed to be decreased within the
metastatic sites of malignant melanoma. In hepatocellular
carcinoma, the methylation of the SOCS1 gene coexists with a
deletion mutation (32). The reason
for this discrepancy in the expression of SOCS1 is yet to be
definitively answered, but its expression in the majority of
malignancies is consistent. The SOCS1 gene is highly expressed in
breast cancer and inhibits the growth of tumor cells (28). Likewise, Shimada et al
(20) observed that the
overexpression of the SOCS1 gene is effective for antitumor therapy
by suppressing the JAK/STAT, focal adhesion kinase and epidermal
growth factor receptor signaling pathways in non-small cell lung
cancer. Numerous cytokines and hormones are able to affect tumor
proliferation, activation and apoptosis by activating the JAK-STAT
signaling pathway. For example, Yun et al (33) observed that IL-6 inhibits senescence
through the activation of the JAK1-STAT3 signaling pathway. In
addition, Cokic et al (34)
observed that EPO-mediated proliferation and survival of erythroid
progenitors occurs mainly through the modulation of JAK-STAT
pathway. Likewise, the inhibition of the cytokine-dependent
activation of the JAK/STAT3 pathway may also afford orthogonal
treatment opportunities for other oncogene-dependent cancer cells.
Among these cytokines and hormones, STAT3 serves various role in
signal transduction, which has been reported in numerous cancer
types including breast (35),
prostate (36) and pancreatic cancer
(37). The expression of SOCS1 may
be induced by the activation of STAT3. In a study by Akira
(27), it was revealed that ovarian
cancer cell lines and clinical specimens exhibited sustained
activation of STAT3, which is able to inhibit apoptosis and promote
the proliferation or metastasis of cells involved in the occurrence
and progression of ovarian cancer via the JAK/STAT signaling
pathway (38). Therefore, the
overexpression of SOCS1 may be the result of the sustained
activation of STAT3, and is able to control the development of a
tumor by inhibiting JAK/STAT signaling.
At present, the mechanism of SOCS1 in ovarian cancer
remains unclear. In the present study, the expression of SOCS1 in
ovarian cancer specimens was increased and its methylation rate was
decreased. This study did not analyze the association between SOCS1
expression and histological grading of ovarian cancer. Furthermore,
the methylation rate of the SOCS1 gene negatively correlated with
the relative expression of SOCS1 in ovarian cancer specimens and
normal ovarian specimens. Therefore, it was deduced that the
reduction of SOCS1 methylation in ovarian cancer specimens could
increase the expression of SOCS1, which is to inhibit the JAK/STAT
signaling pathway, and may be involved in the development and
progression of ovarian cancer. It may also be that the sustained
activation of STAT3 in ovarian cancer causes the high expression of
SOCS1 (39), which could be adopted
by the host as a protective mechanism against disease.
Additionally, analysis of the TCGA gene expression datasets for a
large cohort of patients with ovarian cancer was performed in the
present study, where the SOCS1 expression level was associated with
overall survival time (Fig. 2B),
suggesting that SOCS1 may be used to determine the prognosis of
patients with ovarian cancer. Due to the small sample size, this
study was unable to investigate the association between SOCS1
expression level and the different clinical grades in the ovarian
cancer and normal tissues. A larger sample size is required in
future studies to investigate this further. In addition, in
vitro investigations using ovarian cancer cell lines to detect
differences in the expression of SOCS1, and further analyze the
role of SOCS1 in the development and progression of ovarian cancer
will also be performed. In a recent study, Nakagawa et al
(40) demonstrated that the
adenoviral delivery of SOCS1 inhibited tumor growth through the
downregulation of programmed death-ligand 1 expression, resulting
in the activation of tumor-infiltrating T cells. Thus, the SOCS1
gene may be a novel molecular marker and therapeutic target for
ovarian cancer. Notably, the demethylation of SOCS1 CpG islands may
restore the function of tumor suppressor genes, which could be a
novel starting point for developing therapeutic drugs against
ovarian cancer; however, further investigation into the underlying
mechanisms are required.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Research
Progress of SOCS1 Gene in Ovarian Cancer from the National Natural
Science Foundation (grant no. 81401660) and the Heilongjiang
Natural Science Foundation (grant no. H201351).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
CM and WL analyzed and interpreted data on patients
with ovarian cancer. CK, YF and JL conceived and designed the
experiments. AL, AZ, WS, XH and BY performed the experiments, and
XL participated in the whole process of experimental research and
contributed in the writing of the manuscript. All authors have read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from patients and
their families, and approval was granted from the HMU Medical
Science Ethics Committee (approval no. HMUIRB20180006).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Ahmedin J: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Babon JJ, Varghese LN and Nicola NA:
Inhibition of IL-6 family cytokines by SOCS3. Semin Immunol.
26:13–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liau N, Laktyushin A, Lucet IS, Murphy JM,
Yao S, Whitlock E, Callaghan K, Nicola NA, Kershaw NJ and Babon JJ:
The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun.
9:15582018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liang YB, Tang H, Chen ZB, Zeng LJ, Wu JG,
Yang W, Li ZY and Ma ZF: Downregulated SOCS1 expression activates
the JAK1/STAT1 pathway and promotes polarization of macrophages
into M1 type. Mol Med Rep. 16:6405–6411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan SR, Rickert CG, Vermi W, Sheehan KC,
Arthur C, Allen JA, White JM, Archambault J, Lonardi S, Mcdevitt
TM, et al: Dysregulated STAT1-SOCS1 control of JAK2 promotes
mammary luminal progenitor cell survival and drives ERα(+)
tumorigenesis. CDD. 21:234–246. 2014.
|
|
6
|
Wingelhofer B, Neubauer HA, Valent P, Han
X, Constantinescu SN, Gunning PT, Müller M and Moriggl R:
Implications of STAT3 and STAT5 signaling on gene regulation and
chromatin remodeling in hematopoietic cancer. Leukemia.
1:1713–1726. 2018. View Article : Google Scholar
|
|
7
|
Scutti JA, Matsuo AL, Pereira FV, Massaoka
MH, Figueiredo CR, Moreira DF, Belizário JE and Travassos LR: Role
of SOCS1 gene on melanoma cell growth and tumor development. Transl
Oncol. 4:101–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saelee P, Chuensumran U, Wongkham S,
Chariyalertsak S, Tiwawech D and Petmitr S: Hypermethylation of
suppressor of cytokine signaling 1 in hepatocellular carcinoma
patients. Asian Pac J Cancer Prev. 13:3489–3493. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zalewski K, Doniec J, Włodzimierz W and
Bidziński M: Revised FIGO staging systems for gynecologic
malignancies-2009 update. Ginekol Pol. 81:778–782. 2010.(In
Polish). PubMed/NCBI
|
|
10
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li C, Yang W, Zhang J, Zheng X, Yao Y, Tu
K and Liu Q: SREBP-1 has a prognostic role and contributes to
invasion and metastasis in human hepatocellular carcinoma. Int J
Mol Sci. 15:7124–7138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Therneau T: Survival: Survival analysis
including penalised likelihood. https://CRAN.R-project.org/package=survivalOctober
2–2018
|
|
14
|
R Core Team: R, . A Language and
Environment for Statistical Computing. R Foundation for Statistical
Computing; Vienna, Austria: http://www.R-project.orgOctober 2–2018
|
|
15
|
Chim CS, Fung TK, Cheung WC, Liang R and
Kwong YL: SOCS1 and SHP1 hypermethylation in multiple myeloma:
Implications for epigenetic activation of the Jak/STAT pathway.
Blood. 103:4630–4635. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Endo TA, Masuhara M, Yokouchi M, Suzuki R,
Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H,
et al: A new protein containing an SH2 domain that inhibits JAK
kinases. Nature. 387:921–924. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yasukawa H, Misawa H, Sakamoto H, Masuhara
M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN
and Yoshimura A: The JAK-binding protein JAB inhibits Janus
tyrosine kinase activity through binding in the activation loop.
EMBO J. 18:1309–1320. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Babon JJ, Kershaw NJ, Murphy JM, Varghese
LN, Laktyushin A, Young SN, Lucet IS, Norton RS and Nicola NA:
Suppression of cytokine signaling by SOCS3: Characterization of the
mode of inhibition and the basis of its specificity. Immunity.
36:239–250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kazi JU, Kabir NN, Flores-Morales A and
Rönnstrand L: SOCS proteins in regulation of receptor tyrosine
kinase signaling. CMLS. 71:3297–3310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shimada K, Serada S, Fujimoto M, Nomura S,
Nakatsuka R, Harada E, Iwahori K, Tachibana I, Takahashi T,
Kumanogoh A, et al: Molecular mechanism underlying the
antiproliferative effect of suppressor of cytokine signaling-1 in
non-small-cell lung cancer cells. Cancer Sci. 104:1483–1491. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kamizono S, Hanada T, Yasukawa H,
Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M,
Morita S, et al: The SOCS box of SOCS1 accelerates
ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem.
276:12530–12538. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoshikawa H, Matsubara K, Qian GS, Jackson
P, Groopman JD, Manning JE, Harris CC and Herman JG: SOCS1, a
negative regulator of the JAK/STAT pathway, is silenced by
methylation in human hepatocellular carcinoma and shows
growth-suppression activity. Nat Genet. 28:29–35. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Galm O, Yoshikawa H, Esteller M, Osieka R
and Herman JG: SOCS1, a negative regulator of cytokine signaling,
is frequently silenced by methylation in multiple myeloma. Blood.
101:2784–2788. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin YC, Lin CK, Tsai YH, Weng HH, Li YC,
You L, Chen JK, Jablons DM and Yang CT: Adenovirus-mediated SOCS3
gene transfer inhibits the growth and enhances the radiosensitivity
of human non-small cell lung cancer cells. Oncol Rep. 24:1605–1612.
2010.PubMed/NCBI
|
|
25
|
Watanabe D, Ezoe S, Fujimoto M, Kimura A,
Saito Y, Nagai H, Tachibana I, Matsumura I, Tanaka T, Kanegane H,
et al: Suppressor of cytokine signalling-1 gene silencing in acute
myeloid leukaemia and human haematopoietic cell lines. Br J
Haematol. 126:726–735. 2015. View Article : Google Scholar
|
|
26
|
Sutherland KD, Lindeman GJ, Choong DY,
Wittlin S, Brentzell L, Phillips W, Campbell IG and Visvader JE:
Differential hypermethylation of SOCS genes in ovarian and breast
carcinomas. Oncogene. 23:7726–7733. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Burke WM, Jin X, Lin HJ, Huang M, Liu R,
Reynolds RK and Lin J: Inhibition of constitutively active Stat3
suppresses growth of human ovarian and breast cancer cells.
Oncogene. 20:7925–7934. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sasi W, Jiang WG, Sharma A and Mokbel K:
Higher expression levels of SOCS 1,3,4,7 are associated with
earlier tumour stage and better clinical outcome in human breast
cancer. BMC Cancer. 10:1782010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Flowers LO, Subramaniam PS and Johnson HM:
A SOCS1 peptide mimetic inhibits both constitutive and IL-6 induced
activation of STAT3 in prostate cancer cells. Oncogene.
24:2114–2120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Griffiths EA, Gore SD, Hooker CM, Mohammad
HP, Mcdevitt MA, Smith BD, Karp JE, Herman JG and Carraway HE:
Epigenetic differences in cytogenetically normal versus abnormal
acute myeloid leukemia. Epigenetics. 5:590–600. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Z, Metze D, Nashan D, Müller-Tidow C,
Serve HL, Poremba C, Luger TA and Böhm M: Expression of SOCS1,
suppressor of cytokine signalling-1, in Human Melanoma. J Invest
Dermatol. 123:737–745. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nagai H, Kim YS, Konishi N, Baba M, Kubota
T, Yoshimura A and Emi M: Combined hypermethylation and chromosome
loss associated with inactivation of SSI-1/SOCS1/JAB gene in human
hepatocellular carcinomas. Cancer Lett. 186:59–65. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yun UJ, Park SE, Jo YS, Kim J and Shin DY:
DNA damage induces the IL-6/STAT3 signaling pathway, which has
anti-senescence and growth-promoting functions in human tumors.
Cancer Lett. 323:155–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cokic VP, Bhattacharya B, Beleslin-Cokic
BB, Noguchi CT, Puri RK and Schechter AN: Jak-STAT and AKT
pathway-coupled genes in erythroid progenitor cells through
ontogeny. J Transl Med. 10:1162012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Haricharan S and Li Y: STAT signaling in
mammary gland differentiation, cell survival and tumorigenesis. Mol
Cell Endocrinol. 382:560–569. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dallavalle C, Albino D, Civenni G, Merulla
J, Ostano P, Mellogrand M, Rossi S, Losa M, D'Ambrosio G, Sessa F,
et al: MicroRNA-424 impairs ubiquitination to activate STAT3 and
promote prostate tumor progression. JCI. 126:4585–4602. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang X, Ren D, Wu X, Lin X, Ye L, Lin C,
Wu S, Zhu J, Peng X and Song L: miR-1266 contributes to pancreatic
cancer progression and chemoresistance by STAT3 and NF-κB signaling
pathways. Mol Ther Nucleic Acids. 11:142–158. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saydmohammed M, Joseph D and Syed V:
Curcumin suppresses constitutive activation of STAT-3 by
up-regulating protein inhibitor of activated STAT-3 (PIAS-3) in
ovarian and endometrial cancer cells. J Cell Biochem. 110:447–456.
2010.PubMed/NCBI
|
|
39
|
Cai L, Zhang G, Tong X, You Q, An Y, Wang
Y, Guo L, Wang T, Zhu D and Zheng J: Growth inhibition of human
ovarian cancer cells by blocking STAT3 activation with small
interfering RNA. Eur J Obstet Gynecol Reprod Biol. 148:73–80. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nakagawa S, Serada S, Kakubari R,
Hiramatsu K, Sugase T, Matsuzaki S, Matsuzaki S, Ueda Y, Yoshino K,
Ohkawara T, et al: Intratumoral delivery of an adenoviral vector
carrying the SOCS1 gene enhances T cell-mediated anti-tumor
immunity by suppressing PD-L1. Mol Cancer Ther. 17:1941–1950. 2018.
View Article : Google Scholar : PubMed/NCBI
|