Introduction
Cervical cancer (CC) is the third most prevalent
gynecological malignancy and the second leading cause of
cancer-associated mortality amongst females worldwide with an
estimated 530,000 mortalities per year (1,2). Despite
developments in radiotherapy, chemotherapy and surgery for the
treatment of CC, the 5-year survival rate for patients with CC is
still low (2,3). Therefore, the underlying molecular
mechanisms of the initiation and progression of CC must be
explored, and potential therapeutic strategies should be
identified.
MicroRNAs (miRNAs) are small non-coding RNA
sequences with 18–21 nucleotides that modulate translational
efficiency or stability by targeting the 3′-untranslated regions
(3′-UTRs) of mRNAs (3). miRNAs serve
a key role in the progression and oncogenesis of a variety of
cancers, including CC (4,5). In CC, numerous miRNAs are involved in
cancer initiation, promotion and progression (4). The aberrant expression of miR-758 is
closely related to glioma, hepatocellular carcinoma and non-small
lung cancer (6–8). miR-758 may serve as a tumor suppressor
and inhibit CC metastasis (9).
However, the biological function and molecular mechanism of miR-758
in CC have not been well illustrated.
In the present study, miR-758 was markedly decreased
in primary CC tumor tissues and cell lines. In vitro
analysis demonstrated that miR-758 inhibited cell proliferation,
migration and invasion in CC cells. High mobility group box 3
(HMGB3) was identified as a direct target of miR-758, and it was
involved in miR-758-regulated cell progression. The present study
revealed that miR-758 could negatively regulate the WNT/β-catenin
signaling pathway. The present study was the first to provide novel
clues regarding the role of miR-758 as a tumor suppressor gene by
regulating HMGB3 in CC.
Materials and methods
Tissue collection
Human cervical cancer tissues and paired normal
cervical tissues were collected from 20 patients (stage I,5
patients; stage II,10 patients; stage III,5 patients) with cervical
cancer who were admitted to the Department of Gynecology, Weifang
Maternity and Child Care Hospital between January 2017 to December
2017, and written informed consent was obtained from all patients.
Patients age range was between 42 and 71 years; mean age
52.23±20.13 years. Seventeen patients were diagnosed with squamous
cell carcinoma; while the other 3 patients were adenocarcinoma. No
patients previously received anticancer treatment, including
chemotherapy and radiotherapy. This study involving human samples
was approved by the Medical Ethics Committee of Weifang Maternity
and Child Care Hospital (Weifang, China).
Cell culture
CC cell lines (HeLa, C33A, CaSki and SiHa) and a
normal cervical normal cervical cell line Ect1/E6E7 were obtained
from the American Type Culture Collection (Manassas, VA, USA). 293T
cells were purchased from the Cell Biology of the Chinese Academy
of Sciences (Shanghai, China). Cell identity was confirmed by STR
analysis. All cell lines were cultured in Dulbecco's modified
Eagle's medium (DMEM) (HyClone; GE Healthcare Life Sciences, Logan,
UT, USA) with 10% fetal bovine serum (FBS; HyClone; GE Healthcare
Life Sciences) in a humidified atmosphere containing 5%
CO2 at 37°C.
Cell transfection
The miR-758 mimic (5′-UUUGUGACCUGGUCCACUAACC-3′),
corresponding controls (miR-NC) (5′-TTCTCCGAACGTGTCACGT-3′),
miR-758 inhibitor (5′-GGUUAGUGGACCAGGUCACAAA-3′), inhibitor control
(anti-NC) (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). A HMGB3
overexpression plasmid and control vector were obtained from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). During cell
transfection, the cells were seeded into 6-well plates, and then
cultured until 50–70% confluency was reached in 1 day. Transient
transfection was performed using Lipofectamine 2000 Reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
referring to the manufacturer's protocols.
Cell viability assay
Cell viability was assessed with a Cell Counting
Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology, Haimen,
China). Briefly, HeLa and C33A cells were plated onto 96-well
plates at a density of 3,000 cells/well. Following culture for the
indicated time-points (0, 24, 48 and 72 h), 10 µl CCK-8 solution
was added into each well and incubated at 37°C. After 3 h, the
absorbance of each well was measured using a Multiskan MK3
spectrophotometer (Thermo Fisher Scientific, Inc.) at a wavelength
of 450 nm.
Colony formation assay
Cell proliferation was analyzed by colony formation
assay. Cells (500) were seeded into a 6-well plate and cultured for
10 days. Cells were fixed with 100% methanol and stained with 0.1%
crystal violet for 30 min. The number of the colonies was
counted.
Western blotting
Total proteins were extracted from cells using a
radioimmunoprecipitation lysis buffer (Thermo Fisher Scientific,
Inc.) and protein concentrations were determined using a BCA
Protein Assay kit (Beyotime Institute of Biotechnology). Protein
samples (30 µg/lane) were fractionated using sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% gels),
transferred to polyvinylidene fluoride (PVDF) membranes (EMD
Millipore, Billerica, MA, USA), and blocked for 1 h with 5% skimmed
milk. Membranes were then incubated at room temperature overnight
with the following primary antibodies: HMGB3 (1:500; cat. no.
AF5507; R&D Systems, Inc., Minneapolis, MN, USA), matrix
metalloproteinase (MMP)7 (1:500; cat. no. sc-80205; Santa Cruz
Biotechnology, Inc.), β-catenin (1:500; cat. no. 9562S; Cell
Signaling Technology, MA, USA), c-MYC (1:500; cat. no. sc-373712;
Santa Cruz Biotechnology, Inc.) and GAPDH (1:1,000; cat. no.
AF0006; Beyotime Institute of Biotechnology). GAPDH served as a
control. Subsequently, the membranes were incubated with
horseradish peroxidase-conjugated goat anti-rabbit secondary
antibodies (1:2,000; cat. no. sc-2004; Santa Cruz Biotechnology,
Inc.) at room temperature for 1 h. Protein expression levels were
detected with enhanced chemiluminescence (ECL) detection solution
(Beyotime Institute of Biotechnology).
Quantitative real-time reverse
transcription-PCR (qPCR)
The expression of miR-758 and 4 genes (HMGB3,
β-catenin, MMP7 and c-MYC) was analyzed with SYBR-Green II (Takara
Biotechnology Co., Ltd., Dalian, China) and a RT-qPCR system (MJ
Research; Bio-Rad Laboratories, Inc., Hercules, CA, USA). RNA was
isolated from cells and tissues using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol. The
cDNA was synthesized using the PrimeScript™ RT Reagent Kit (Takara
Biotechnology Co., Ltd.). Reverse transcription was performed on a
GeneAmp PCR System 9700 (Applied Biosystems; Thermo Fisher
Scientific, Inc.), and qPCR was performed on an ABI 7500 Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
All samples were processed at the same time to avoid
inter-experiment variance. The thermocycling conditions were as
follows: A holding step at 95°C for 30 sec, and 40 cycles at 95°C
for 5 sec and 60°C for 30 sec. The relative mRNA expression of
miR-758 was analyzed as the inverse log of ΔΔCq and normalized to
the reference gene U6 (10). The
relative mRNA expression of HMGB3, MMP7, β-catenin, c-MYC was
analyzed as the inverse log of ΔΔCq and normalized to the reference
gene GAPDH (10). The primers were
designed as follows: miR-758 forward, 5′-ACACTCCAGCTGGGAACGATG3′
and reverse, 5′-CTCAACTGGTGTCGTGGAGTCGGCA3′; U6 forward,
5′-TGCGGGTGCTCGCTTCGCAGC-3′, and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; HMGB3 forward,
5′-CAGCTTGATACCTGTGAATGGG-3′ and reverse,
5′-TATCTGTGGTCGTGTGGGACT-3′; MMP7 forward,
5′-GTCTCTGGACGGCAGCTATG-3 and reverse, 5′-GATAGTCCTGAGCCTGTTCCC-3′;
β-catenin forward, 5′-ACCTCCCAAGTCCTGTAT-3′ and reverse,
5′-CCTGGTCCTCGTCATTTA-3′; c-MYC forward, 5′-CACAGCAAACCTCCTCACAG-3′
and reverse, 5′-GGATAGTCCTTCCGAGTGGA-3′; and GAPDH forward,
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Transwell assay
For cell migration assays, 1×105
transfected cells in serum-free medium were added into the upper
separate compartment of a Transwell chamber (Corning Incorporated,
Corning, NY, USA). Medium containing 10% FBS that was placed into
the bottom chamber was used as a chemoattractant. For cell invasion
assays, transfected cells were seeded into the upper chamber of the
Transwell after adding diluted Matrigel. After 24 h of incubation
for migration assays and 48 h of incubation for invasion assays at
37°C of a 5% CO2 atmosphere, cells on the top surface of
the filters that did not pass through the pores were removed from
the upper chamber using a cotton swab, while cells on the bottom
surface of the membrane that migrated or invaded through the pores
were fixed with methanol and stained with 0.1% crystal violet for
30 min. Images from 5 different fields were captured and counted
under a light microscope.
Bioinformatic prediction
To investigate the possible target gene of miR-758,
the online prediction system, TargetScan (http://www.targetscan.org), was applied. TargetScan
target gene prediction software identified the 2784–2791 site at
the 3′-end of the 3′-UTR of HMGB3 mRNA as a possible site of action
of miR-758.
Dual-luciferase reporter assay
Wild-type (Wt) and mutated (Mut) putative
miR-758-binding sites in the HMGB3 3′-UTR region were cloned into
the downstream region of the luciferase gene in the psiCHECK-2™
Vector (Promega Corporation, Madison, WI, USA). For the reporter
assay, 293T cells were co-transfected with Wt or mut HMGB3-3′-UTR
vectors and miR-758 mimics or inhibitor. Luciferase activities were
assessed with a Dual-Luciferase Reporter Assay Kit (Promega
Corporation) following manufacturer's protocols. Data were
normalized against the activity of the Renilla luciferase
gene.
Statistical analysis
Statistical analyses were performed using SPSS
version 16.0 (SPSS, Inc., Chicago, IL, USA). All experiments were
performed in triplicate. Unless otherwise indicated, the data were
evaluated as the mean ± standard deviation. Differences between 2
groups were assessed using two-tailed Student's t-test. Data of
>2 groups were assessed using one-way analysis of variance with
post hoc Tukey's test. The correlations between miR-758 expression
levels and the mRNA expression of HMGB3 levels in CC tissues were
analyzed using Spearman's rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
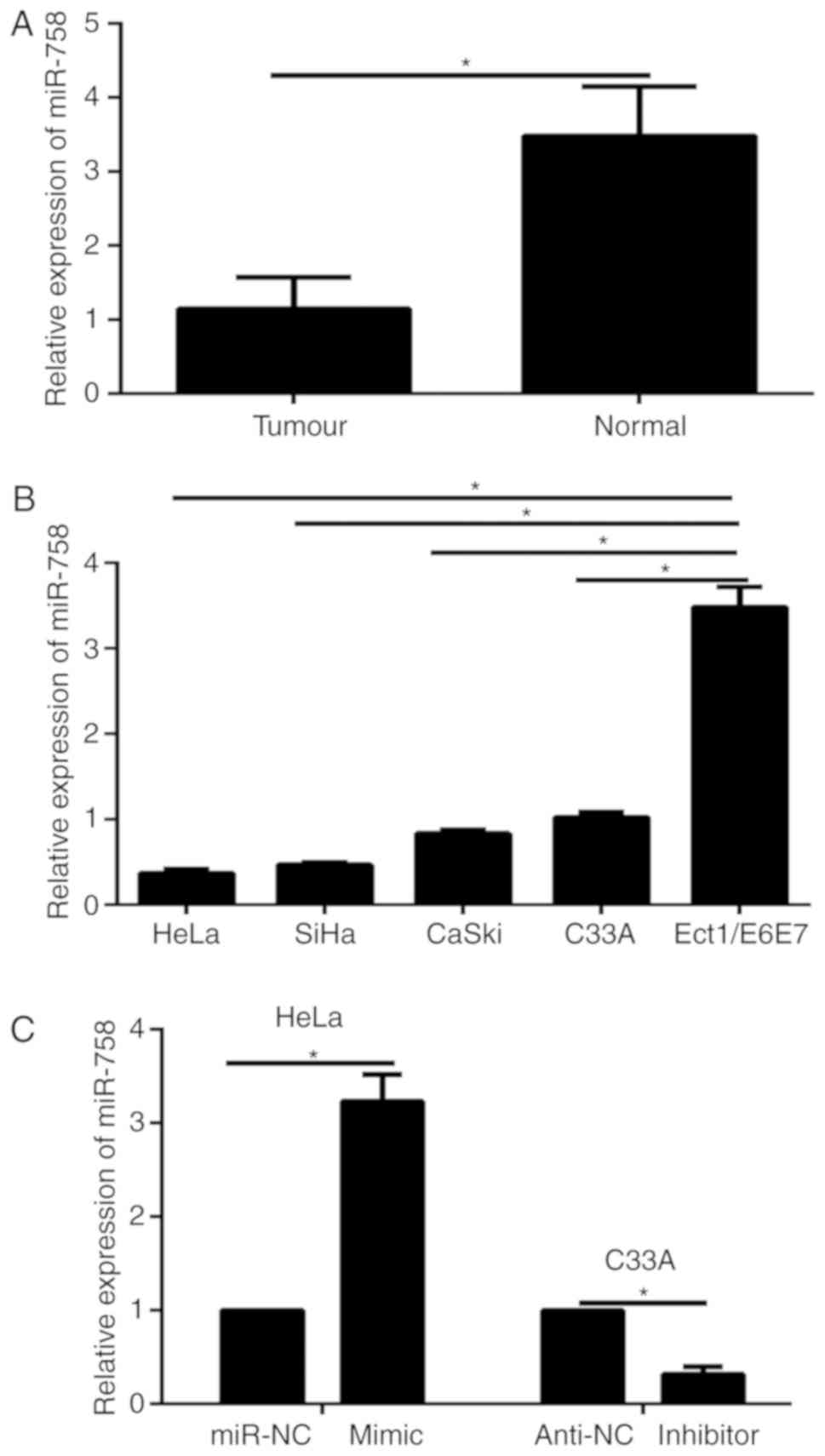
The expression of miR-758 is
downregulated in clinical samples and CC cells
The expression of miR-758 in tumor tissues and
paired adjacent normal tissues isolated from 20 CC patients was
detected via qPCR. The results indicated that the expression of
miR-758 was significantly reduced in CC tissues compared to
adjacent normal tissues (Fig. 1A).
Furthermore, we monitored miR-758 levels in CC cell lines (HeLa,
C33A, CaSki and SiHa) and a normal human cervical epithelial cell
line (Ect1/E6E7). Results of qPCR revealed that the expression
level of miR-758 was significantly decreased in CC cells compared
to that of Ect1/E6E7 cells (Fig.
1B). These data illustrated that miR-758 may serve as a tumor
suppressive gene in CC. The highest expression levels of miR-758
were detected in the C33A cells and the lowest expression was
detected in the HeLa cells. Then, these two cell lines were
selected for the further study. To discover the biological function
of miR-758 in the progression of CC cells, gain- or
loss-of-function assays were carried out by transfection of the
miR-758 mimic or inhibitor in HeLa or C33A cells, respectively.
Results of qPCR revealed that transfection of the miR-758 mimic
significantly increased miR-758 expression in HeLa cells, while the
miR-758 inhibitor significantly decreased the expression of miR-758
in C33A cells (Fig. 1C).
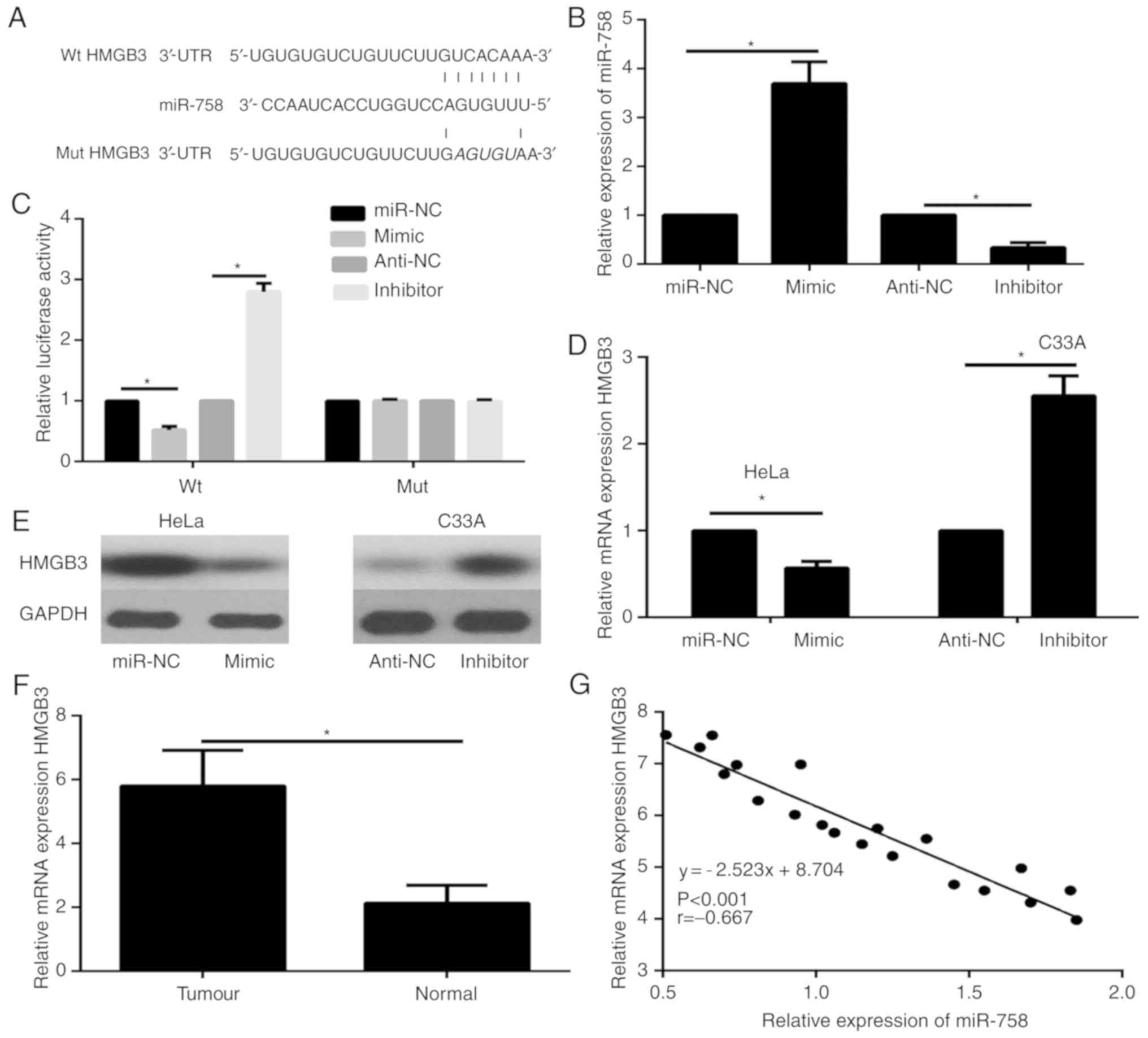
HMGB3 is a direct target gene of
miR-758
Analysis with the predictive database TargetScan
suggested that HMGB3 is a putative target of miR-758 (Fig. 2A). Previous research has revealed
that HMGB3 is a central player associated with cellular metastasis
in some types of cancer. To illustrate that HMGB3 was a direct
target gene of miR-758, 293T cells were co-transfected with Wt or
mut HMGB3-3′-UTR vectors and miR-758 mimics or inhibitor. The
expression of miR-758 in 293T cells was revealed using qPCR. The
results revealed that transfection of the miR-758 mimic
significantly increased miR-758 expression in 293T cells compared
to the miR-NC group, while the miR-758 inhibitor markedly decreased
the expression of miR-758 in 293T cells compared to the anti-NC
group (Fig. 2B). Luciferase reporter
assays were carried out to explore whether miR-758 targets HMGB3 by
binding to its 3′-UTR. Results of luciferase reporter assay
demonstrated that the miR-758 mimic significantly decreased the
luciferase activity of the wild-type 3′-UTR of HMGB3, while the
miR-758 inhibitor significantly increased the luciferase activity
of the wild-type HMGB3 3′-UTR (Fig.
2C). Furthermore, the results of qPCR and western blotting
revealed that both the mRNA (Fig.
2D) and protein (Fig. 2E) levels
of HMGB3 in the miR-758 mimic group were significantly decreased
compared with the negative control group. However, the mRNA
(Fig. 2D) and protein (Fig. 2E) expression level of HMGB3 were
significantly upregulated in the miR-758 inhibitor group.
Furthermore, the mRNA expression levels of HMGB3 in CC tumor
tissues and paired adjacent normal tissues was analyzed via qPCR.
The results revealed that the expression level of HMGB3 was
significantly enhanced in CC tissues compared with adjacent normal
tissues (Fig. 2F). In addition,
Spearman's correlation analysis revealed that the expression levels
of miR-758 were negatively correlated with HMGB3 mRNA in CC tissues
(Fig. 2G). In summary, these data
indicated that miR-758 directly targeted HMGB3 by binding to its
3′-UTR region in CC cells.
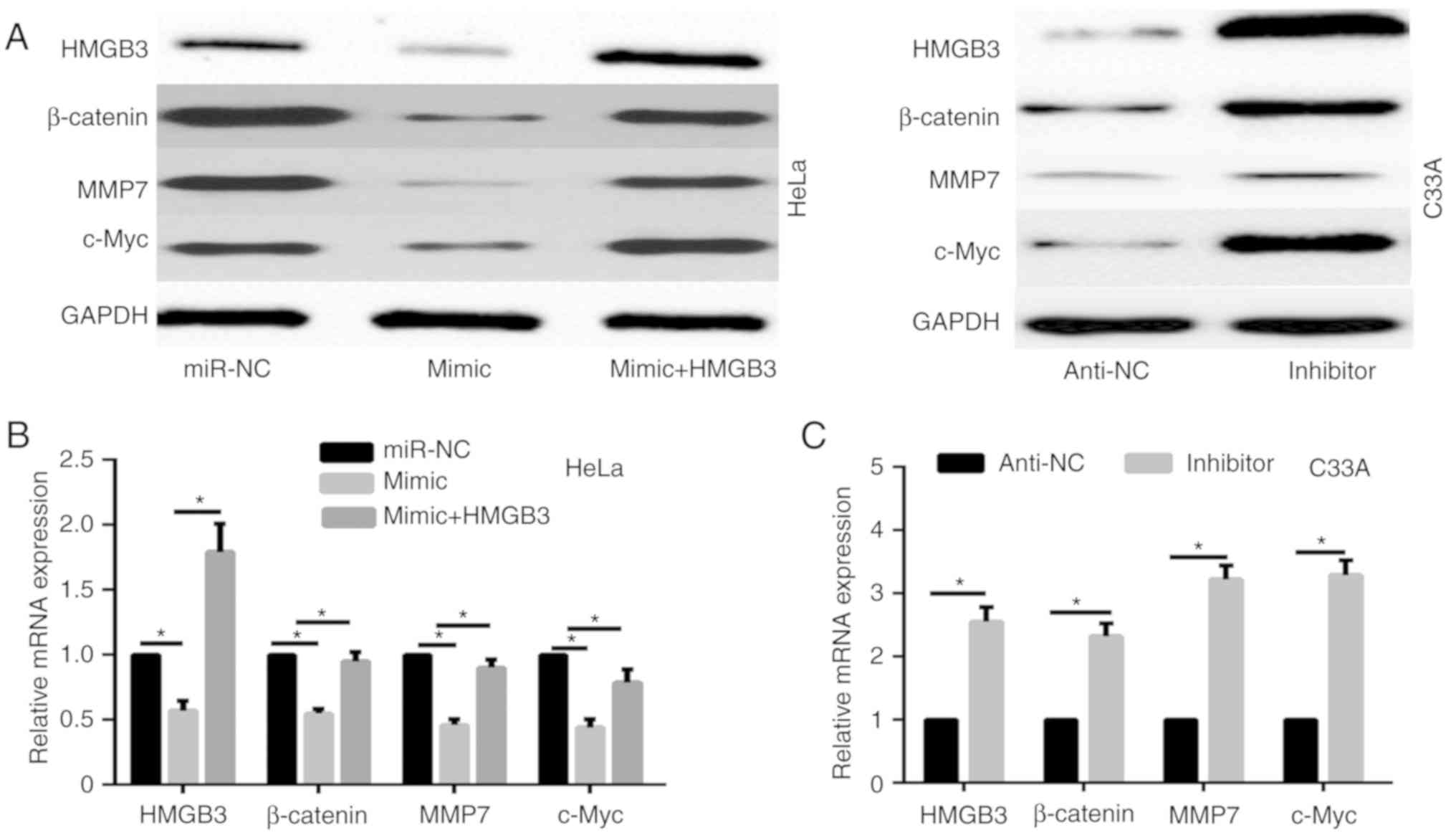
miR-758 regulates the WNT/β-catenin
signaling pathway via HMGB3
To illustrate the mechanism of miR-758 in CC
progression, we focused on the correlation of miR-758 and
WNT/β-catenin signaling pathway. Results of western blotting and
qPCR revealed that miR-758 mimic significantly decreased the
expression of β-catenin and its target gene c-Myc, and MMP7 in HeLa
cells (Fig. 3A and B), while miR-758
inhibitor markedly enhanced the expression of β-catenin, c-Myc, and
MMP7 in C33A cells (Fig. 3A and C).
A previous study revealed that HMGB3 negatively regulated the
WNT/β-catenin signaling pathway (11). Thus, whether miR-758 regulated the
WNT/β-catenin signaling pathway via HMGB3 was investigated. HeLa
cells were co-transfected with miR-758 mimic and HMGB3
overexpression plasmid. The protein expression level of HMGB3 was
detected by western blot assay (Fig.
3A). The results revealed that HMGB3 overexpression
significantly enhanced the expression of β-catenin, c-Myc, and MMP7
decreased by the miR-758 mimic both at the protein and mRNA level
(Fig. 3A and B). These data
demonstrated that miR-758 regulated the WNT/β-catenin signaling
pathway via HMGB3.
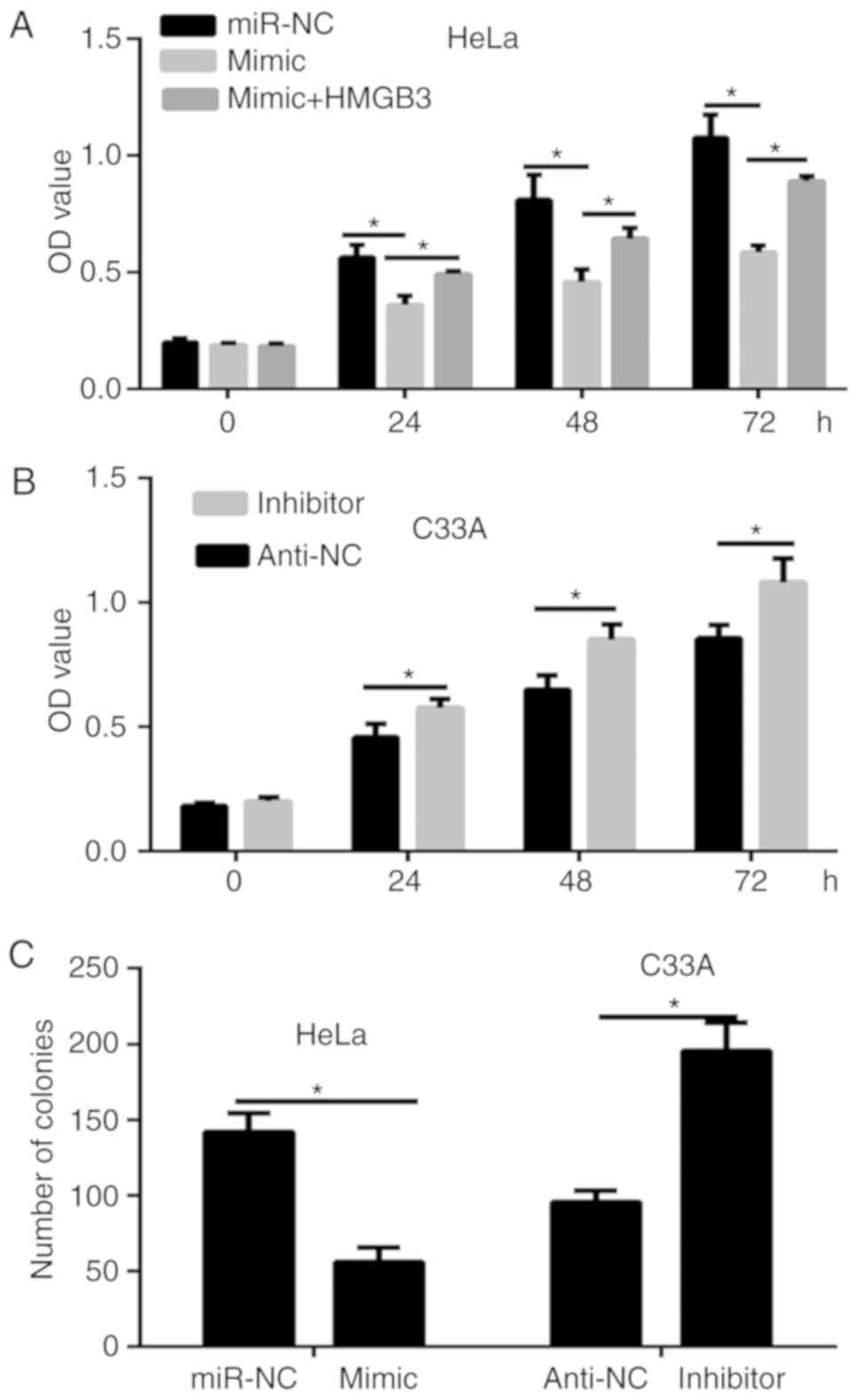
miR-758 inhibits the proliferation of
CC cells via HMGB3
To discover the biological function of miR-758 in
the progression of CC cells, CCK-8 and colony formation assays were
performed. Results of the CCK-8 assay revealed that the viability
of HeLa cells was significantly decreased by the miR-758 mimic
(Fig. 4A), while opposite results
were obtained in C33A cells transfected with the miR-758 inhibitor
(Fig. 4B). Furthermore, the colony
formation assay revealed that the transfection of the miR-758 mimic
significantly decreased the proliferation of HeLa cells compared
with the control group, while the miR-758 inhibitor significantly
enhanced the proliferation of C33A cells (Fig. 4C). To ascertain whether HMGB3 could
regulate the cell viability of miR-758 in CC cells, HeLa cells were
co-transfected with miR-758 mimic and HMGB3 overexpression plasmid.
The results of the CCK-8 assay (Fig.
4A) demonstrated that HMGB3 overexpression partially rescued
the inhibitory effects of the miR-758 mimic on the viability of
HeLa cells.
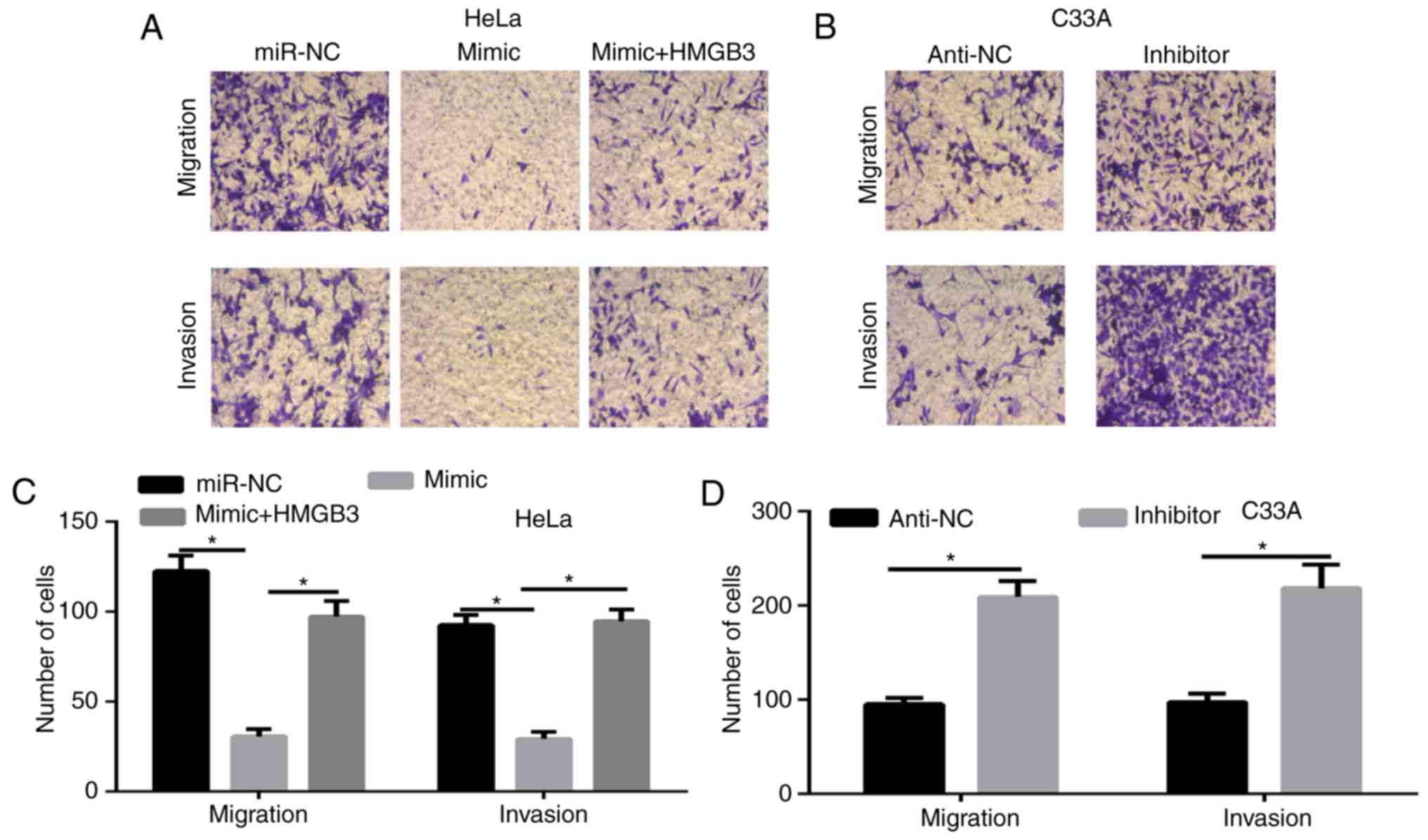
miR-758 inhibits the migration and
invasion of CC cells
Cell migration and invasion are critical events in
tumor progression (12). Thus,
Transwell assays were employed to assess the function of the
miR-758 on the migration and invasion of CC cells. The results
revealed that the overexpression of miR-758 significantly decreased
cell migration and invasion in HeLa cells, in contrast, the miR-758
inhibitor significantly enhanced cell migration and invasion in
C33A cells (Fig. 5A-D). To ascertain
whether HMGB3 regulated the cell migration and invasion of miR-758
in CC cells, HeLa cells were co-transfected with miR-758 mimic and
HMGB3 overexpression plasmid. The results of the Transwell assays
(Fig. 5A and C) demonstrated that
HMGB3 overexpression partially rescued the inhibitory effects of
miR-758 mimic on the migration and invasion of HeLa cells.
Discussion
Aberrant miR-758 expression has been discovered in
glioblastoma, hepatocellular carcinoma and non-small lung cancer;
furthermore, miR-758 has been revealed to inhibit the
proliferation, migration and invasion of these cancer cells
(6–8). miR-758 expression has been revealed to
be significantly decreased in CC (9). However, the function and molecular
mechanism of miR-758 in CC progression have not been well
elucidated. Therefore, the molecular mechanism of miR-758 in the
progression of CC must be further revealed. In the present study,
we demonstrated that miR-758 was downregulated in CC tissues and
cell lines, and it may serve as a novel tumor suppressor in CC.
miR-758 upregulation significantly suppressed tumor growth,
migration and invasion, whereas miR-758 silencing enhanced tumor
progression. In the mechanism study, we illustrated that miR-758
acted as a tumor suppressor by negatively regulating the
WNT/β-catenin signaling pathway by directly targeting HMGB3.
miR-758 serves as a tumor suppressor by regulating
different targets in various types of cancers. In the progression
of glioblastoma, miR-758 inhibited tumor progression by directly
targeting BTB domain-containing protein 20 (6). In hepatocellular carcinoma, miR-758
served as a tumor suppressor and played a crucial role in
inhibiting proliferation, migration and invasion by targeting MDM2
and mTOR (8). In the CC tissues,
miR-758 may regulate the infiltration and invasion of CC by
targeting matrix extracellular phosphoglycoprotein (9). However, miR-758 regulates the
expression of multiple genes. Therefore, the molecular mechanism of
miR-758 in the inhibitory function of CC progression was
investigated. In the present study, the correlation of miR-758 and
HMGB3 was demonstrated. HMGB3 belongs to the high-mobility group
box family, which includes HMGB1, HMGB2, HMGB3 and HMGB4 (13). The high-mobility group box family
plays an important role in the progression of several types of
cancer (11,14–17).
HMGB3 has been regulated by some different miRNAs in several types
of cancer. However, the correlation of miRNA and HMGB3 has not been
illustrated in CC. Therefore, this correlation was determined. In
the present study, it was revealed that HMGB3 expression was
significantly decreased after downregulation of miR-758 in CC
cells, but enhanced by the miR-785 inhibitor. Moreover, miR-758
expression was negatively correlated with HMGB3 mRNA expression in
CC cell tissues. Finally, it was demonstrated that HMGB3
overexpression rescued the inhibitory function role of the miR-758
mimic. In summary, our results demonstrated that HMGB3 is a
molecular and functional target of miR-758. The WNT/β-catenin
signaling pathway has been revealed to promote cancer progression
in some types of cancer, including CC (18). HMGB3 can regulate the WNT/β-catenin
signaling pathway in colorectal cancer (11). In the present study, it was
demonstrated that miR-758 negatively regulated the WNT/β-catenin
signaling pathway. In the rescue experiments, it was demonstrated
that HMGB3 overexpression enhanced the expression of β-catenin and
its target genes MMP7 and c-Myc. These data demonstrated that
miR-758 promoted CC growth, migration and invasion by negatively
regulating the WNT/β-catenin signaling pathway by directly
targeting HMGB3.
In summary, the present study demonstrated that
miR-758 was downregulated in CC tissues and cell lines. miR-758
also functioned as a tumor suppressor of CC growth by targeting
HMGB3. This newly identified miR-758/HMGB3 link provides new
insight into the mechanisms underlying CC development and suggested
that targeting the miR-758/HMGB3 axis may represent a promising
therapeutic strategy for CC treatment. However, further studies are
required to determine the exact mechanism of decreased miR-758
expression during the progression of CC and to further explore
other possible molecular mechanisms of miR-758 in CC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
TS and XHH conceived and designed the experiments.
TS, XHH and BL conducted all of the experiments. XHH and BL wrote
and revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study involving human samples was
approved by the Medical Ethics Committee of Weifang Maternity and
Child Care Hospital (Weifang, China) and written informed consent
was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith RA, Brooks D, Cokkinides V, Saslow D
and Brawley OW: Cancer screening in the United States, 2013: A
review of current American cancer society guidelines, current
issues in cancer screening, and new guidance on cervical cancer
screening and lung cancer screening. CA Cancer J Clin. 63:88–105.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dai S, Lu Y, Long Y, Lai Y, Du P, Ding N
and Yao D: Prognostic value of microRNAs in cervical carcinoma: A
systematic review and meta-analysis. Oncotarget. 7:35369–35378.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kanekura K, Nishi H, Isaka K and Kuroda M:
MicroRNA and gynecologic cancers. J Obstet Gynaecol Res.
42:612–617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kwan JY, Psarianos P, Bruce JP, Yip KW and
Liu FF: The complexity of microRNAs in human cancer. J Radiat Res.
57 (Suppl 1):i106–i111. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J, Jiang J, Hui X, Wang W, Fang D and
Ding L: Mir-758-5p suppresses glioblastoma proliferation, migration
and invasion by targeting ZBTB20. Cell Physiol Biochem.
48:2074–2083. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang S and Jiang M: The long non-coding
RNA-DANCR exerts oncogenic functions in non-small cell lung cancer
via miR-758-3p. Biomed Pharmacother. 103:94–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang D, Cho W, Li Z, Xu X, Qu Y, Jiang Z,
Guo L and Xu G: MiR-758-3p suppresses proliferation, migration and
invasion of hepatocellular carcinoma cells via targeting MDM2 and
mTOR. Biomed Pharmacother. 96:535–544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meng X, Zhao Y, Wang J, Gao Z, Geng Q and
Liu X: Regulatory roles of miRNA-758 and matrix extracellular
phosphoglycoprotein in cervical cancer. Exp Ther Med. 14:2789–2794.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Z, Chang Y, Zhang J, Lu Y, Zheng L,
Hu Y, Zhang F and Li X, Zhang W and Li X: HMGB3 promotes growth and
migration in colorectal cancer by regulating WNT/β-catenin pathway.
PLoS One. 12:e01797412017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vu M, Yu J, Awolude OA and Chuang L:
Cervical cancer worldwide. Curr Probl Cancer. 42:457–465. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nemeth MJ, Curtis DJ, Kirby MR,
Garrett-Beal LJ, Seidel NE, Cline AP and Bodine DM: Hmgb3: An
HMG-box family member expressed in primitive hematopoietic cells
that inhibits myeloid and B-cell differentiation. Blood.
102:1298–1306. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X and Zeng L: Ginkgo biloba extract
761 enhances 5-fluorouracil chemosensitivity in colorectal cancer
cells through regulation of high mobility group-box 3 expression.
Am J Transl Res. 10:1773–1783. 2018.PubMed/NCBI
|
|
15
|
Yamada Y, Nishikawa R, Kato M, Okato A,
Arai T, Kojima S, Yamazaki K, Naya Y, Ichikawa T and Seki N:
Regulation of HMGB3 by antitumor miR-205-5p inhibits cancer cell
aggressiveness and is involved in prostate cancer pathogenesis. J
Hum Genet. 63:195–205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao J, Zou Z, Gao J, Zhang H, Lin Z, Zhang
Y, Luo X, Liu C, Xie J and Cai C: Increased expression of HMGB3: A
novel independent prognostic marker of worse outcome in patients
with esophageal squamous cell carcinoma. Int J Clin Exp Pathol.
8:345–352. 2015.PubMed/NCBI
|
|
17
|
Song N, Liu B, Wu JL, Zhang RF, Duan L, He
WS and Zhang CM: Prognostic value of HMGB3 expression in patients
with non-small cell lung cancer. Tumour Biol. 34:2599–2603. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bahrami A, Hasanzadeh M, ShahidSales S,
Yousefi Z, Kadkhodayan S, Farazestanian M, Joudi Mashhad M, Gharib
M, Mahdi Hassanian S and Avan A: Clinical significance and
prognosis value of Wnt signaling pathway in cervical cancer. J Cell
Biochem. 118:3028–3033. 2017. View Article : Google Scholar : PubMed/NCBI
|