Introduction
Glioblastoma is the most common primary brain
malignancy in adults, with an average annual incidence of ~3-10 in
every 100,000 adults in the United States (1,2).
Glioblastoma is defined as any tumor that is produced by glial
cells or precursor cells, including astrocytoma, glioblastoma,
ependymoma, mixed glioblastoma, and other rare histologies
(3). Grade IV glioblastomas have a
poor prognosis, with a median survival time of 14.6–16 months and a
5-year survival rate of 3.4% (4,5). The
standard first-line treatment for glioblastoma involves a variety
of modalities, including surgical resection, radiotherapy and
temozolomide-adjuvant chemotherapy (6). Despite surgical resection and
chemoradiation, tumors in almost all patients are likely to
relapse, with a 2-year survival rate of only 26% following relapse
(7). There is currently no standard
and uniform management method for recurrent glioblastoma.
Therefore, the search for novel glioblastoma treatment pathways is
particularly important. In recent years, with the development of
molecular biology techniques, tumor-related genetic research has
increased (8). At present, gene
therapies and molecular-targeted drugs are a novel area of study
(9,10). The most important aspect of these
treatments is identifying a specific molecular therapy target for
different tumors.
microRNAs (miRNAs/miRs) have been identified as
promising therapeutic targets in a number of types of cancer
(11,12). miRNAs are short endogenous
single-stranded RNA molecules that transcriptionally inhibit gene
expression by targeting 3′-untranslated regions (3′-UTRs) of
messenger RNAs (mRNAs) by binding to complementary sequences
(13,14). miRNAs regulate a number of important
cellular functions, including angiogenesis, proliferation,
apoptosis and invasion, and are deregulated in numerous types of
cancer (15). A growing body of
research indicates that miRNAs regulate the expression of
chemokines, further effecting disease progression and prognosis
(16). miRNA-181 has been found to
inhibit the progression of cervical cancer by targeting a
transcription factor, yin-yang-1 (17,18). The
expression of miR-181 and its impact on CCL8 in the progression of
glioblastoma has not been studied.
The present study aimed to evaluate the potential of
miRNA-181 and CLL8 as biomarkers or treatment targets in
glioblastoma by investigating their expression in patients, and to
determine the effects of altered miRNA-181 and CCL8 expression in
glioblastoma cell lines.
Materials and methods
Patient tissues
The present study was approved by the Institutional
Review Board and Ethics Committee of The First Affiliated Hospital
of Zhengzhou University (Zhengzhou, Henan, China). Tissue samples
of glioblastoma, including 23 grade II tumors, 26 grade III tumors
and 31 grade IV tumors, and adjacent non-cancerous tissues samples
were obtained from 80 patients with glioblastoma undergoing surgery
at The First Affiliated Hospital of Zhengzhou University between
February and August 2013 (46 men and 34 women; age range, 28–73
years). None of the patients had received radiotherapy,
chemotherapy or other anticancer treatment prior to surgery. All
participants had provided written informed consent. The patients
were followed up for 3 years. The tumor tissues, including 42
tumors with diameter <5 cm and 38 tumors with diameter ≥5 cm,
and adjacent non-tumor tissues were collected at surgery,
immediately frozen in liquid nitrogen and stored at −80°C until RNA
or protein extraction.
Cell culture
Human glioblastoma cell lines, U251, A172 and U87
cells (cat. no. HTB-14, glioblastoma of unknown origin) and normal
human astrocytes (NHA) were obtained from the American Type Culture
Collection (ATCC; Manassas, VA, USA) and cultured in Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.). All cell lines were
routinely passaged as monolayer cultures at 37°C in a humidified
atmosphere of 5% CO2.
Vector construction and
transfection
The wild-type 3′UTR of CCL8 mRNA and a mutated
version flanked by KpnI and HindIII restriction sites
were synthesized and ligated into the pGL3 luciferase reporter
vector (Sangon Biotech Co., Ltd., Shanghai, China). The cDNA
sequence of CCL-8 was ligated into a pcDNA3.1 (+) vector (Thermo
Fisher Scientific, Inc.). The CCL-8-containing vector (10 µM) and
an miR-181 mimic (100 nM) (Shanghai GenePharma Co., Ltd., Shanghai,
China) were transfected into U87 and A172 cells using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The sequences were as
follows: miR-181 mimic, 5′-AACAUUCAACGCUGUCGGUGAGUUCACCGACAGCG-3′
and miR-negative control (NC), 5′-UUCUCCGAACGUGUCACGUTT-3′.
Subsequent experiments were performed 48 h following
transfection.
Dual-luciferase reporter assay
Bioinformatics analysis was conducted to predict the
target gene using TargetScan (version 6.2; http://genes.mit.edu/targetscan). A luciferase
reporter gene assay was performed in cells 48 h following
transfection with the pGL3 vector containing the wild-type or
mutated 3′-UTR of CCL8. The Dual-Luciferase® Reporter
Assay System (Promega Corporation, Madison, WI, USA) was used
according to the manufacturer's instructions. Renilla
luciferase was co-transfected as a control for normalization.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from all samples and cell lines was
extracted using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.). Isolated RNA was reverse-transcribed using
MultiScribe reverse transcriptase (Applied Biosystems; Thermo
Fisher Scientific, Inc). Briefly, samples containing l µg total RNA
were incubated with 1 µl gDNA Eraser, 2 µl 5X gDNA eraser buffer
and RNase free dH2O at 42°C for 2 min. After adding the
enzyme mix, the reaction was incubated at 37°C for 15 min. cDNA was
diluted and stored at −20°C prior to use. mRNA expression was
detected with SYBR® Premix Ex Taq II (Takara
Biotechnology Co., Ltd., Dalian, China). The PCR primers were
designed as follows: miR-181, forward
5′-AACATTCAACGCTGTCGGTGAAGT-3′, reverse
5′-ACTTCACCGACAGCGTTGAATGTT-3′; U6, forward
5′-CTCGCTTCGGCAGCACA-3′, reverse 5′-AACGCTTCACGAATTTGCGT-3′; CCL-8
forward 5′-AGATGAAGGTTTCTGCAGCGC-3′, reverse
5′-TGGAAACTGAATCTGGCTGAG-3′; and GAPDH, forward
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse 5′-TGGTGAAGACGCCAGTGGA-3′.
RT-qPCR reactions were performed as follows: 95°C for 10 min
followed by 40 cycles of 95°C for 25 sec and 60°C for 30 sec. The
relative expression levels of miR-181 and CCL-8 relative to GAPDH
and U6 were calculated using the relative quantification
2−ΔΔCq method (19). Each
sample was assayed in triplicates. The high and low expression of
miR-181 was defined according to the average expression levels of
miR-181 in patients with glioblastoma.
Western blot analysis
Total protein was extracted from the tissues and
cells using radioimmunoprecipitation lysis buffer (Beyotime
Institute of Biotechnology, Jiangsu, China) and separated using
SDS-PAGE (10% gel), and then transferred to polyvinylidene
difluoride membranes. The membranes were then incubated with 5%
non-fat milk in Tris-buffered saline (TBS; 20 mM Tris-HCl, pH 7.5,
150 mM NaCl) containing 0.1% Tween 20 at room temperature.
Subsequently, the membranes were incubated at room temperature with
rabbit antibodies against CCL-8 (cat. no. ab39625; 1:500; Abcam,
Cambridge, UK;) and GAPDH (cat. no. sc-47724; 1:500; Santa Cruz,
Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Membranes
were washed with TBST and incubated at room temperature with
horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (cat. no. 7074; 1:5,000; Cell Signaling Technology, Inc.,
Danvers, MA, USA) for 40 min. Protein bands were detected by
enhanced chemiluminescence (ECL) using Pierce ECL Western Blot
Substrate (Thermo Fisher Scientific, Inc.) and exposed to an X-ray
film using an ECL detection system (Thermo Fisher Scientific,
Inc.). GAPDH served as a loading control. All assays were performed
in triplicate.
Cell proliferation assay
A total of ~1×103 cells/well were
cultured in 96-well plates and a Cell Counting Kit-8 (CCK-8;
Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to
determine proliferation in each treatment group (6 wells per group)
and in the blank controls. Cells were incubated with 10 µl CCK-8 at
different time points (0, 24, 48 and 72 h) for 3 h at 37°C and 5%
CO2. The absorbance value at 450 nm was measured. The
cell growth curves were plotted using the absorbance value at each
time point. All assays were performed in triplicate.
Migration and invasion assay
Transwell chambers (24-wells; Corning Inc., Corning,
NY, USA) were used to evaluate the migratory and invasive ability
of glioblastoma cells. Transfected U87 and A172 cells in DMEM were
added to the top chambers of the Transwell plate that were
pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA) for
the invasion assay, and all other steps were identical. A total of
4×104 transfected cells without FBS were placed in the
top chamber, and the bottom chamber was filled with 20% FBS. The
cells were incubated at 37°C for 24 h to allow migration or
invasion through the membrane. The cells were then stained at room
temperature with crystal violet (Beyotime Institute of
Biotechnology, Shanghai, China) for 10 min. All assays were
performed in triplicates.
Flow cytometry
Cells were cultured in 6-well plates at a
concentration of 3×105 cells/well. Cells were washed
with PBS and harvested with 0.25% trypsin without EDTA. Cells were
then washed with PBS and stained with 5 µl Annexin V-fluorescein
isothiocyanate (BD Biosciences) and 5 µl propidium iodide (BD
Biosciences) for 15 min at room temperature in the dark according
to the manufacturer's instructions. Data acquisition and analyses
were performed on a BD FACSCalibur flow cytometer (Becton,
Dickinson and Company, Franklin Lakes, NJ, USA) using CellQuest
software (version 5.2.1; BD Biosciences). All assays were performed
in triplicate.
Statistical analysis
Data are shown as the mean ± standard deviation
where applicable. Graphpad Prism (version 5.0; Graphpad Software
Inc., La Jolla, CA, USA) was used for all statistical analysis. The
differences between groups were evaluated by one way analysis of
variance followed by Dunnett's test. Kaplan-Meier analysis and a
log-rank test were used to compare patient survival. The
correlation between the expression levels of miR-181 and CCL8 was
assessed using Spearman's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
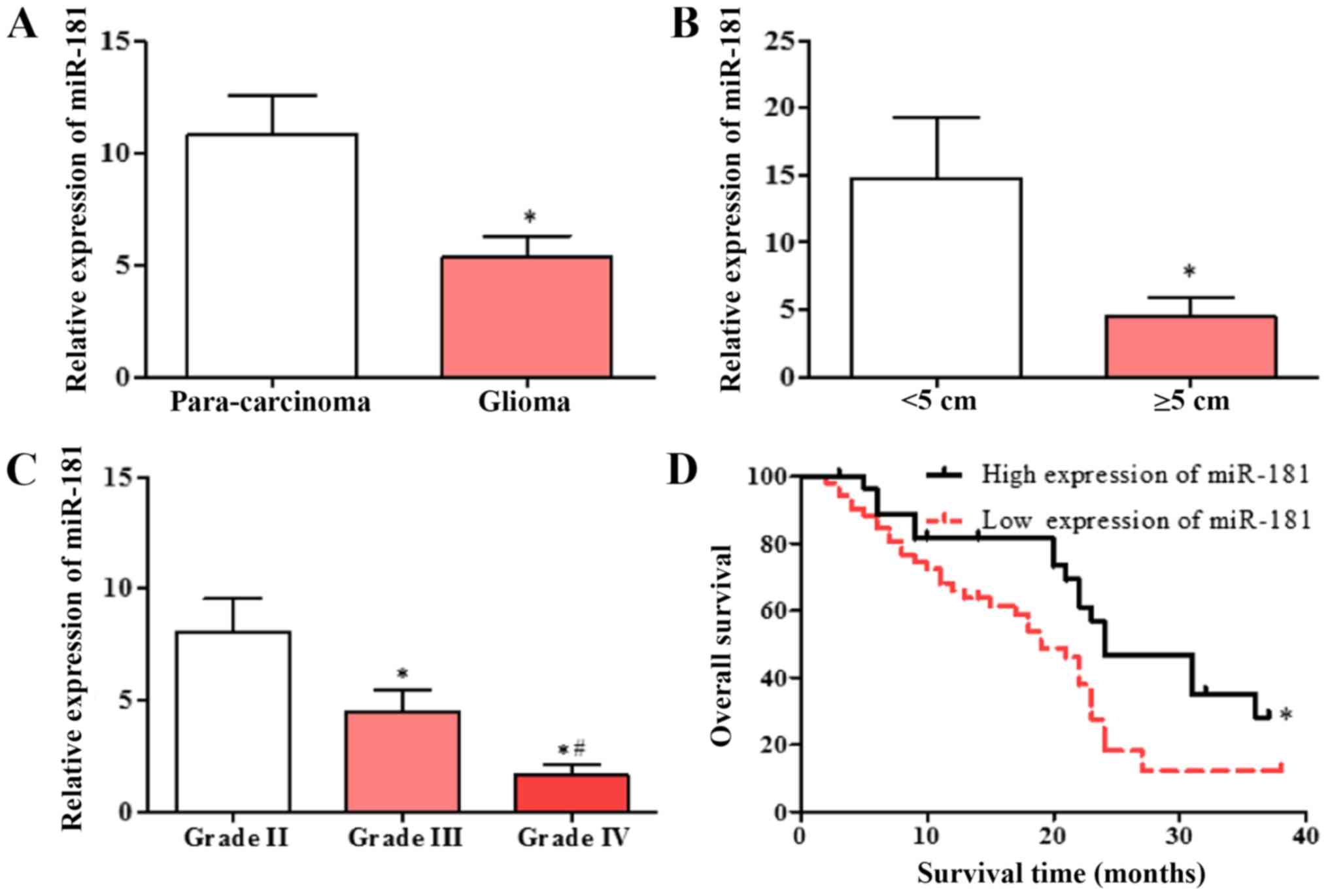
miR-181 is downregulated in patients
with glioblastoma and is associated with poor prognosis
Expression of miR-181 in glioblastoma and adjacent
tissues was detected using RT-qPCR. miR-181 expression in
glioblastoma tissues was significantly decreased compared with that
in adjacent tissues (Fig. 1A), and
the expression of miR-181 in glioblastoma with tumor diameter ≥5 cm
was decreased compared with that in glioblastoma with tumor
diameter <5 cm (Fig. 1B). The
expression of miR-181 in patients with different diagnostic grades
was also evaluated. The expression of miR-181 in patients with
grade III and IV glioblastoma was significantly lower compared with
that in patients with grade II, and the expression of miR-181 in
patients with grade IV glioblastoma was significantly lower
compared with that in patients with grade III (Fig. 1C). The patient group with low
expression of miR-181 had a significantly lower survival rate
compared with the group with high expression of miR-181 (Fig. 1D). The results indicate that
decreased expression of miR-181 is associated with poor
prognosis.
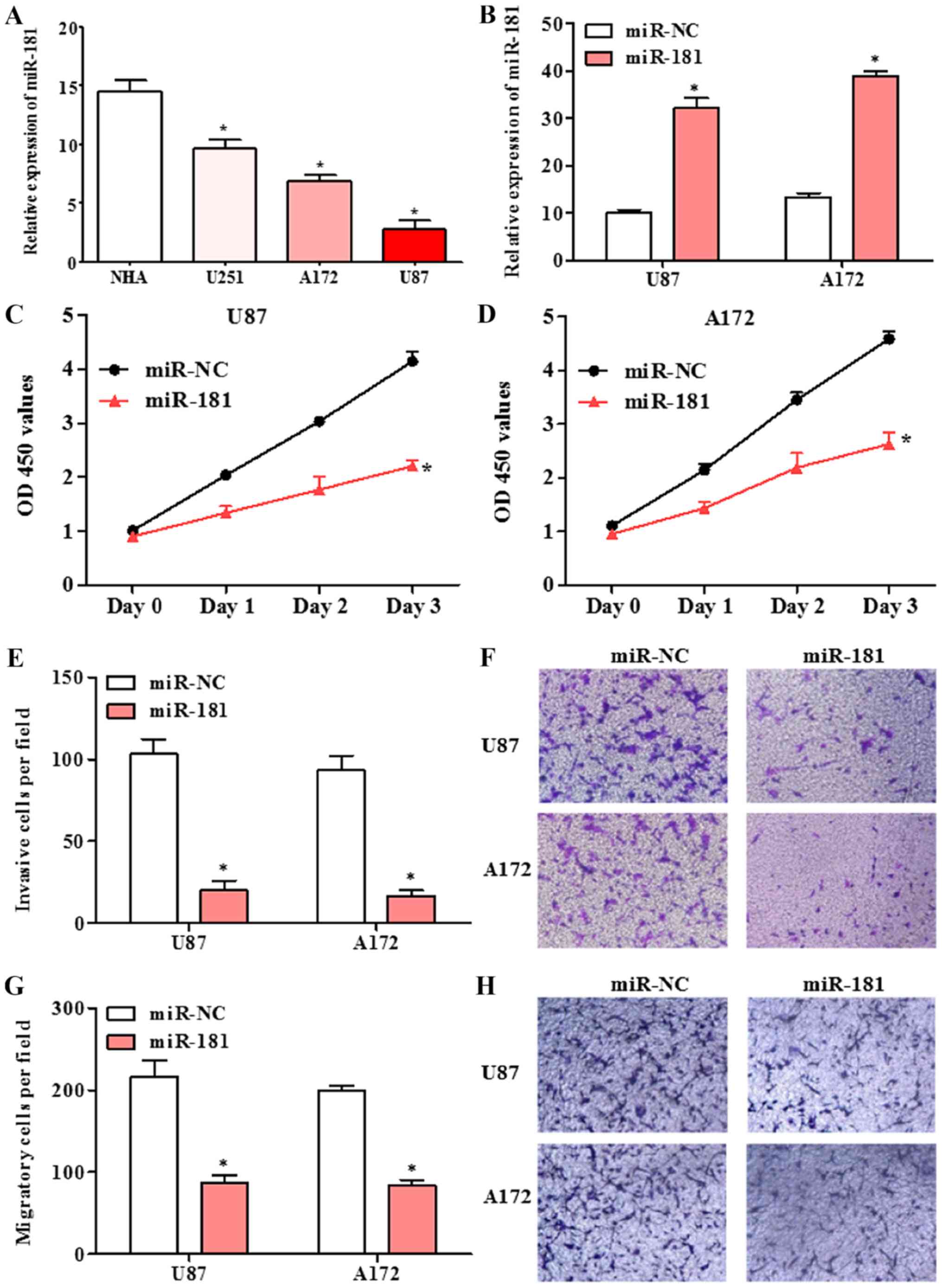
Overexpression of miR-181 inhibited
cell proliferation, invasion and migration in glioblastoma cell
lines
The expression of miR-181 in human glioblastoma cell
lines U251, A172 and U87, and normal human astrocytes NHA was
detected by RT-qPCR. The expression of miR-181 in glioblastoma cell
lines was significantly lower compared with that in normal human
astrocytes NHA cells (Fig. 2A).
Following transfection of A172 and U87 cells with miR-181 mimic,
miR-181 expression was significantly increased compared with that
in cells transfected with miR-NC (Fig.
2B). The effect of miR-181 on proliferation in glioblastoma
cells, was investigated using the CCK-8 assay. The proliferation of
A172 and U87 cells transfected with miR-181 was decreased compared
with that of cells transfected with miR-NC (Fig. 2C and D). A Transwell assay revealed
that the ability of A172 and U87 cells transfected with miR-181 to
invade or migrate across a membrane was decreased compared with
that of cells transfected with miR-NC (Fig. 2E-H). The results indicate that
upregulation of miR-181 inhibits glioblastoma cell growth, invasion
and migration.
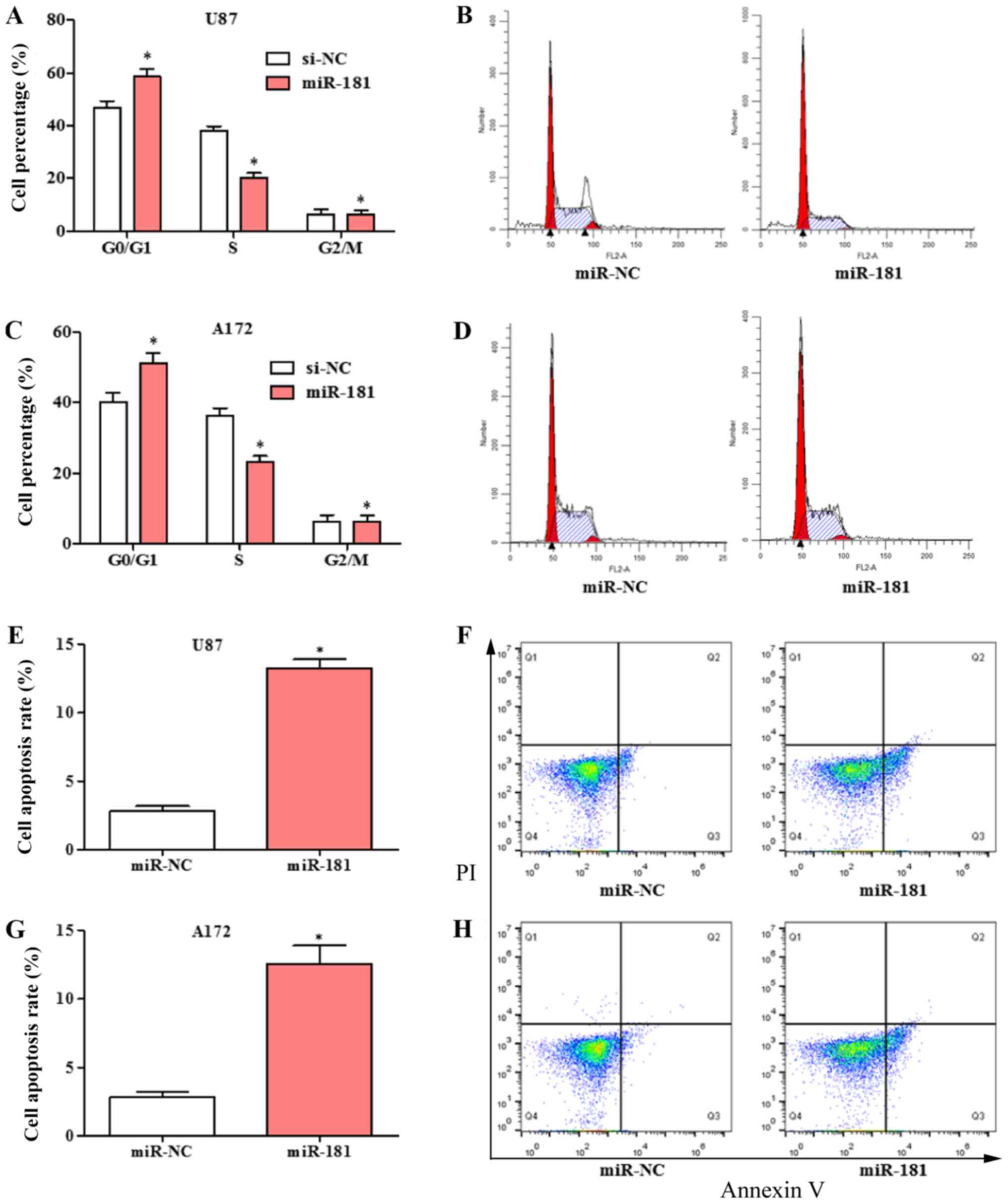
Overexpression of miR-181 arrested
cell cycle in the G1 phase and induced cell apoptosis in
glioblastoma cell lines
The effects of overexpression of miR-181 on cell
cycle and apoptosis in glioblastoma cell was evaluated. The cell
cycle was significantly shifted from S phase to G1 phase in U87 and
A172 cells transfected with miR-181 compared with those transfected
with miR-NC (Fig. 3A-D). The
percentage of cells in the G1 phase was increased and the
percentage in the S phase was decreased in the
miR-181-overexpressing groups. Overexpression of miR-181 promoted
apoptosis in U87 (Fig. 3E and F) and
A172 cells (Fig. 3G and H).
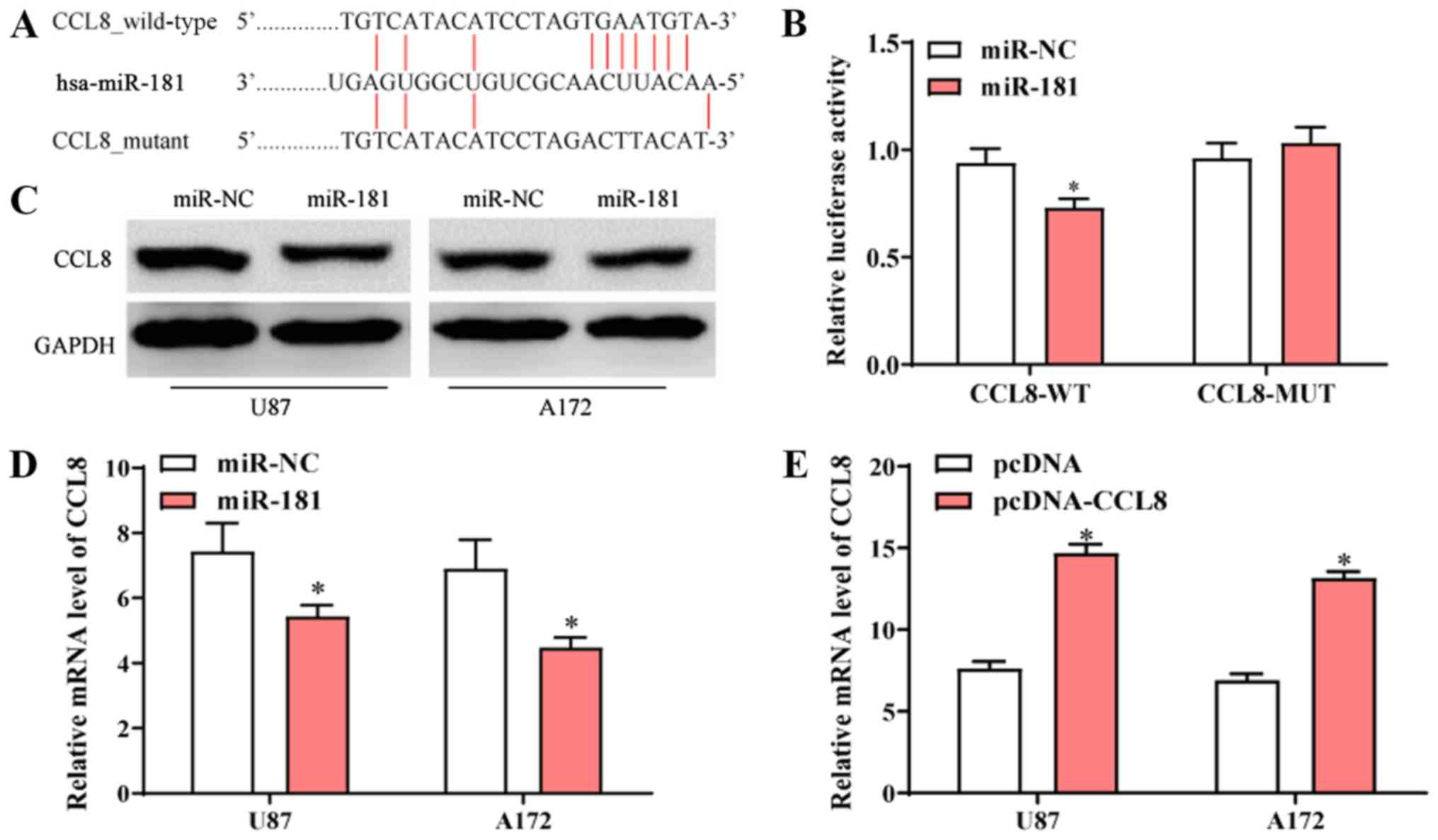
CCL8 is a target of miR-181
The sequence alignment of miR-181 and the 3′-UTR of
CCL-8 mRNA is presented in Fig. 4A.
To further determine the interaction between miR-181 and CCL-8,
luciferase vectors containing wide-type or mutated CCL8 3′-UTR were
co-transfected into 293 cells (ATCC) with miR-NC or miR-181. The
dual-luciferase reporter assay revealed that transfection with
miR-181 reduced the activity of the luciferase reporter fused to
the wild-type 3′-UTR of CCL-8, but did not suppress that of the
reporter fused to the mutated version (Fig. 4B). Following transfection with
miR-181, protein and mRNA expression of CCL8 in U87 and A172 cells
was decreased compared to those of cells transfected with miR-NC
(Fig. 4C and D). Following
transfection of A172 and U87 cells with pcDNA-CCL8, CCL8 mRNA
expression was significantly higher compared with cells that were
transfected with pcDNA (Fig. 4E).
The results indicated that CCL8 was a direct target of miR-181 and
overexpression of miR-181 reduced the expression of CCL8.
Overexpression of CCL8 partially
reversed the effects of miR-181 overexpression in glioblastoma cell
lines
To confirm that the effects of miR-181 on cell
proliferation, invasion and migration are mediated by CCL8
downregulation, U87 and A172 cells were transfected with miR-NC,
miR-181 and co-transfected with CCL8. Overexpression of CCL8
partially reversed the inhibition of proliferation by miR-181 in
U87 and A172 cells (Fig. 5A and B).
The inhibition of invasion and migration by miR-181 in the two cell
lines was also partially rescued by overexpression of CCL8
(Fig. 5C and D). Flow cytometric
analysis revealed that overexpression of CCL8 partially reversed
the apoptotic effect of miR-181 in U87 and A172 cells (Fig. 5E and F). These results suggested that
miR-181 promoted glioblastoma cell proliferation, invasion and
migration and inhibited apoptosis partially through downregulation
of CCL8.
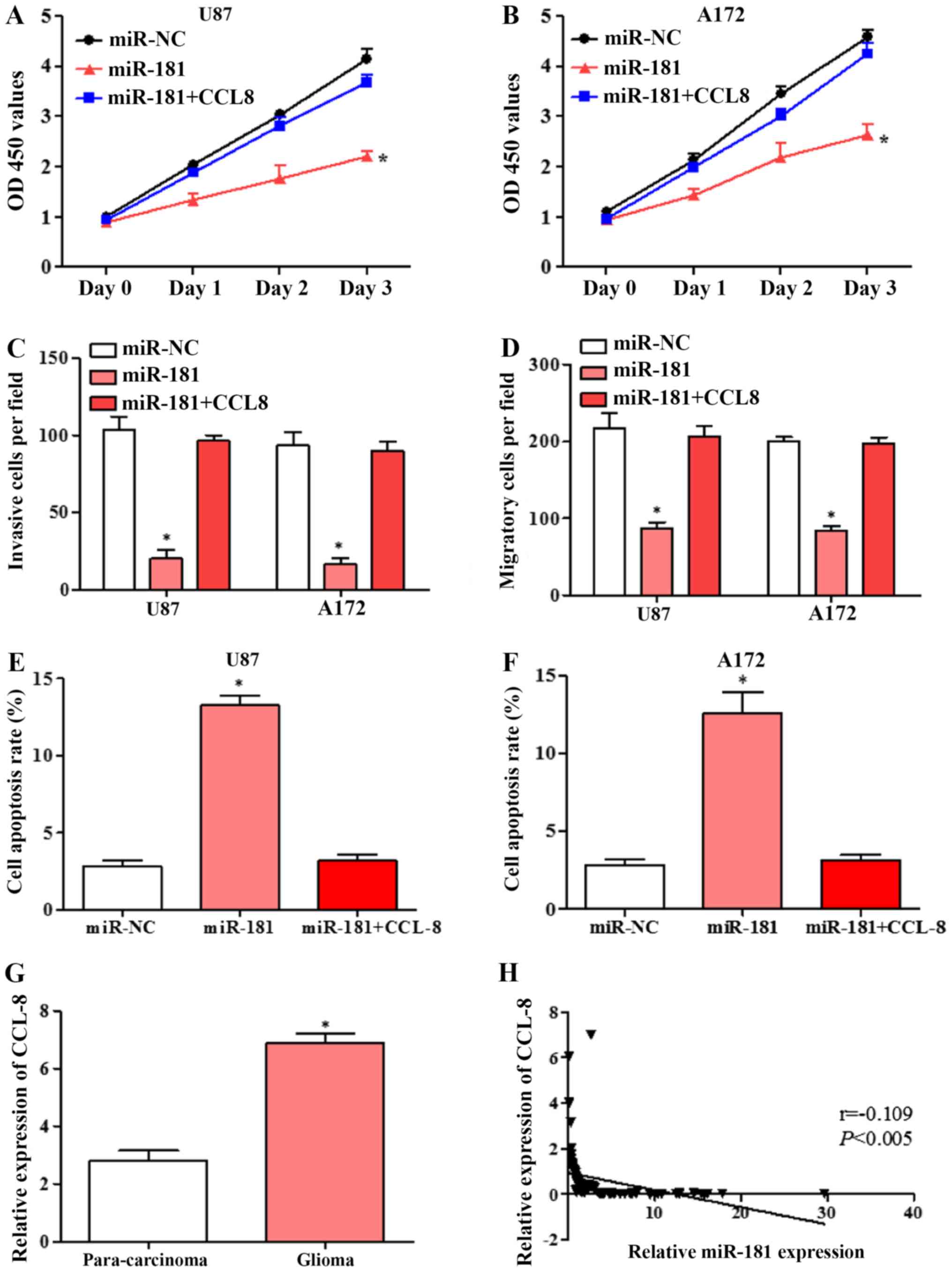 | Figure 5.Overexpression of CCL8 reverses the
miR-181-mediated inhibition of proliferation, invasion and
migration and induction of apoptosis in glioblastoma cell lines.
(A) Proliferation of U87 cells transfected with miR-NC, miR-181 and
co-transfected with miR-181 and CCL8. (B) Proliferation of A172
cells transfected with miR-NC, miR-181 and co-transfected with
miR-181 and CCL8. (C) Invasion of U87 and A172 cells transfected
with miR-NC, miR-181 and co-transfected with miR-181 and CCL8. (D)
Migration of U87 and A172 cells transfected with miR-NC, miR-181
and co-transfected with miR-181 and CCL8. (E) Apoptosis rate of U87
cells transfected with miR-NC, miR-181 and co-transfected with
miR-181 and CCL8; (F) apoptosis rate of A172 cells transfected with
miR-NC, miR-181 and co-transfected with miR-181 and CCL8. (G)
Expression of CCL8 in glioblastoma and para-carcinoma tissue
detected by RT-qPCR (n=80). (H) Expression of CCL8 in glioblastoma
negatively correlated with miR-181 expression, analyzed by
Spearman's correlation analysis (n=80). All data are shown as the
mean ± standard deviation and based on at least three independent
experiments. *P<0.05. miR-181, microRNA-181; CCL8, C-C motif
chemokine ligand 8; OD, optical density; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; miR-NC,
miR-negative control. |
Expression of CCL8 in glioblastoma
tissues was negatively correlated with miR-181 expression
The expression of CCL8 in glioblastoma and adjacent
tissues were detected by RT-qPCR. CCL8 expression in glioblastoma
tissues was significantly higher compared with that in adjacent
tissues (Fig. 5G). Spearman's
correlation analysis showed that CCL8 was negatively correlated
with miR-181 expression (Fig. 5H).
The results further suggested that miR-181 may inhibit glioblastoma
cell growth and induces apoptosis by directly targeting CCL8.
Discussion
In the present study, miR-181 expression was
revealed to be low in glioblastoma tissue and associated with poor
prognosis in patients with glioblastoma. Overexpression of miR-181
inhibited glioblastoma cell proliferation, invasion and migration
and arrested glioblastoma cell cycle in the G1 phase and induced
glioblastoma cell apoptosis. CCL8 was upregulated in glioblastoma
tissues and was negatively correlated with miR-181 expression. CCL8
was confirmed as a direct target of miR-181 using a dual luciferase
assay. In addition, overexpression of CCL8 reversed the inhibition
of proliferation, invasion and migration and induction of apoptosis
by miR-181 in glioblastoma cells. The results suggested that
downregulated miR-181 may promote glioblastoma cell proliferation,
invasion and migration and inhibit apoptosis partially through
binding and inhibiting CCL8.
miRNAs can regulate cell proliferation, invasion,
migration, the cell cycle and apoptosis (20), as well as regulating transcription
through RNA polymerase, to impact tumor development (21,22).
Previous studies indicated that aberrant expression of miRNAs has
been identified in a number of types of cancer, including stomach,
liver, colorectal, breast and cervical cancer, as well as glioma
and others (23,24). miR-181 is an important miRNA that is
involved in the development of multiple tumor types (25,26).
miR-181 was found to regulate cisplatin-resistant non-small cell
lung cancer via the PTEN/PI3K/AKT pathway (27). In the present study, downregulation
of miR-181 was identified in patients with glioblastoma, and was
associated with poor prognosis. The detailed mechanism remains
unknown and further research is required.
To explore the function of miR-181 in glioblastoma,
the proliferation, invasion and migratory abilities of A172 and U87
cells transfected with miR-181 were investigated and found to be
lower compared with cells transfected with miR-NC. In addition, the
cell cycle was significantly shifted from S phase to G1 phase, and
apoptosis was promoted in U87 and A172 cells transfected with
miR-181, compared with cells transfected with miR-NC. Su et
al (28) reported that miR-181
is aberrantly overexpressed in patients with acute myeloid leukemia
and can inhibit granulocytic and macrophage-like differentiation by
directly targeting and downregulating the expression of PRKCD,
CTDSPL and CAMKK1.
CCL8 has been reported to be a CCR5 agonist with a
similar affinity to the receptor as CCL3 (MIP-1a), which in
combination with CCR5 stimulates receptor internalization and
intracellular calcium release (29,30). It
has been reported that in lung cancer, CCL8 further promotes tumor
progression by recruiting CCR5+ Treg cells to the tumor site
(27). In patients with malignant
melanoma, CCL8 also promotes tumorigenesis and progression
(31). At present, the expression of
CCL8 in glioma tissue has not been explored, and its role in the
progression of glioma and the corresponding molecular mechanisms
remain unclear. In the present study, the dual-luciferase reporter
assay indicated that miR-181 reduced the activity of the luciferase
reporter fused to the 3′-UTR-WT of CCL-8 but did not suppress that
of the reporter fused to the MUT version. Protein and mRNA
expression of CCL8 were significantly decreased in U87 and A172
cells transfected with miR-181. These results suggest that CCL8 is
a direct target of miR-181 and overexpression of miR-181 reduces
the expression of CCL8.
The impact of miR-181 via CCL8 on cell
proliferation, invasion and migration was further confirmed by
overexpression of CCL8. U87 and A172 cells were co-transfected with
miR-181 and CCL8. The results demonstrated that overexpression of
CCL8 partially reversed the proliferation, invasion, migration and
apoptosis effects of miR-181 in U87 and A172 cells. In addition,
CCL8 expression was high in glioblastoma tissues and negatively
correlated with miR-181 expression. The results further suggested
that miR-181 inhibits glioblastoma cell growth and induces
apoptosis by directly targeting CCL8.
In summary, the present study revealed that miR-181
was downregulated in glioblastoma and associated with poor
prognosis. Overexpression of miR-181 inhibited proliferation,
invasion and migration and induced apoptosis in glioblastoma cells
by directly targeting CCL8. miR-181 may serve as a novel prognostic
biomarker glioblastoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
YZ and FZ conceived and designed the study. FZ and
XC performed the majority of experiments. QH, HZ and YH collected
the clinical samples. DW and SL helped to perform the cell culture
experiments. YZ and FZ wrote the manuscript. All authors have read
and approved this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of The First Affiliated Hospital of Zhengzhou
University. Written informed consent was provided by all
patients.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vengoji R, Macha MA, Batra SK and Shonka
NA: Natural products: A hope for glioblastoma patients. Oncotarget.
9:22194–22219. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ouanouki A, Lamy S and Annabi B:
Periostin, a signal transduction intermediate in TGF-β-induced EMT
in U-87MG human glioblastoma cells, and its inhibition by
anthocyanidins. Oncotarget. 9:22023–22037. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu J, Wang WM, Zhang XL, Du QH, Li HG and
Zhang Y: Effect of downregulated lncRNA NBAT1 on the biological
behavior of glioblastoma cells. Eur Rev Med Pharmacol Sci.
22:2715–2722. 2018.PubMed/NCBI
|
|
4
|
Jayachandran A, Jonathan GE, Patel B and
Prabhu K: Primary spinal cord glioblastoma metastasizing to the
cerebellum: A missed entity. Neurol India. 66:854–857. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang L, Boling W and Zhang JH: Hyperbaric
oxygen therapy as adjunctive strategy in treatment of glioblastoma
multiforme. Med Gas Res. 8:24–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paulmurugan R, Afjei R, Sekar TV, Babikir
HA and Massoud TF: A protein folding molecular imaging biosensor
monitors the effects of drugs that restore mutant p53 structure and
its downstream function in glioblastoma cells. Oncotarget.
9:21495–21511. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tanaka H, Yamaguchi T, Hachiya K, Miwa K,
Shinoda J, Hayashi M, Ogawa S, Nishibori H, Goshima S and Matsuo M:
11C-methionine positron emission tomography for target
delineation of recurrent glioblastoma in re-irradiation planning.
Rep Pract Oncol Radiother. 23:215–219. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Puchalski RB, Shah N, Miller J, Dalley R,
Nomura SR, Yoon JG, Smith KA, Lankerovich M, Bertagnolli D, Bickley
K, et al: An anatomic transcriptional atlas of human glioblastoma.
Science. 360:660–663. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schulze M, Hutterer M, Sabo A, Hoja S,
Lorenz J, Rothhammer-Hampl T, Herold-Mende C, Floßbach L, Monoranu
C and Riemenschneider MJ: Chronophin regulates active vitamin B6
levels and transcriptomic features of glioblastoma cell lines
cultured under non-adherent, serum-free conditions. BMC Cancer.
18:5242018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Sebastiano AR, Deweyert A, Benoit S,
Iredale E, Xu H, De Oliveira C, Wong E, Schmid S and Hebb MO:
Preclinical outcomes of intratumoral modulation therapy for
glioblastoma. Sci Rep. 8:73012018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Sun S, Tong X, Ma Q, Di H, Fu T,
Sun Z, Cai Y, Fan W, Wu Q, et al: MiRNA-154-5p inhibits cell
proliferation and metastasis by targeting PIWIL1 in glioblastoma.
Brain Res. 1676:69–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hinske LC, Heyn J, Hübner M, Rink J,
Hirschberger S and Kreth S: Intronic miRNA-641 controls its host
Gene's pathway PI3K/AKT and this relationship is dysfunctional in
glioblastoma multiforme. Biochem Biophys Res Commun. 489:477–483.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Zhang X, Hu S, Zheng M, Zhang J,
Zhao J, Zhang X, Yan B, Jia L, Zhao J, et al: Identification of
miRNA-7 by genome-wide analysis as a critical sensitizer for
TRAIL-induced apoptosis in glioblastoma cells. Nucleic Acids Res.
45:5930–5944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li LY, Xiao J, Liu Q and Xia K: Parecoxib
inhibits glioblastoma cell proliferation, migration and invasion by
upregulating miRNA-29c. Biol Open. 6:311–316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bing ZT, Yang GH, Xiong J, Guo L and Yang
L: Identify signature regulatory network for glioblastoma prognosis
by integrative mRNA and miRNA co-expression analysis. IET Syst
Biol. 10:244–251. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu HQ, Sun LG and Guo WJ: Decreased
miRNA-146a in glioblastoma multiforme and regulation of cell
proliferation and apoptosis by target Notch1. Int J Biol Markers.
31:e270–e275. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu W, Li P, Xu Q and Wang C: Regulation
of miR-181 on chemotherapy-resistant cervical cancer cells. Int J
Clin Experimental Pathol. 9:4560–4565. 2016.
|
|
18
|
Zhou WY, Chen JC, Jiao TT, Hui N and Qi X:
MicroRNA-181 targets Yin Yang 1 expression and inhibits cervical
cancer progression. Mol Med Rep. 11:4541–4546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Xin Z, Xu Y and Ma J: Upregulated
miRNA-622 inhibited cell proliferation, motility, and invasion via
repressing Kirsten rat sarcoma in glioblastoma. Tumour Biol.
37:5963–5970. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bo LJ, Wei B, Li ZH, Wang ZF, Gao Z and
Miao Z: Bioinformatics analysis of miRNA expression profile between
primary and recurrent glioblastoma. Eur Rev Med Pharmacol Sci.
19:3579–3586. 2015.PubMed/NCBI
|
|
22
|
Kouri FM, Ritner C and Stegh AH: miRNA-182
and the regulation of the glioblastoma phenotype-toward miRNA-based
precision therapeutics. Cell Cycle. 14:3794–3800. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
LeBlanc VC and Morin P: Exploring
miRNA-associated signatures with diagnostic relevance in
glioblastoma multiforme and breast cancer patients. J Clin Med.
4:1612–1630. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu SL, Fu X, Huang J, Jia TT, Zong FY, Mu
SR, Zhu H, Yan Y, Qiu S, Wu Q, et al: Genome-wide analysis of
YB-1-RNA interactions reveals a novel role of YB-1 in miRNA
processing in glioblastoma multiforme. Nucleic Acids Res.
43:8516–8528. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Das S, Kohr M, Dunkerly-Eyring B, Lee DI,
Bedja D, Kent OA, Leung AK, Henao-Mejia J, Flavell RA and
Steenbergen C: Divergent effects of miR-181 family members on
myocardial function through protective cytosolic and detrimental
mitochondrial microRNA targets. J Am Heart Assoc. 6(pii):
e0046942017.PubMed/NCBI
|
|
26
|
Strotbek M, Schmid S, Sànchezgonzàlez I,
Boerries M, Busch H and Olayioye MA: miR-181 elevates Akt signaling
by co-targeting PHLPP2 and INPP4B phosphatases in luminal breast
cancer. Int J Cancer. 140:2310–2320. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu J, Xing Y and Rong L: miR-181
regulates cisplatin-resistant non-small cell lung cancer via
downregulation of autophagy through the PTEN/PI3K/AKT pathway.
Oncol Rep. 39:1631–1639. 2018.PubMed/NCBI
|
|
28
|
Su R, Lin HS, Zhang XH, Yin XL, Ning HM,
Liu B, Zhai PF, Gong JN, Shen C, Song L, et al: MiR-181 family:
Regulators of myeloid differentiation and acute myeloid leukemia as
well as potential therapeutic targets. Oncogene. 34:3226–3239.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ge B, Li J, Wei Z, Sun T, Song Y and Khan
NU: Functional expression of CCL8 and its interaction with
chemokine receptor CCR3. BMC Immunol. 18:542017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee JU, Cheong HS, Shim EY, Bae DJ, Chang
HS, Uh ST, Kim YH, Park JS, Lee B, Shin HD and Park CS: Gene
profile of fibroblasts identify relation of CCL8 with idiopathic
pulmonary fibrosis. Respir Res. 18:32017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Halvorsen EC, Hamilton MJ, Young A,
Wadsworth BJ, LePard NE, Lee HN, Firmino N, Collier JL and
Bennewith KL: Maraviroc decreases CCL8-mediated migration of
CCR5(+) regulatory T cells and reduces metastatic tumor growth in
the lungs. Oncoimmunology. 5:e11503982016. View Article : Google Scholar : PubMed/NCBI
|