Introduction
Glioblastoma, also known as glioblastoma multiforme
(GBM), is the most common cancer of the central nervous system,
accounting for >80% of all brain tumours (1). GBM has an annual incidence of 5.26
cases per 100,000 individuals, with 17,000 new cases being
diagnosed each year (2). Due to
rapid proliferative rate and a characteristic invasive nature, the
survival rate of patients with GBM remains low (1). Typically, patients with GBM survive for
12–15 months after diagnosis, with only 3–5% of individuals
surviving >5 years (3). The
underlying cause of the majority of cases is unclear. Identifying
biomarkers of GBM may improve the diagnosis and prognosis of
patients, and provide novel therapeutic targets for treating
patients with GBM.
The contactin (CNTN) proteins are a subgroup of
proteins, which belong to the immunoglobulin (Ig) superfamily of
proteins, and are primarily expressed in the nervous system. CNTNs
consist of six members: CNTN1, CNTN2 [transient axonal glycoprotein
1 (TAG-1)], CNTN3 [brefeldin A-inhibited guanine
nucleotide-exchange protein 1 (BIG)-1; plasmacytoma-associated
neuronal glycoprotein (PANG)], CNTN4 (BIG-2), CNTN5 (neural
recognition molecule NB-2) and CNTN6 (neural recognition molecule
NB-3) (4). The expression of
CNTN3 mRNA is developmentally regulated and reaches its
highest level in the adult brain (5). CNTN3 is also termed PANG or BIG-1, and
was discovered in the endoplasmic reticulum of plasmacytomas
(6). CNTN3 is a membrane protein
anchored by glycosylphosphatidylinositol, with six Ig-like domains
and four-fibronectin type III repeats, and it belongs to the
TAG-1/F3 subgroup of the Ig superfamily of proteins (5). The CNTN3 gene is located at 3p26
in the genome (7) and its expression
is restricted to certain subsets of neurons, including the
cerebellar Purkinje cells, the granule cells of the dentate gyrus
and the neurons in the superficial layers of the cerebral cortex
(1). CNTN3 may function in the
formation and maintenance of specific neuronal networks (6,8–11). Bouyain and Watkins (12) found that CNTN3 interacts with the
receptor protein tyrosine phosphatases γ and is involved in the
construction of neural networks. The expression and function of
CNTN3 in different types of cancer have not been thoroughly
investigated. A previous study suggested that CNTN3 is a potential
target gene of hsa-miR-3675b in breast cancer, and by using gene
ontology analysis, it was demonstrated that CNTN3 may be associated
with cell proliferation, apoptosis and cell cycle progression
(13). However, to the best of our
knowledge, the biological role of CNTN3 in GBM remains unknown.
In the present study, microarrays and sequencing
datasets were analysed to elucidate the clinical value of CNTN3 in
GBM and the associated molecular mechanisms. The expression of
CNTN3 in GBM was compared with that in normal tissues using
SAGE Anatomical viewer, Gene Expression Omnibus (GEO), Oncomine and
The Cancer Genome Atlas (TCGA) databases. The expression of the
CNTN3 protein was examined in microarrays from GBM tissues. The
clinical data were used to determine the prognostic value of CNTN3
in patients with GBM. The pathways associated with CNTN3 were
determined using gene set enrichment analysis (GSEA) and
interaction networks were constructed using Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) database and
Cytoscape analysis.
Materials and methods
SAGE anatomical viewer
The SAGE Anatomical Viewer (cgap.nci.nih.gov/sage/anatomicviewer) is a
website established by the National Cancer Institute (Bethesda, MD,
USA) that displays gene expression in normal and malignant human
tissues by shading a given organ in one of 10 colours, depicting
differing levels of gene expression. CNTN3 expression (the
NM_020872 transcript) was found to be present in both normal and
cancer tissues. The tag with the highest rank and the highest
frequency was selected for CNTN3 gene, and the sequence was
AGATATGCAC.
GEO and TCGA datasets
GEO is a public functional genomics data repository
in which gene array and sequence-based data are accepted
(https://www.ncbi.nlm.nih.gov/geo/).
GSE4290 (14) and GSE7696 (15) microarray datasets were obtained from
the GEO database. GSE4290 contained data from 81 GBM samples and 23
samples from patients with epilepsy, which were used as the
non-tumour samples. GSE7696 contained samples from 80 patients with
GBM whom participated in clinical trials and 4 samples of normal
brain tissue. Transcription and clinical data for patients with GBM
were also downloaded from the TCGA website (https://cancergenome.nih.gov/), and consisted of 156
GBM samples and 5 normal tissues.
Data processing
Raw data were downloaded and normalised using the
multi-array average analysis method (16) in R version 3.4.4 (17). Differentially expressed genes were
identified with the Limma package (18), and a fold change of >2 and
P<0.05 were set as the cut-offs.
Oncomine database analysis
CNTN3 mRNA levels in normal and GBM tissues
were compared in the Oncomine database (www.oncomine.org). CNTN3 mRNA levels were
downregulated in the GBM samples from the ‘Sun Brain’ (14) and ‘Murat Brain’ (15) datasets in the Oncomine database.
CNTN3 levels were also compared across these two datasets.
The rank for a gene was the median rank for that gene across each
of the analyses. The P-value for a gene was its P-value for the
median-ranked analysis.
Immunohistochemistry
A GBM tissue microarray was purchased from Shanghai
Outdo Biotech Co., Ltd., which contained 67 GBM and 13 adjacent
normal brain tissue samples. All of the patients had been
pathologically diagnosed with GBM. The thickness of tissue sections
was 4 µm. Two serial tissue sections were prepared for CNTN3 and
EGFR detection. The tissue slides were deparaffinised in xylene and
rehydrated gradually in 100, 85 and 75% alcohol. Next, the slides
were treated with 3% H2O2 for 15 min,
autoclaved in 10 mM citric sodium (pH 6.0) for 30 min to
permeabilize the samples, rinsed in PBS and then incubated with a
primary antibody against CNTN3 (Abcam; cat. no. ab203592; 1:100) or
EGFR (Proteintech; cat. no. 18986-1-AP; 1:500) at 4°C overnight.
This was followed by incubation with biotinylated anti-rabbit IgG
(OriGene Technologies, Inc.; cat. no. TA130016; 1:200) for 30 min
at 37°C and then peroxidase-labelled streptavidin (OriGene
Technologies, Inc.; cat. no. AR100017; 1:100) for 15 min at 37°C.
The signals were visualised using a diaminobenzidine substrate kit
(ZSGB-BIO; OriGene Technologies, Inc.) according to the
manufacturer's protocol. CNTN3 immunostaining score was determined
using a semi-quantitative approach. The percentage of stained cells
in each sample was scored as follows: 0, 0; 1, 1–25; 2, 26–50; 3,
51–75; and 4, 76–100%. The intensity of staining was scored as
follows: 0, negative; 1, weak staining; 2, moderate staining; and
3, strong staining. The final staining score was the intensity
score multiplied by the percentage score. A final staining score
<6 was classified as negative CNTN3/EGFR expression and a score
≥6 was classified as positive CNTN3/EGFR expression.
Statistical analysis
The mRNA expression and protein expression between
different groups was compared respectively using t-test and Mann
Whitney U test, and correlations were evaluated using Pearson's
correlation analyses. The associations between the CNTN3 expression
levels and the clinicopathological features were evaluated using
χ2 test. Kaplan-Meier estimator curves and log-rank test
were used for the survival analysis. Univariate and multivariate
survival analyses were performed using Cox's proportional hazards
model. The data are presented as the mean ± the standard error the
mean. P<0.05 was considered to indicate a statistically
significant difference. All of the statistical analyses were
performed using SPSS version 21.0 (IBM Corp.).
Identifying potential molecular
mechanisms of CNTN3 expression in GBM
GSEA 3.0 (Broad Institute, Inc.) (19) was used to detect the expression
changes of gene sets associated with CNTN3. The annotated gene sets
c2.cp.kegg.v6.0.symbols.gmt were selected as the reference gene
sets. A false-discovery-rate of <0.05 was used as the cut-off
criteria. STRING (http://string-db.org) was used to identify the
potential interaction networks between proteins encoded by these
genes. Cytoscape version 2.8.3 (20)
software was used to construct interaction networks between the
proteins in GBM.
Results
CNTN3 gene expression in GBM and
normal tissues
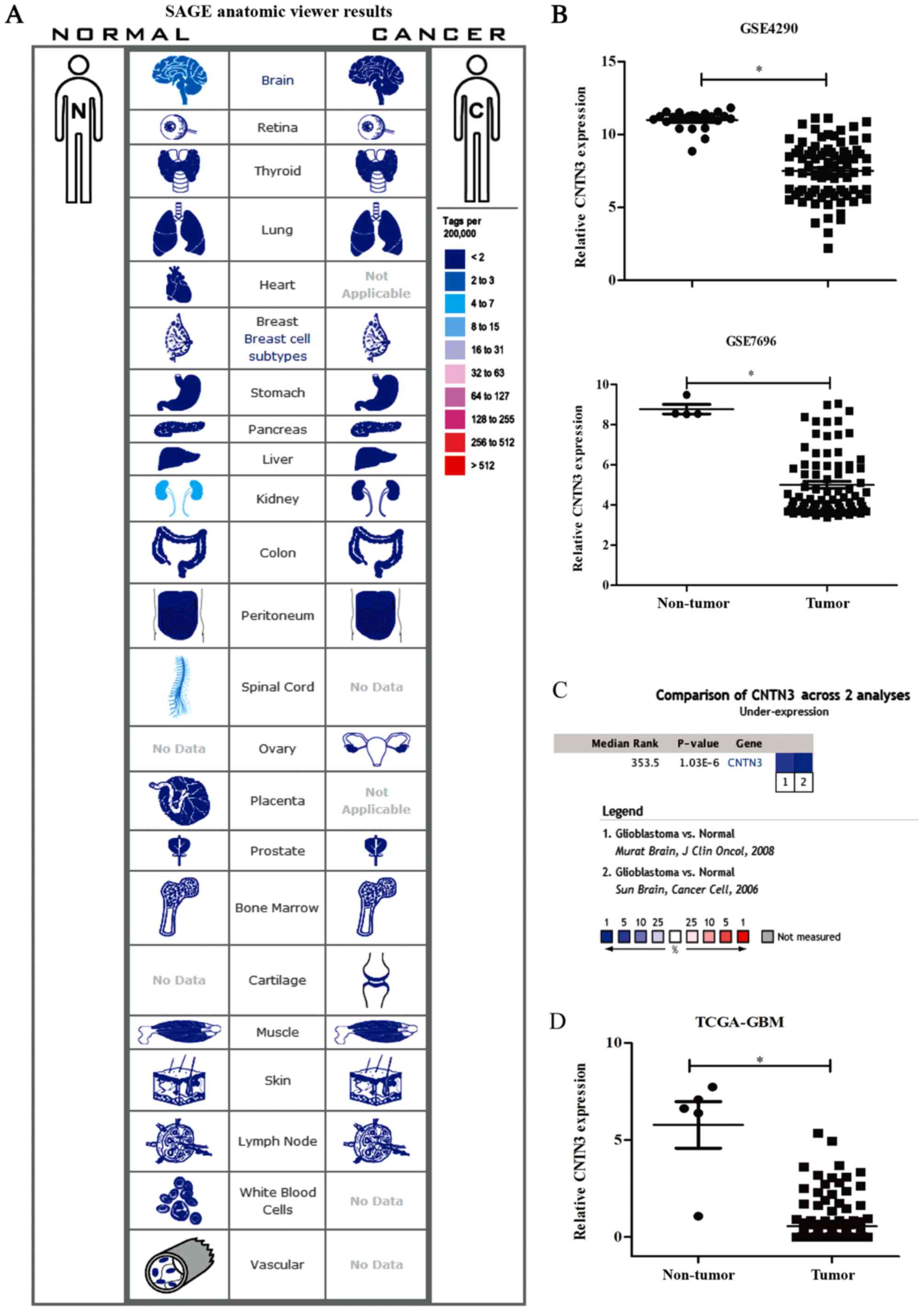
The gene expression levels of CNTN3 between
cancer and matched normal tissues were analysed using SAGE
anatomical viewer with tag. CNTN3 mRNA expression was
downregulated in brain and kidney cancer tissues when compared with
that in normal tissues (Fig. 1A).
Analysing the GEO datasets revealed that the gene expression level
of CNTN3 was significantly lower in GBM compared with that
in normal tissue in the two datasets (both P<0.05; Fig. 1B). CNTN3 expression was
downregulated in the GSE4290 (fold-change, −12.218;
P=4.64×10−26) and GSE7696 (fold-change, −13.6;
P=2.07×10−06) datasets. The median rank of CNTN3
was 353.3 based on a meta-analysis across the two datasets using
Oncomine algorithms (P=1.03×10−6; Fig. 1C). Similarly, CNTN3 was also
downregulated in GBM in the dataset obtained from TCGA
(fold-change, −5.855; P=1.32×10−24; Fig. 1D).
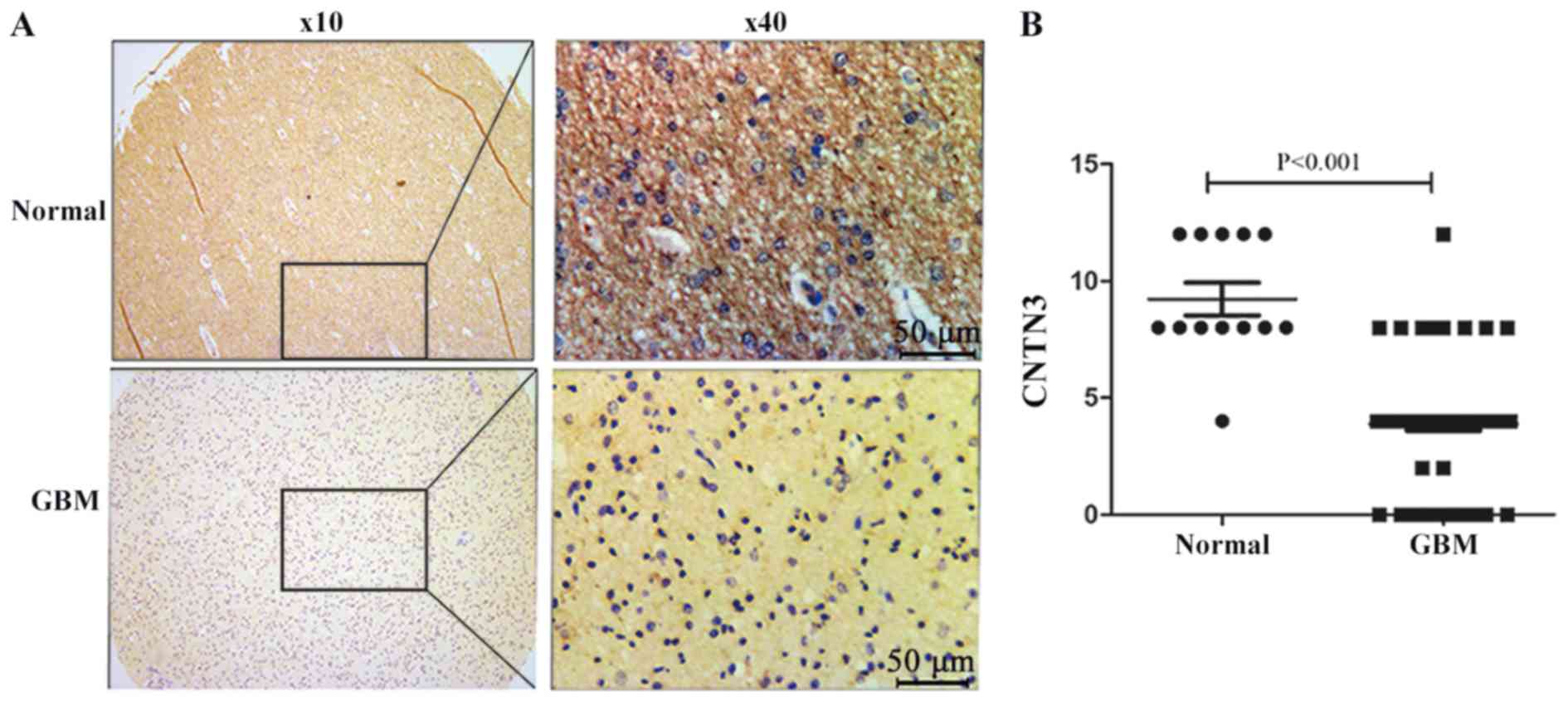
Immunohistochemistry staining was performed on the
GBM tissue microarray. CNTN3 was primarily located in the cytoplasm
of neuronal cells and its expression was downregulated in GBM
tissues (Fig. 2A). Positive CNTN3
expression was observed in 92.3% (12/13) of the adjacent normal
brain tissues and in 13.4% (9/67) in the GBM tissues (P<0.0001;
Fig. 2B).
Clinical and prognostic value of CNTN3
in GBM patients
The association between CNTN3 expression and the
clinicopathological features of patients was statistically analysed
in the GSE7696 dataset as it had survival data and other clinical
data. Clinical parameters consisted of sex, age, treatment method
(temozolomide with radiotherapy or radiotherapy alone) and
O6-methylguanine-DNA methyltransferase (MGMT) methylation status.
There was no significant association between CNTN3 expression and
any of the parameters (all P>0.05; Table I).
 | Table I.Association of CNTN3 expression with
the clinicopathological characteristics of patients with GBM. |
Table I.
Association of CNTN3 expression with
the clinicopathological characteristics of patients with GBM.
|
|
| CNTN3
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Total, n | Low, n | High, n | P-value |
|---|
| Sex |
|
|
| 0.799 |
|
Male | 59 | 29 | 30 |
|
|
Female | 21 | 11 | 10 |
|
| Age, years |
|
|
| 0.264 |
|
≤60 | 64 | 30 | 34 |
|
|
>60 | 16 | 10 | 6 |
|
| Treatment |
|
|
| 0.348 |
| TMZ +
radiotherapy | 52 | 24 | 28 |
|
|
Radiotherapy | 28 | 16 | 12 |
|
| MGMT |
|
|
| 0.171 |
|
Methylation | 44 | 19 | 25 |
|
| No
methylation | 34 | 20 | 14 |
|
|
Unknown | 2 | 1 | 1 |
|
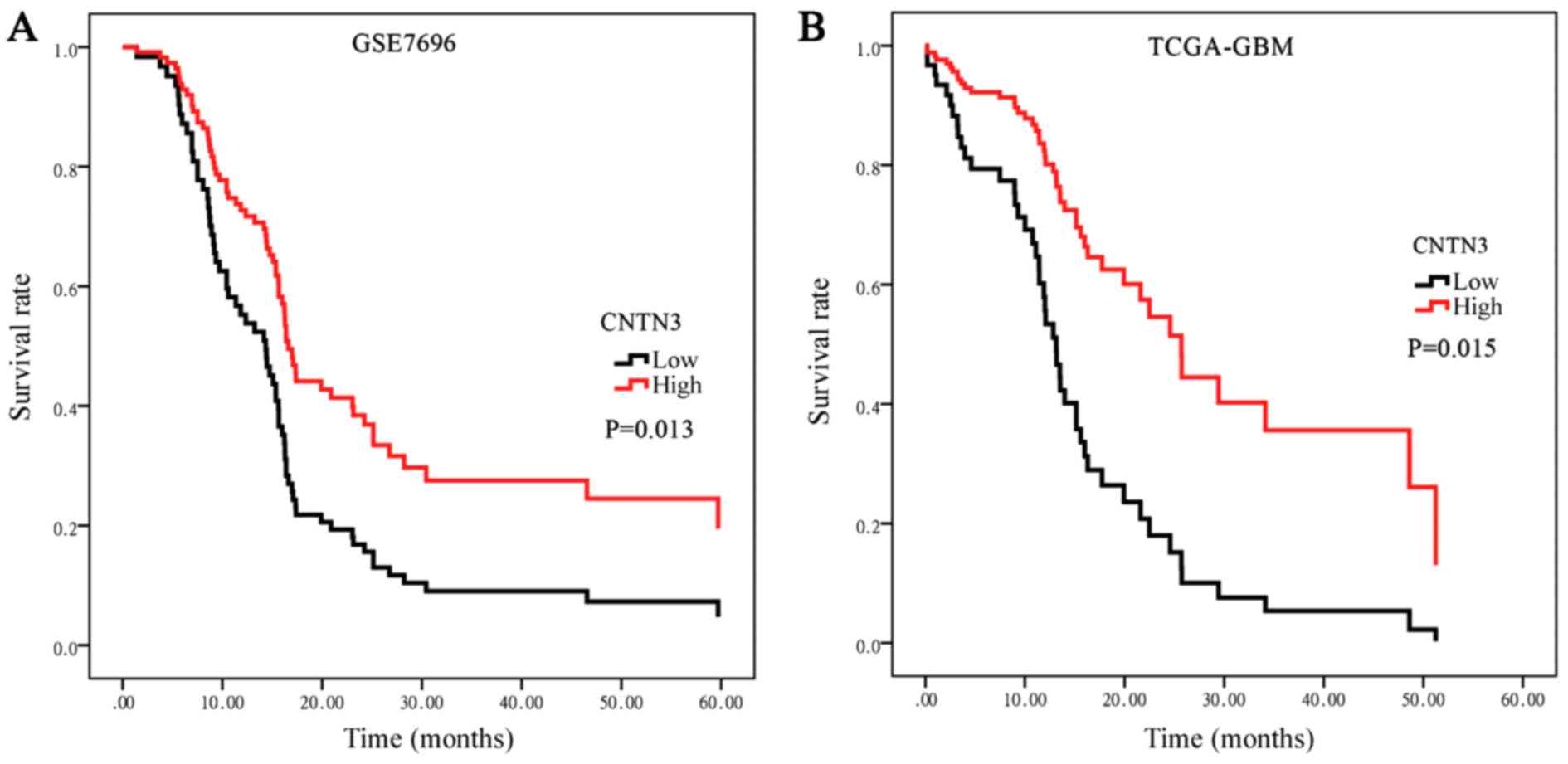
To determine the prognostic significance of CNTN3
expression in patients with GBM, Kaplan-Meier survival analysis was
used to evaluate the association between CNTN3 expression and
survival in the GS7696 dataset (Fig.
3A). The data was divided into high- and low-expression
subgroups based on the median CNTN3 expression, 15.58. The median
overall survival (OS) time was 14.140 months [95% confidence
interval (CI), 10.933–17.347] in the low expression group and
17.070 months (95% CI, 15.288–18.852) in the high expression group.
A log-rank test demonstrated that patients with low CNTN3
expression had a significantly shorter OS time compared with those
with high CNTN3 expression (P=0.013; Fig. 3A). Survival analysis was additionally
performed on the TCGA-GBM dataset. The median OS time was 12.833
months (95% CI, 10.897–14.770) in the low expression group and 25.7
months (95% CI, 17.176–40.950) in the high expression group, with
survival being significantly shorter in the low expression group
(P=0.015; Fig. 3B).
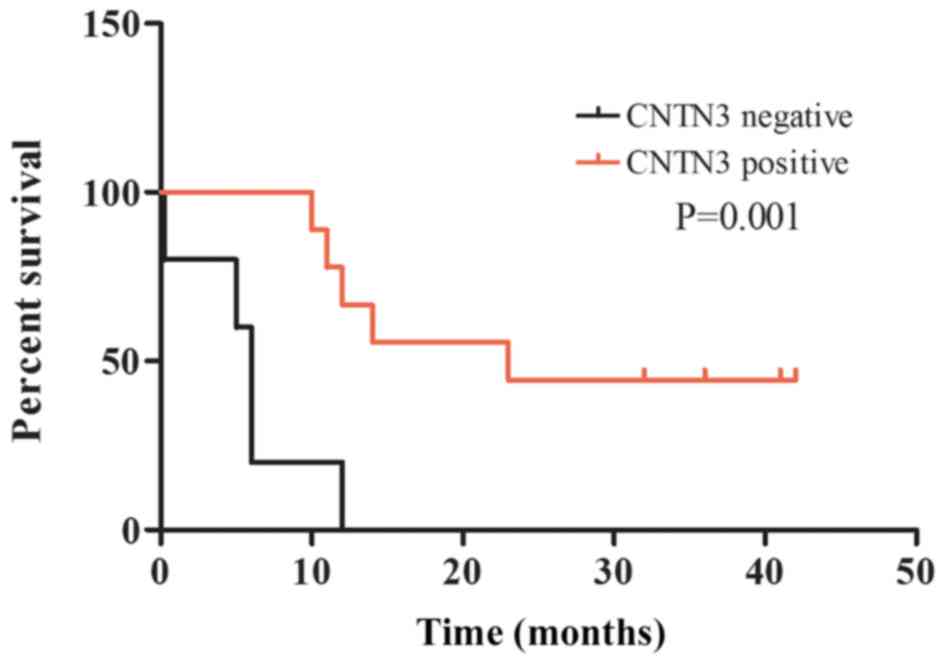
Kaplan-Meier survival analysis was also performed to
investigate the prognostic value of CNTN3 protein expression on the
survival rate of patients with GBM. The OS rate of patients with
tumours that did not express CNTN3 was significantly lower compared
with that of patients with tumours positive for CNTN3 expression
(P=0.001; Fig. 4).
Univariate and multivariate Cox regression analyses
were performed to confirm the possibility that CNTN3 may be useful
as an independent prognostic factor in patients with GBM.
Univariate analysis demonstrated that a low expression level of
CNTN3 (P=0.027), age ≥60 (P=0.031), treatment with radiotherapy
(P=0.023) and MGMT methylation (P<0.001) were significantly
associated with a poor clinical outcome in GBM cases (Table II). Multivariate analysis further
identified that low expression levels of CNTN3 (P=0.026), treatment
with radiotherapy (P=0.024) and MGMT methylation (P<0.001) were
independent prognostic factors in patients with GBM (Table II).
 | Table II.Univariate and multivariate Cox
proportional hazard regression analysis. |
Table II.
Univariate and multivariate Cox
proportional hazard regression analysis.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, ≥60 vs. <60
years | 1.033 | 1.003–1.065 | 0.031a | 1.019 | 0.990–1.048 | 0.212 |
| Sex, male vs.
female | 0.864 | 0.496–1.505 | 0.605 | – | – | – |
| CNTN3, high vs. low
expression | 0.823 | 0.693–0.978 | 0.027a | 0.822 | 0.691–0.977 | 0.026a |
| Treatment, TMZ +
radiotherapy vs. radiotherapy | 0.549 | 0.328–0.919 | 0.023a | 0.534 | 0.310–0.920 | 0.024a |
| MGMT, methylation
vs. no methylation | 3.971 | 2.273–6.937 | <0.001 | 4.324 | 2.401–7.788 | <0.001 |
Potential molecular mechanisms
involving CNTN3 in patients with GBM
To gain insight into the molecular mechanism
involving CNTN3 in patients with GBM, GSEA was used to determine
whether the CNTN3 gene and associated genes were involved in
any oncogenic pathways. The results from the two GEO datasets
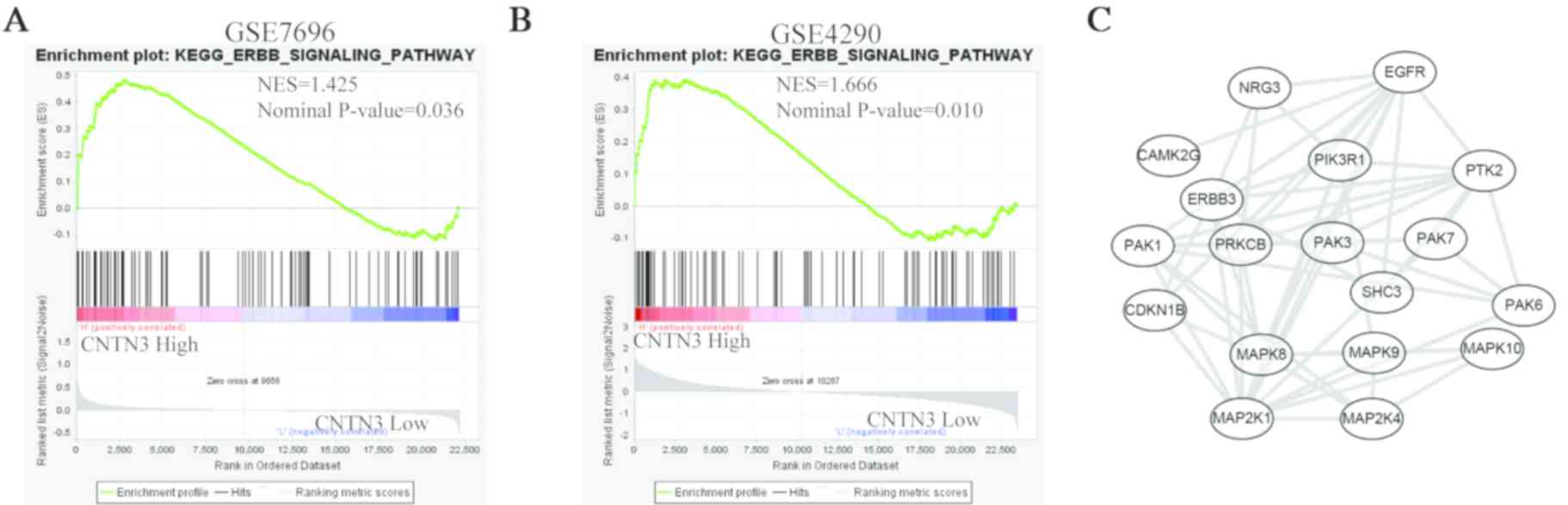
revealed that CNTN3 was associated with the ErbB signalling
pathway (Fig. 5A and B). The 18
genes in this pathway that were associated with CNTN3 were
EGFR, NRG3, CAMK2G, PIK3R1, PTK2, ERBB3, PAK1, PRKCB, PAK3,
PAK7, CDKN1B, SHC3, PAK6, MAPK8, MAPK9, MAPK10, MAP2K1 and
MAP2K4. The PPI network showed that these genes exhibited
complex interactivity with each other (Fig. 5C). EGFR was amplified in
40–50% of primary GBMs and is thought to participate in tumour cell
invasion, angiogenesis and proliferation (21). Therefore, the correlation between the
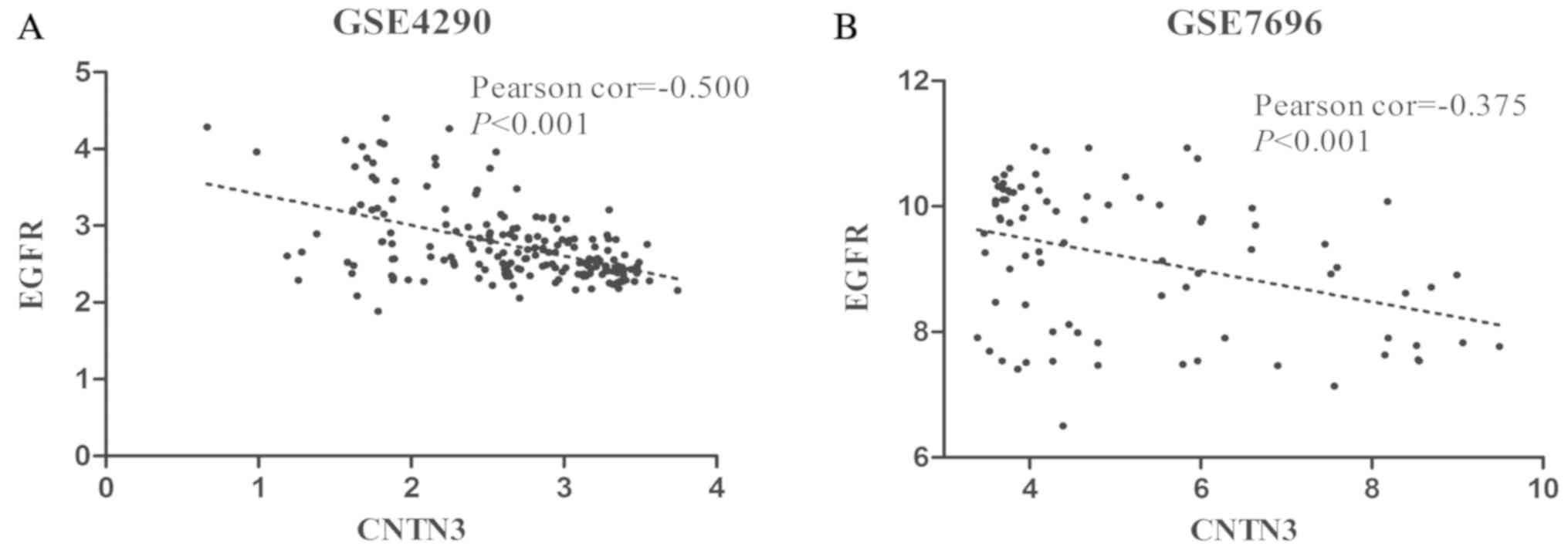
expression of EGFR and CNTN3 was analysed.
EGFR expression levels were negatively correlated with
CNTN3 expression in the GSE4290 (P<0.001; Fig. 6A) and GSE7696 (P<0.001; Fig. 6B) datasets. Additionally, the
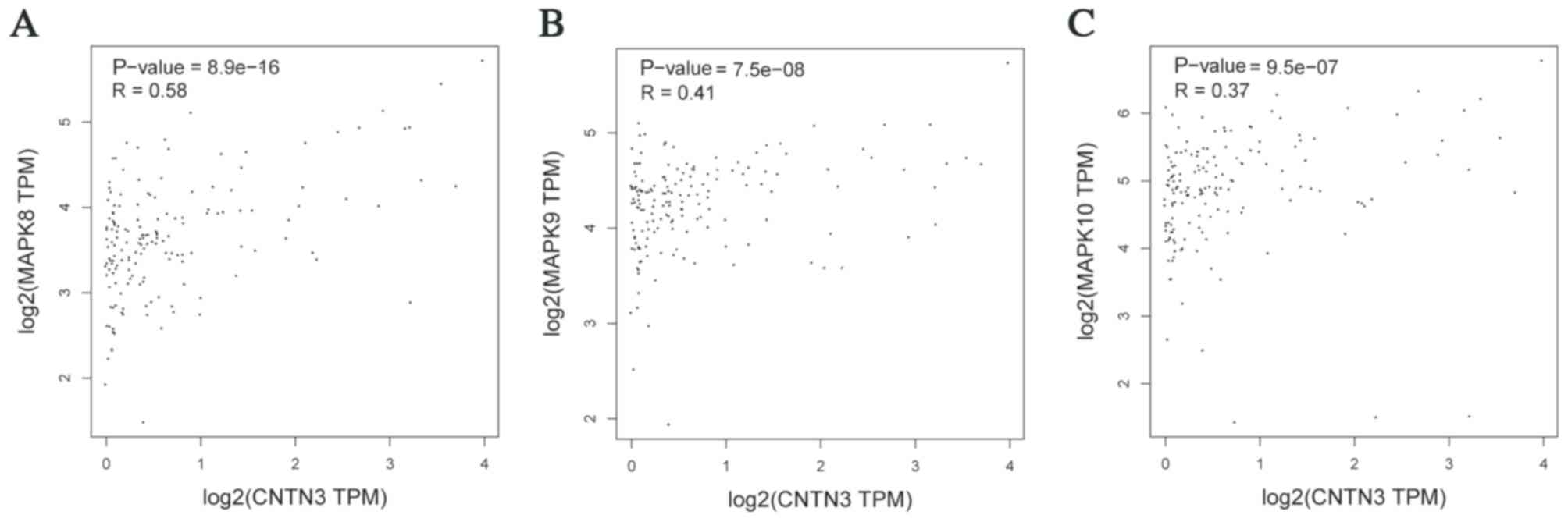
expression of CNTN3 was positively correlated with the
expression of MAPK8, MAPK9 and MAPK10, which are
downstream of EGFR (22)
(Fig. 7).
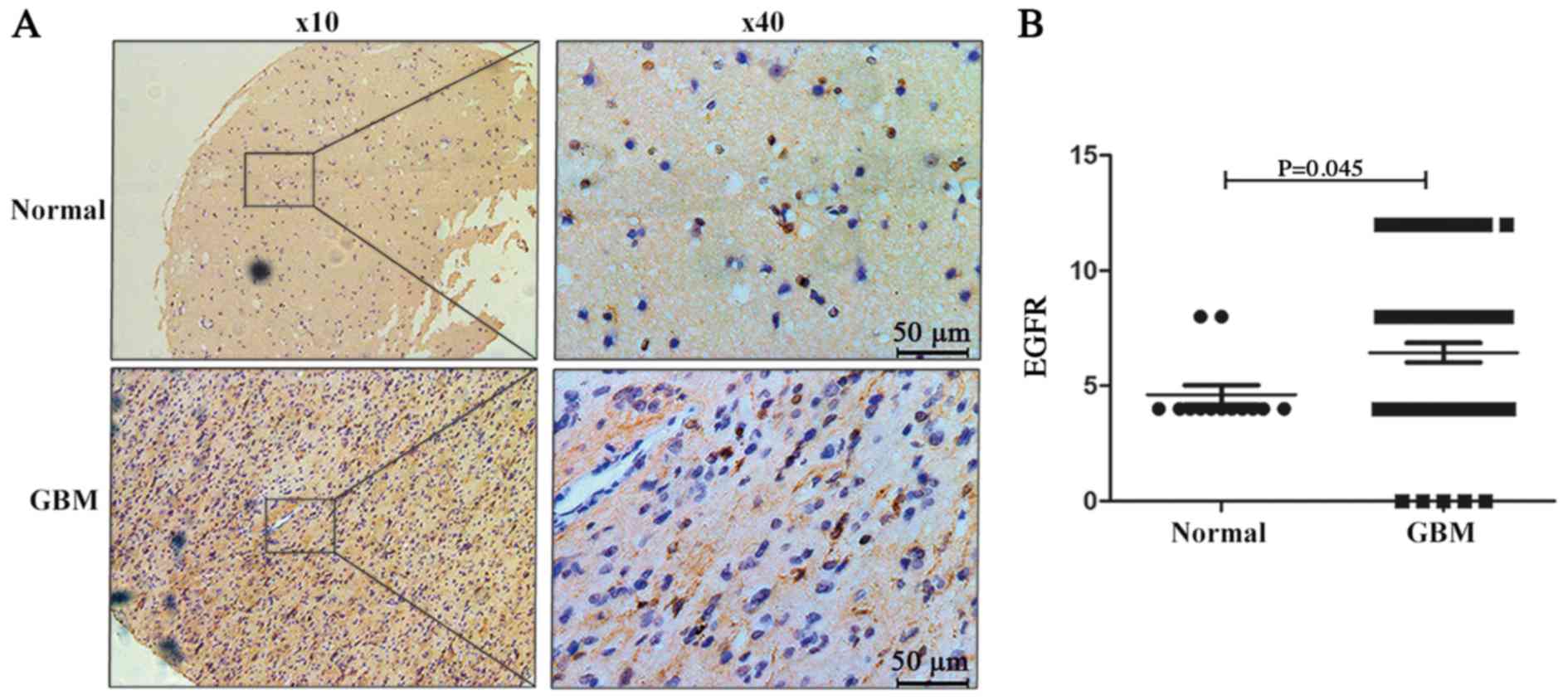
EGFR expression in the tissue microarray was also
detected. EGFR was primarily located in the cytoplasm and nucleus
of neuronal cells (Fig. 8A) and its
expression was upregulated in GBM tissues (Fig. 8B). Positive EGFR expression was
observed in 15.4% (2/13) of adjacent normal brain tissues, and in
50.7% (34/67) of GBM samples (P=0.037, Fig. 8B).
A similar negative correlation was observed between
CNTN3 and EGFR expression in the tissue microarray; 52.5% (31/59)
of brain tissue with low CNTN3 expression showed high EGFR
expression and 76.2% (16/21) of brain tissue with high CNTN3
expression showed low EGFR expression (P=0.045; Table III).
 | Table III.Association between CNTN3 and EGFR
expression in the tissue microarray. |
Table III.
Association between CNTN3 and EGFR
expression in the tissue microarray.
|
| CNTN3
expression |
|
|---|
|
|
|
|
|---|
| EGFR
expression | Negative, n
(%) | Positive, n
(%) | P-value |
|---|
| Negative | 28 (47.5) | 16 (76.2) | 0.023a |
| Positive | 31 (52.5) | 5 (23.8) |
|
Discussion
In the present study, the expression level of the
CNTN3 gene was compared in silico between patients
with GBM and non-tumour tissues (normal brain tissues and
non-tumour tissues from patients with epilepsy) across independent
datasets. Expression of the CNTN3 gene was decreased in the
GBM samples, and was negatively correlated with OS. Univariate and
multivariate analysis revealed that CNTN3 expression was an
independent prognostic factor in patients with GBM. These analyses
suggest that CNTN3 may serve as a potentially protective factor in
GBM. To gain further insights into the role of CNTN3, GSEA analysis
was used. CNTN3 was associated with the ErbB signalling pathway. As
the most commonly altered gene, EGFR exhibited expression
that was negatively correlated with CNTN3. To the best of our
knowledge, this is the first study on the clinicopathological and
prognostic significance of CNTN3 in GBM.
The function of CNTN3 has not been thoroughly
investigated. Cell adhesion/recognition molecules of the
immunoglobulin (Ig) superfamily serve vital roles in forming and
maintaining the nervous system (8).
CNTN3 belongs to the Ig superfamily of proteins and its function is
primarily associated with the properties of this family. Akane
et al (23), illustrated that
the rat ortholog of CNTN3, Cntn3, functions in neurite
outgrowth-promotion, and when treated with glycidol at 1,000 ppm,
axonopathy occurred and the expression of CNTN3 was altered.
Morales et al (10), found an
isolated cryptic proximal interstitial 3p deletion, del(3)
(p12.3p13) in a patient with neurodevelopmental delay. Since this
deletion site is where the CNTN3 gene is located, it was
hypothesized that CNTN3 may serve a role in neuronal development.
Subsequently, Hu et al (9)
found that deletion or duplication of CNTN6 were associated
with neurodevelopmental disorders. However, Monies et al
(24) found that possessing
additional copies of CNTN3 was not associated with the
aforementioned phenotypes. The present study highlights the
potential role of CNTN3 in GBM and its use as a prognostic
biomarker.
The ErbB receptor family of tyrosine kinases is
comprised of four members, EGFR (ErbB1/HER1), ErbB2 (HER2/neu),
ErbB3 (HER3) and ErbB4 (HER4) (25).
Aberrations in EGFR are the most common oncogenic alterations in
patients with GBM, and are associated with tumour malignancy and a
poor prognosis (26). As EGFR
expression was negatively correlated with the expression of
CNTN3, CNTN3 may inhibit EGFR expression
physiologically. When CNTN3 is downregulated, such as in GBM, EGFR
may be aberrantly activated and initiate the ErbB signalling
pathway to promote tumour growth and invasion. The signalling
pathways mediated by EGFR activation include the
RAS/mitogen-activated protein kinase (MAPK)/extracellular
signal-regulated kinase, PI3K/protein kinase B, Janus kinase/signal
transducer and activator of transcription and protein kinase C
pathways (27). In the present
study, the expression of CNTN3 correlated with the expression of
MAPK8, MAPK9 and MAPK10, which are downstream of
EGFR, and therefore these findings suggest that CNTN3 may
regulate the properties of GBM by ErbB-MAPK pathway. As the present
study was based on results from bioinformatic analyses and tissue
microarrays, additional studies are required to determine the
association between CNTN3 and the malignant properties of GBM in
vitro and in vivo, such as cell proliferation,
self-renewal, angiogenesis and drug-resistance.
In conclusion, the present study demonstrates that
CNTN3 is downregulated in patients with GBM, and CNTN3
expression is negatively correlated with OS. CNTN3 may be
functionally associated with the EGFR signalling pathway, as the
expression of EGFR and CNTN3 were negatively correlated. CNTN3 may
be used as a prognostic marker and a potential therapeutic target
for patients with GBM.
Acknowledgements
Not applicable.
Funding
The present study was funded by The Technology
Innovation Project of the Chengdu Science and Technology Bureau
(grant no. 2018-YF05-00669-SN), The Project of Health and Family
Planning Commission of the Sichuan Province (grant no. 18PJ126) and
The Project of Health and Family Planning Commission of the Chengdu
City (grant no. 2018056).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YFZ designed the research, analyzed the data,
drafted and revised the paper. YBG and LL analyzed the data and
revised the manuscript. PY, TTZ, BRP and DFW performed the
experiments and re-checked the results. HYZ designed the research.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shergalis A, Bankhead A III, Luesakul U,
Muangsin N and Neamati N: Current challenges and opportunities in
treating glioblastoma. Pharmacol Rev. 70:412–445. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lefranc F, Le Rhun E, Kiss R and Weller M:
Glioblastoma quo vadis: Will migration and invasiveness reemerge as
therapeutic targets? Cancer Treat Rev. 68:145–154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gallego O: Nonsurgical treatment of
recurrent glioblastoma. Curr Oncol. 22:e273–e281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shimoda Y and Watanabe K: Contactins:
Emerging key roles in the development and function of the nervous
system. Cell Adh Migr. 3:64–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yoshihara Y, Kawasaki M, Tani A, Tamada A,
Nagata S, Kagamiyama H and Mori K: BIG-1: A new TAG-1/F3-related
member of the immunoglobulin superfamily with neurite
outgrowth-promoting activity. Neuron. 13:415–426. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Connelly MA, Grady RC, Mushinski JF and
Marcu KB: PANG, a gene encoding a neuronal glycoprotein, is
ectopically activated by intracisternal A-type particle long
terminal repeats in murine plasmacytomas. Proc Natl Acad Sci USA.
91:1337–1341. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo C, Li Y, Zhang X, Zhang Y, Zhang H,
Chen C, Xu Z, Cui P, Hu S, Yang H and Dong W: DNA sequence
comparative analysis of the 3pter-p26 region of human genome. Sci
China C Life Sci. 48:34–40. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mohebiany AN, Harroch S and Bouyain S: New
insights into the roles of the contactin cell adhesion molecules in
neural development. Adv Neurobiol. 8:165–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu J, Liao J, Sathanoori M, Kochmar S,
Sebastian J, Yatsenko SA and Surti U: CNTN6 copy number variations
in 14 patients: A possible candidate gene for neurodevelopmental
and neuropsychiatric disorders. J Neurodev Disord. 7:262015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morales C, Mademont-Soler I, Armengol L,
Milà M, Badenas C, Andrés S, Soler A and Sánchez A:
Characterization of a 5.8-Mb interstitial deletion of chromosome 3p
in a girl with 46, XX, inv(7)dn karyotype and phenotypic
abnormalities. Cytogenet Genome Res. 125:334–340. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshihara Y, Kawasaki M, Tamada A, Nagata
S, Kagamiyama H and Mori K: Overlapping and differential expression
of BIG-2, BIG-1, TAG-1, and F3: Four members of an axon-associated
cell adhesion molecule subgroup of the immunoglobulin superfamily.
J Neurobiol. 28:51–69. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bouyain S and Watkins DJ: The protein
tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of
the contactin family of neural recognition molecules. Proc Natl
Acad Sci USA. 107:2443–2448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Q, Wang C, Guo L, Ge Q and Lu Z:
Identification and characterization of novel microRNA candidates
from deep sequencing. Clin Chim Acta. 415:239–244. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun L, Hui AM, Su Q, Vortmeyer A,
Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey
R, et al: Neuronal and glioma-derived stem cell factor induces
angiogenesis within the brain. Cancer Cell. 9:287–300. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murat A, Migliavacca E, Gorlia T, Lambiv
WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven
MC, et al: Stem cell-related ‘self-renewal’ signature and high
epidermal growth factor receptor expression associated with
resistance to concomitant chemoradiotherapy in glioblastoma. J Clin
Oncol. 26:3015–3024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia L, Su X, Shen J, Meng Q, Yan J, Zhang
C, Chen Y, Wang H and Xu M: ANLN functions as a key candidate gene
in cervical cancer as determined by integrated bioinformatic
analysis. Cancer Manag Res. 10:663–670. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Team RC, . R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: ISBN 3-900051-07-0. http://www.R-project.org/2012
|
|
18
|
Yu J, Wu X, Huang K, Zhu M, Zhang X, Zhang
Y, Chen S, Xu X and Zhang Q: Bioinformatics identification of
lncRNA biomarkers associated with the progression of esophageal
squamous cell carcinoma. Mol Med Rep. 19:5309–5320. 2019.PubMed/NCBI
|
|
19
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Talasila KM, Soentgerath A, Euskirchen P,
Rosland GV, Wang J, Huszthy PC, Prestegarden L, Skaftnesmo KO,
Sakariassen PØ, Eskilsson E, et al: EGFR wild-type amplification
and activation promote invasion and development of glioblastoma
independent of angiogenesis. Acta Neuropathol. 125:683–698. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wee P and Wang Z: Epidermal growth factor
receptor cell proliferation signaling pathways. Cancers (Basel).
9(pii): E522017.PubMed/NCBI
|
|
23
|
Akane H, Saito F, Shiraki A, Imatanaka N,
Akahori Y, Itahashi M, Wang L and Shibutani M: Gene expression
profile of brain regions reflecting aberrations in nervous system
development targeting the process of neurite extension of rat
offspring exposed developmentally to glycidol. J Appl Toxicol.
34:1389–1399. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Monies D, Abouelhoda M, AlSayed M,
Alhassnan Z, Alotaibi M, Kayyali H, Al-Owain M, Shah A, Rahbeeni Z,
Al-Muhaizea MA, et al: The landscape of genetic diseases in Saudi
Arabia based on the first 1000 diagnostic panels and exomes. Hum
Genet. 136:921–939. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berezowska S and Schlegel J: Targeting
ErbB receptors in high-grade glioma. Curr Pharm Des. 17:2468–2487.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang L, Liu Q, Zhao H, Cui K, Yao L, Nie
F, Jin G, Hao A and Wong ST: Differential effects of low- and
high-dose GW2974, a dual epidermal growth factor receptor and HER2
kinase inhibitor, on glioblastoma multiforme invasion. J Neurosci
Res. 91:128–137. 2013.PubMed/NCBI
|
|
27
|
An Z, Aksoy O, Zheng T, Fan QW and Weiss
WA: Epidermal growth factor receptor and EGFRvIII in glioblastoma:
Signaling pathways and targeted therapies. Oncogene. 37:1561–1575.
2018. View Article : Google Scholar : PubMed/NCBI
|