Introduction
Since ancient times, China has been well known for
its tasty food. There are large differences in the dietary habit
among different regions in China due to large population, vast
territory and different nationalities (1–3), which
greatly enriches material life. However, the extremely complicated
dietary habits also lead to high incidence rate of diet-related
diseases, of which oral carcinoma is a malignant tumor with a high
incidence rate, mainly oral squamous cell carcinoma (OSCC) in China
(4,5).
According to statistical data, OSCC accounts for
more than 90% of oral carcinoma, so enhancing the research on OSCC
has important theoretical and practical significance in China.
Currently, the operation, including radiotherapy and chemotherapy,
is dominated in the treatment of OSCC. However, metastasis occurs
more easily in OSCC cells than general tumor cells, so there is no
effective treatment method at present (6,7). In
recent years, related studies have found that the phosphatase and
tensin homolog deleted on chromosome ten (PTEN) has close
correlations with the incidence of a variety of tumors, including
breast cancer and melanoma, and its expression declines obviously
in the above tumor cells (8,9).
On this basis, the present study investigated
whether there is a correlation between PTEN gene and OSCC, and
further explored whether there is also a correlation between PTEN
gene polymorphism and OSCC, aiming to provide a certain theoretical
and experimental basis for the treatment of OSCC.
Patients and methods
General data
In the experiment, 33 OSCC patients treated in the
Affiliated Hospital of Taishan Medical University (Taian, China)
from January 2016 to January 2018 were selected as the experimental
group, including 18 males and 15 females, aged 45 years on average,
while 33 healthy subjects were selected as the control group,
including 17 males and 16 females, aged 45 years on average. The
experimental scheme was discussed and approved by the Academic
Committee, and agreed by the family members.
This study was approved by the Ethics Committee of
Affiliated Hospital of Taishan Medical University. Patients who
participated in this research had complete clinical data. The
signed informed consents were obtained from the patients or the
guardians.
The following experimental reagents were used:
Fluorescence quantitative polymerase chain reaction (PCR) reagent
and ribonucleic acid (RNA) extraction reagent were purchased from
Takara and animal cell protein extraction kit was purchased from
Thermo Fisher Scientific, Inc. Streptomycin-peroxidase (S-P) kit
and corresponding antibodies were purchased from Thermo Fisher
Scientific, Inc. Genome extraction kit was purchased from AXYGEN
and other reagents and consumables were purchased from Sangon
Biotech Co., Ltd.
Methods
Quantitative PCR
RNA extraction
After 5 ml of peripheral blood was drawn from
healthy subjects and OSCC patients and centrifuged at 1,000 × g at
4°C for 5 min, RNA was extracted according to the instructions of
RNA extraction kit (10).
Quantitative PCR
To detect the messenger RNA (mRNA) expression of
PTEN gene in different samples, SYBR-Green 1 staining was
performed in accordance with the instructions. The reaction system
volume was in total 25 µl, pre-denaturation at 95°C for 5 min,
denaturation at 95°C for 30 sec, annealing at 60° for 45 sec,
extension at 72°C for 3 min, with 35 cycles, and then extension at
72°C for 5 min. PCR products were stored at 4°C. With GAPDH as the
internal control, the relative expression level of miR-204 was
calculated by 2−ΔΔCq method (11). The sequences are shown in Table I.
 | Table I.Primer sequences in quantitative
PCR. |
Table I.
Primer sequences in quantitative
PCR.
| Primers | Sequences |
|---|
| rs2943773RT-F |
ATGCTAGCTGATCGATCAGCTA |
| rs2943773RT-R |
CGTAGCTAGTATACGTACGCTACG |
| rs9651495RT-F |
CGTAGCTAGCTAGCATCGATACG |
| rs9651495RT-R |
CGTAGCTAGCATCGAGGCTACGAGC |
| GAPDH-F |
CGTAGGGCTAGCTAGCTAGATAC |
| GAPDH-R |
CGTAGCTGAGAGTTAGCTAGCATC |
Western blot analysis
The total protein was extracted from the sample
using the AXYGEN animal cell protein extraction kit. According to
the instructions 0.5 mg of different research samples were
accurately taken and quantified via Coomassie blue staining. After
treatment, 20 µl samples were taken for sodium dodecyl sulfate
polyacrylamide gel electrophoresis (10%). Then the protein was
transferred onto a polyvinylidene difluoride membrane, sealed for 2
h at 4°C. Bull serum albumin (BSA), blocking buffer (5%) was used
as the blocking reagent. Then the membrane was incubated with the
primary antibody (shown below) at 20°C for 2 h, incubated again
with the secondary antibody at room temperature for 2 h, and washed
with eluent 5 times (10 min/time). Rabbit monoclonal anti-PTEN
antibody (cat. no. ab32199; dil, 1:500); rabbit polyclonal GAPDH
antibody (cat. no. ab37168; dil, 1:500) and secondary goat
anti-rabbit (HRP) IgG antibody (cat. no. ab6721; dil, 1:2,000) were
all purchased from Abcam. Finally, the color was developed using
the developing solution.
Immunohistochemical assay
In this study, the lesion tissue samples were
routinely incubated with the antibody and stained with S-P. The
immunohistochemical evaluation criteria were: membrane staining
<10% or negative after staining (negative), and only membrane
staining or >10% (positive) (12).
Gene polymorphism detection
Genome extraction
After 5 ml peripheral blood was drawn from healthy
subjects and OSCC patients and centrifuged at 1,000 × g and 4°C for
5 min, and genome was extracted according to the instructions of
the kit (13).
PCR-restriction fragment length polymorphism (RFLP).
The primers used in this study were produced by Sangon Biotech Co.,
Ltd., and the primer sequences are shown in Table II.
 | Table II.Primers in PCR-RFLP. |
Table II.
Primers in PCR-RFLP.
| Primers | Sequences |
|---|
| rs2943773-F |
ATCGTAGCTAGAGCATCGATCGAC |
| rs2943773-R |
CGATGCTACGATCGAGACTAGCTA |
| rs9651495-F |
TGCATCGAGGCGAGCGACTAGATA |
| rs9651495-R |
GCTAGCATGGAGCAGCGATCAGCATG |
The PCR products obtained were collected and
connected according to the gel recovery and T-vector connection
operations in the Molecular Cloning Manual, and added with DH5α,
followed by colony PCR verification and sequencing.
Sequencing
In the present study, the Escherichia coli
transfected with the target plasmid was used as a template for
colony PCR verification, and the gene was sent to Sangon Biotech
Co., Ltd., for sequencing.
Statistical analysis
The experimental data in this study were processed
and analyzed using Statistical Product and Service Solutions (SPSS)
20.0 software (IBM, Armonk, NY, USA). The test level was α=0.05,
P<0.05 indicates that the difference was significant, and
P<0.01 indicates that the difference was very significant.
Results
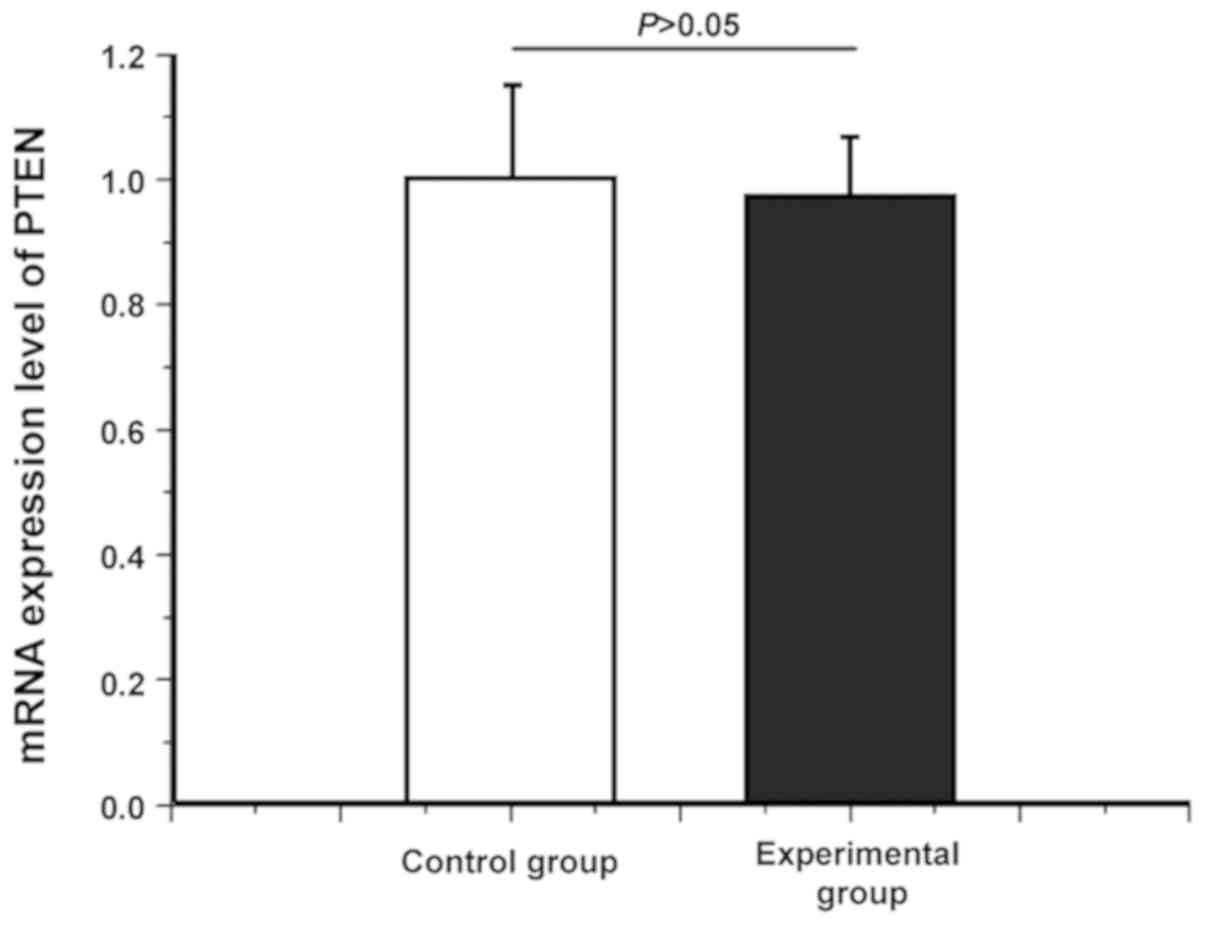
Difference in mRNA expression level of
PTEN between healthy subjects and OSCC patients
Whether there was a difference in the PTEN mRNA
level between healthy subjects and OSCC patients was detected via
quantitative PCR. As shown in Fig.
1, there was no significant difference in the PTEN mRNA
expression level between healthy subjects and OSCC patients
(P>0.05), indicating that the PTEN mRNA level had no difference
between healthy subjects and OSCC patients.
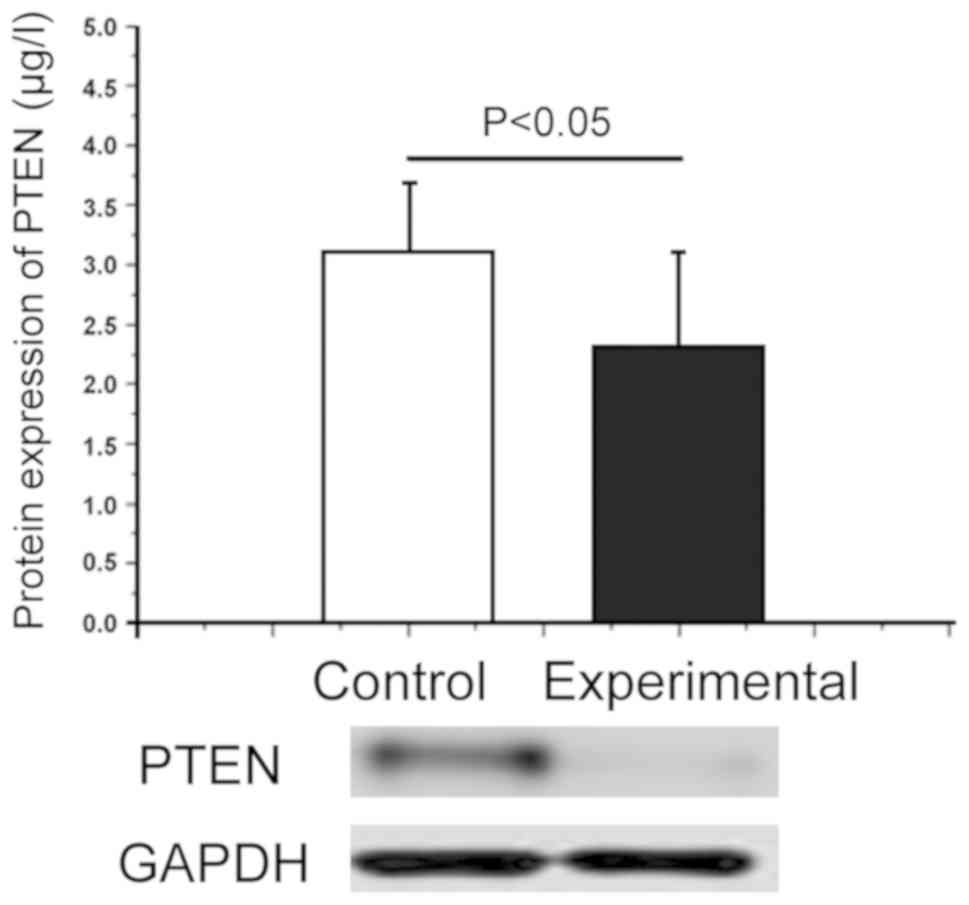
Difference in protein expression level
of PTEN between healthy subjects and OSCC patients
The total protein extracted from the blood in
healthy subjects and OSCC patients were used as the objects of
study, and the difference in PTEN protein expression level in
different subjects was detected via western blot analysis. As shown
in Fig. 2, the PTEN protein
expression level significantly declined in OSCC patients (2.37±1.01
µg/l)compared with that in healthy subjects (3.09±0.95 µg/l), and
there was a significant difference (P<0.05), indicating that the
decrease in the PTEN protein expression level was negatively
correlated with OSCC. At the same time, the results of quantitative
PCR revealed that PTEN was correlated with OSCC at the protein
level, but not at the mRNA level, suggesting that OSCC affects the
translation process of PTEN gene without affecting its gene
transcription process.
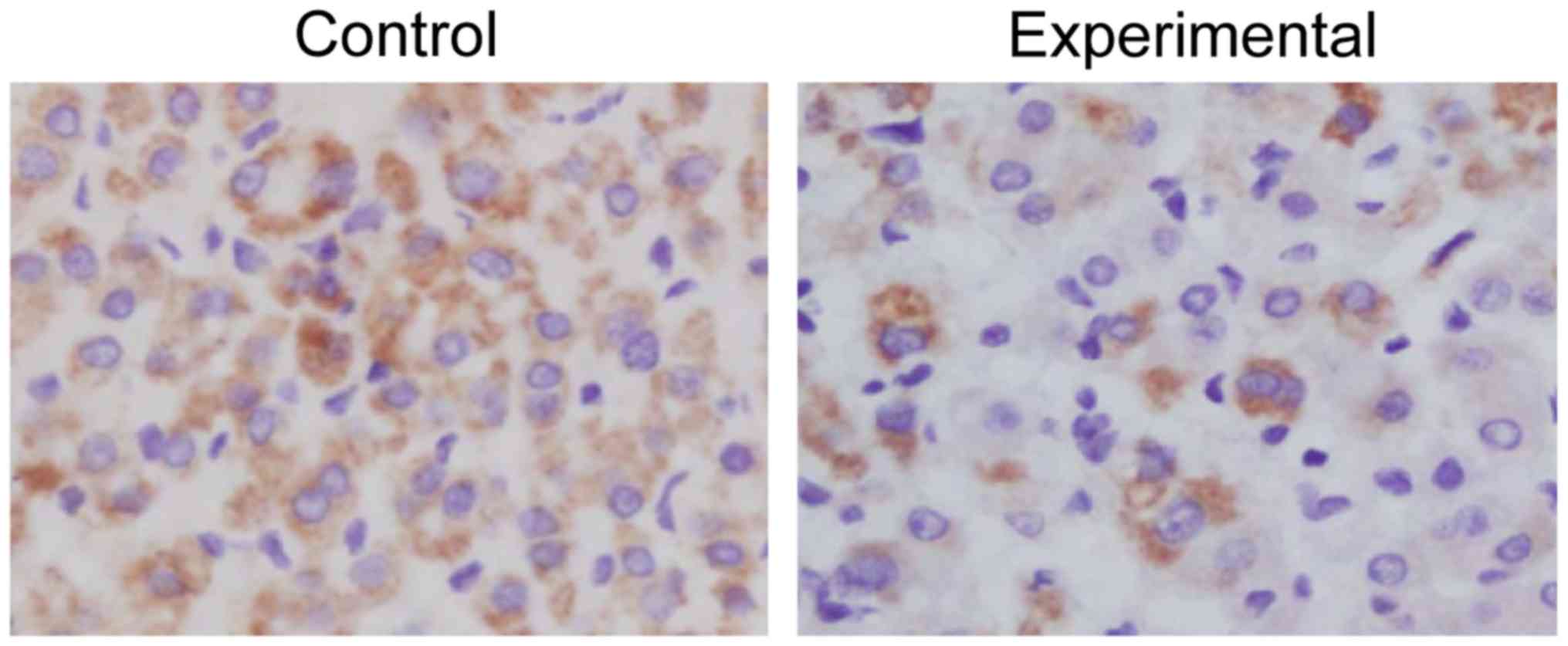
Immunohistochemical detection of PTEN
gene between healthy subjects and OSCC patients
The analysis of immunohistochemical results of
samples from healthy subjects and OSCC patients showed that the
expression level of PTEN in oral cells of healthy subjects was
higher than that of OSCC patients. In other words, the proportion
of PTEN-positive cells in the total in control group (68%) was
significantly higher than that in experimental group (28%), and
there was a significant difference (P=0.015 <0.05), which is
consistent with the protein detection results (Fig. 3 and Table III).
 | Table III.Immunohistochemical results of PTEN
gene between healthy people and OSCC patients. |
Table III.
Immunohistochemical results of PTEN
gene between healthy people and OSCC patients.
| Groups | Number of positive
cells | Ratio of positive
cells in the total (%) | Number of negative
cells |
|---|
| Control | 68/100 cells | 68 | 30/100 cells |
| Experimental | 28/100 cells | 28 | 70/100 cells |
| P-value |
| 0.015 <0.05 |
|
Determination of rs2943773 genotype of
PTEN gene in healthy subjects and OSCC patients
The total DNAs extracted in control and experimental
group were used as templates, and then the PTEN gene was
amplified and sequenced. It was found that there was no significant
difference in the rs2943773 genotype between the control and
experimental groups (χ2=0.863, P=0.712), indicating that
the difference in PTEN protein expression between healthy subjects
and OSCC patients is not caused by the difference in rs2943773
(Table IV).
 | Table IV.Determination of rs2943773 genotype of
PTEN gene in healthy people and OSCC patients. |
Table IV.
Determination of rs2943773 genotype of
PTEN gene in healthy people and OSCC patients.
|
| Genotype |
|---|
|
|
|
|---|
| Groups | C/C (%) | C/T (%) | T/T (%) |
|---|
| Control | 42.5 | 38.4 | 19.1 |
| Experimental | 40.3 | 35.6 | 24.1 |
| P-value | 0.327 >0.05 | 0.291 >0.05 | 0.185 >0.05 |
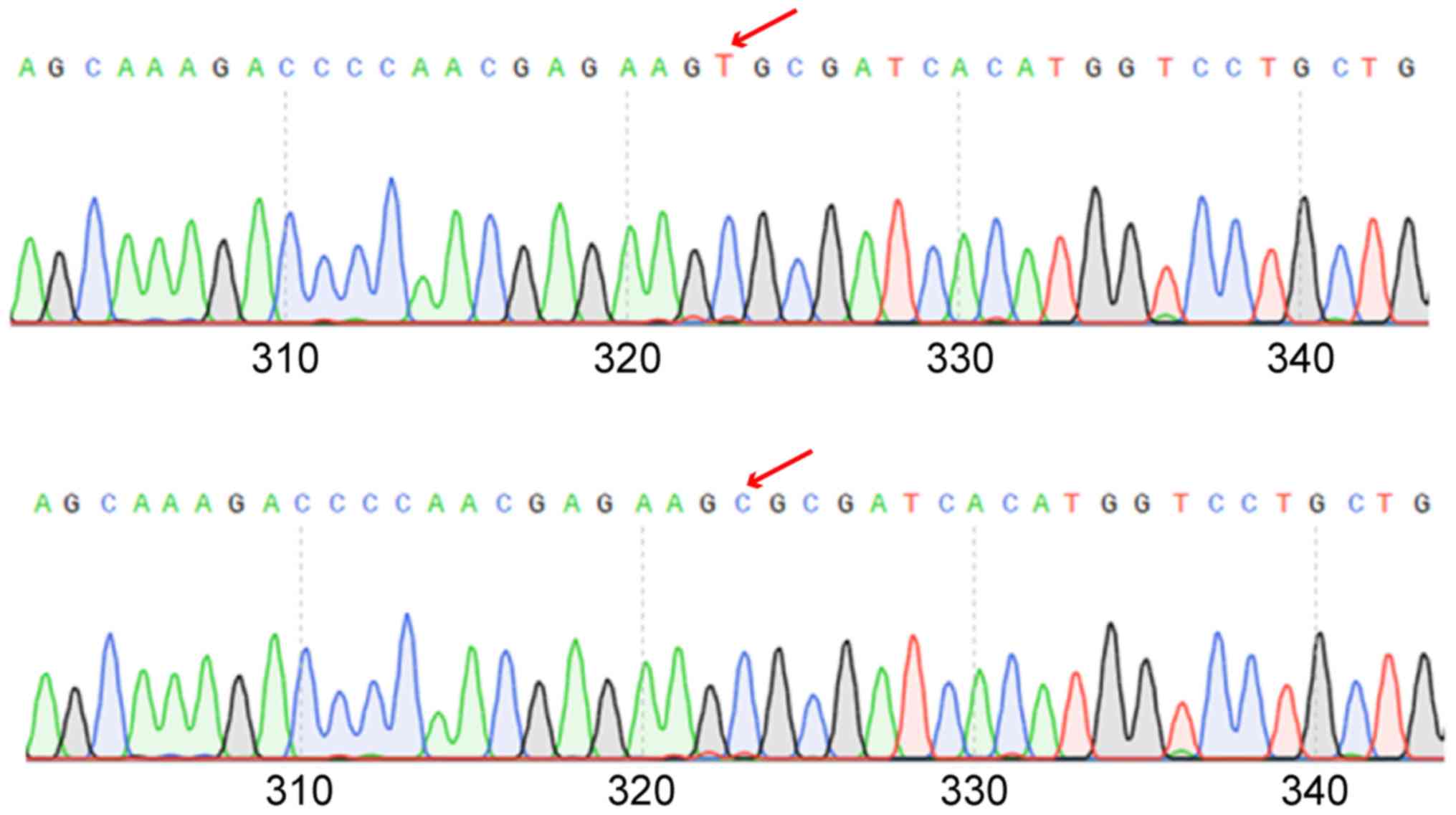
Determination of rs9651495 genotype of
PTEN gene in healthy subjects and OSCC patients
The genome extracted in the control and experimental
groups was used as a template, and then the rs9651495 was amplified
and sequenced. It was found that there was a significant difference
in the rs9651495 genotype between the two groups (P<0.05)
(Fig. 4). The C/C genotype frequency
of rs9651495 in OSCC patients (50.15%) was significantly higher
than that in healthy subjects (23.71%), showing a significant
difference (P<0.05). The C/T genotype frequency of rs9651495 had
no significant difference between the two groups (18.52 vs. 19.01%)
(P>0.05). The T/T genotype frequency of rs9651495 in OSCC
patients (31.33%) was obviously lower than that in healthy subjects
(57.19%), displaying a significant difference (P<0.05),
suggesting the correlation between rs9651495 locus polymorphism of
PTEN gene and OSCC (Table
V).
 | Table V.Determination of rs9651495 genotype of
PTEN gene in healthy subjects and OSCC patients. |
Table V.
Determination of rs9651495 genotype of
PTEN gene in healthy subjects and OSCC patients.
|
| Genotype |
|---|
|
|
|
|---|
| Groups | C/C (%) | C/T (%) | T/T (%) |
|---|
| Control | 23.71 | 19.01 | 57.19 |
| Experimental | 50.15 | 18.52 | 31.33 |
| P-value | 0.012 <0.05 | 0.129 >0.05 | 0.023 <0.05 |
Detection of correlation between PTEN
gene polymorphism and genotype
To explore the correlation between OSCC and PTEN
polymorphism, the figure was plotted with the ratio of C/C of
rs9651495 in PTEN gene as the abscissa and the presence or absence
of OSCC as the ordinate. As shown the proportion of OSCC patients
was significantly increased with the increase of C/C ratio
(Table V), suggesting that there is
a significant correlation between rs9651495 locus polymorphism of
PTEN gene and OSCC.
Discussion
The morbidity rate of OSCC, an oral disease
seriously harming human health, has shown an increasing trend year
by year (14–16). According to statistical data, the
incidence rate of OSCC in China is significantly higher than that
in other countries due to large population and different dietary
habits (17,18). Therefore, enhancing the research on
OSCC has important medical significance. In the present study, OSCC
patients treated in the hospital and healthy subjects were selected
as the objects to investigate the correlation between OSCC and
PTEN gene polymorphism. It was found via quantitative PCR
that there was no significant difference in the expression level of
PTEN gene between healthy subjects and OSCC patients
(P>0.05), indicating that OSCC does not inhibit the PTEN
gene transcription level. Then the PTEN protein expression level
was detected in the experimental and control groups via western
blot analysis. The results showed that the PTEN protein expression
level significantly declined in OSCC patients (2.37±1.01 µg/l)
compared with that in healthy subjects (3.09±0.95 µg/l), and there
was a significant difference (P<0.05), indicating that
PTEN gene is correlated with OSCC. The immunohistochemical
results were consistent with the protein detection results,
suggesting that OSCC can inhibit the translation process of
PTEN gene. However, how this process occurs remains unclear
(19,20). It is evident through the above
experiments that there is a correlation between OSCC and
PTEN gene, namely, PTEN expression is low in OSCC patients.
Based on these results, the genome extracted from healthy subjects
and OSCC patients was used as a template, and then the different
regions of the PTEN gene were amplified and sequenced. It
was found that there was a significant difference in the rs9651495
genotype between the control group and experimental group
(P<0.05). The C/C genotype frequency of rs9651495 in OSCC
patients (50.15%) was significantly higher than that in healthy
subjects (23.71%), showing a significant difference (P<0.05).
The C/T genotype frequency of rs9651495 had no significant
difference between the control and experimental groups (18.52 vs.
19.01%) (P>0.05). The T/T genotype frequency of rs9651495 in
OSCC patients (31.33%) was obviously lower than that in healthy
subjects (57.19%), displaying a significant difference (P<0.05).
The above results demonstrate that there is a positive correlation
between rs9651495 locus polymorphism of PTEN gene and OSCC,
and OSCC is induced more easily in subjects with higher C/C
genotype frequency.
In conclusion, the higher C/C genotype frequency
corresponds to the lower PTEN protein expression level, thus
inducing OSCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Project of Taian
Science and Technology Development Plan (2017NS0134).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ML and DC designed the study and performed the
experiments. ML, HS and ZX collected the data, GL and JL analyzed
the data, and ML and DC prepared the manuscript. All the authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Affiliated Hospital of Taishan Medical University (Taian, China).
Patients who participated in this research had complete clinical
data. The signed informed consents were obtained from the patients
or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they had no competing
interests.
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tsai LL, Yu CC, Chang YC, Yu CH and Chou
MY: Markedly increased Oct4 and Nanog expression correlates with
cisplatin resistance in oral squamous cell carcinoma. J Oral Pathol
Med. 40:621–628. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bhaijee F, Pepper DJ, Pitman KT and Bell
D: Cancer stem cells in head and neck squamous cell carcinoma: A
review of current knowledge and future applications. Head Neck.
34:894–899. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sayed SI, Dwivedi RC, Katna R, Garg A,
Pathak KA, Nutting CM, Rhys-Evans P, Harrington KJ and Kazi R:
Implications of understanding cancer stem cell (CSC) biology in
head and neck squamous cell cancer. Oral Oncol. 47:237–243. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu SJ, Huang SY, Lin CT, Lin YJ, Chang CJ
and Tien HF: The incidence of chronic lymphocytic leukemia in
Taiwan, 1986–2005: A distinct increasing trend with birth-cohort
effect. Blood. 116:4430–4435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun C and Li J: Expression of MiRNA-137 in
oral squamous cell carcinoma and its clinical significance. J BUON.
23:167–172. 2018.PubMed/NCBI
|
|
7
|
Wiestner A: Emerging role of
kinase-targeted strategies in chronic lymphocytic leukemia. Blood.
120:4684–4691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Albers AE, Chen C, Köberle B, Qian X,
Klussmann JP, Wollenberg B and Kaufmann AM: Stem cells in squamous
head and neck cancer. Crit Rev Oncol Hematol. 81:224–240. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
White AC, Tran K, Khuu J, Dang C, Cui Y,
Binder SW and Lowry WE: Defining the origins of Ras/p53-mediated
squamous cell carcinoma. Proc Natl Acad Sci USA. 108:7425–7430.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lapouge G, Youssef KK, Vokaer B, Achouri
Y, Michaux C, Sotiropoulou PA and Blanpain C: Identifying the
cellular origin of squamous skin tumors. Proc Natl Acad Sci USA.
108:7431–7436. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Scmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Van Keymeulen A and Blanpain C: Tracing
epithelial stem cells during development, homeostasis, and repair.
J Cell Biol. 197:575–584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou ZJ, Zhang R, Fan L, Wang L, Fang C,
Zhang LN, Yang S, Li YY, Li JY and Xu W: Low expression level of
phosphatase and tensin homolog deleted on chromosome ten predicts
poor prognosis in chronic lymphocytic leukemia. Leuk Lymphoma.
54:1159–1164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang
YH, Liu P, Hong M, Miao KR, Liu P, et al: miR-181a/b significantly
enhances drug sensitivity in chronic lymphocytic leukemia cells via
targeting multiple anti-apoptosis genes. Carcinogenesis.
33:1294–1301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doupé DP, Klein AM, Simons BD and Jones
PH: The ordered architecture of murine ear epidermis is maintained
by progenitor cells with random fate. Dev Cell. 18:317–323. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hoellenriegel J, Meadows SA, Sivina M,
Wierda WG, Kantarjian H, Keating MJ, Giese N, O'Brien S, Yu A,
Miller LL, et al: The phosphoinositide 3′-kinase delta inhibitor,
CAL-101, inhibits B-cell receptor signaling and chemokine networks
in chronic lymphocytic leukemia. Blood. 118:3603–3612. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fabbri M, Bottoni A, Shimizu M, Spizzo R,
Nicoloso MS, Rossi S, Barbarotto E, Cimmino A, Adair B, Wojcik SE,
et al: Association of a microRNA/TP53 feedback circuitry with
pathogenesis and outcome of B-cell chronic lymphocytic leukemia.
JAMA. 305:59–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jotta PY, Ganazza MA, Silva A, Viana MB,
da Silva MJ, Zambaldi LJ, Barata JT, Brandalise SR and Yunes JA:
Negative prognostic impact of PTEN mutation in pediatric T-cell
acute lymphoblastic leukemia. Leukemia. 24:239–242. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jjingo D, Conley AB, Yi SV, Lunyak VV and
Jordan IK: On the presence and role of human gene-body DNA
methylation. Oncotarget. 3:462–474. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khan H, Vale C, Bhagat T and Verma A: Role
of DNA methylation in the pathogenesis and treatment of
myelodysplastic syndromes. Semin Hematol. 50:16–37. 2013.
View Article : Google Scholar : PubMed/NCBI
|